Evaluation of the Prognostic Value of Low-Frequency KRAS Mutation Detection in Circulating Tumor DNA of Patients with Metastatic Colorectal Cancer
Abstract
1. Introduction
2. Methods
2.1. Patients
2.2. Isolation of Plasma ctDNA
2.3. Library Preparation and Next-Generation Sequencing
2.4. Data Analysis
2.5. Validation of Low-Frequency KRAS Mutation in ctDNA
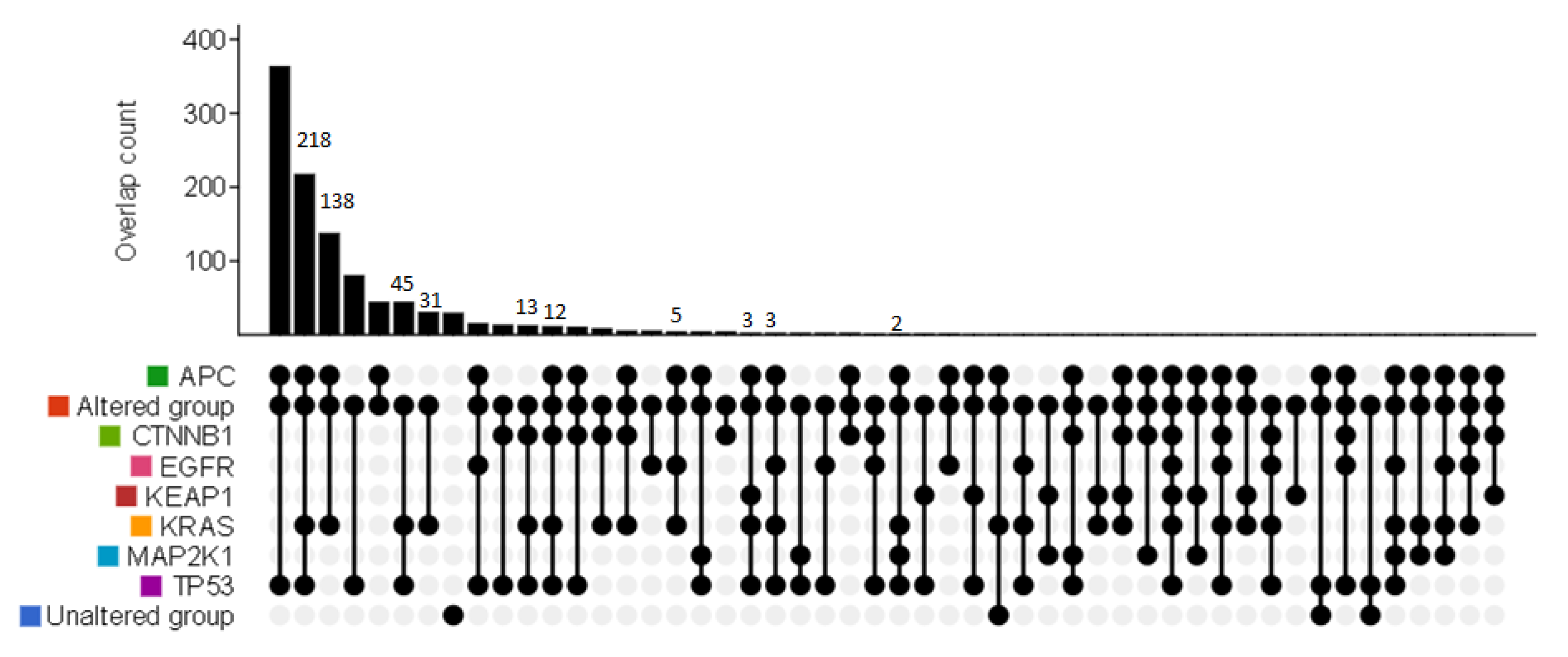
3. Results
3.1. Patient Characteristics
3.2. Mutation Profiling of Plasma ctDNA in mCRC
3.3. Analysis of Treatment Suggestions Generated by Decision Support Platform
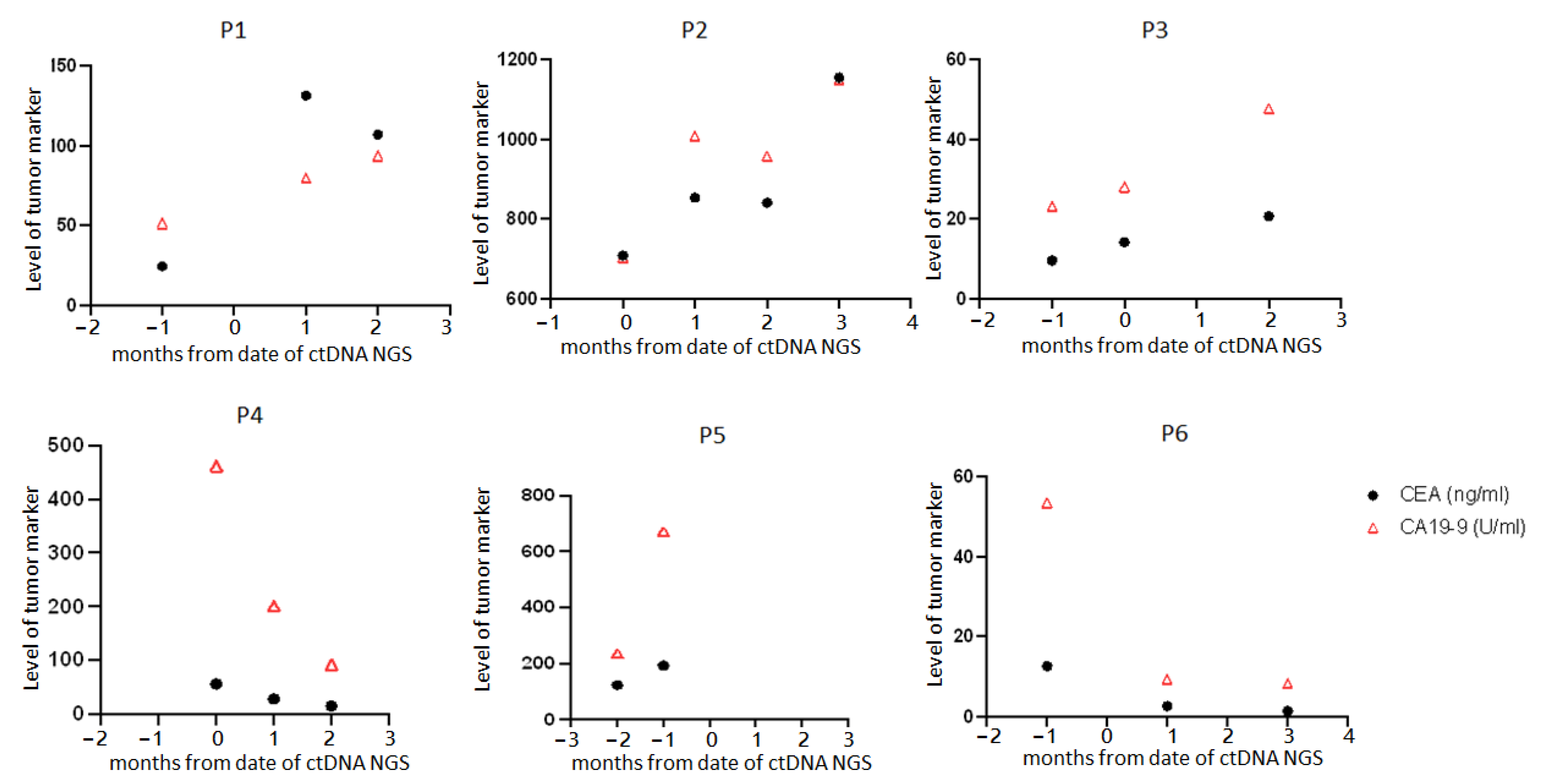
3.4. Clinical Course of the Six Patients
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Waring, P.; Tie, J.; Maru, D.; Karapetis, C.S. RAS Mutations as Predictive Biomarkers in Clinical Management of Metastatic Colorectal Cancer. Clin. Colorectal Cancer 2016, 15, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.H.; Ke, T.W.; Huang, C.W.; Jiang, J.K.; Chen, C.C.; Hsieh, Y.Y.; Teng, H.W.; Lin, B.W.; Liang, Y.H.; Su, Y.L.; et al. Taiwan Society of Colon and Rectal Surgeons Consensus on mCRC Treatment. Front. Oncol. 2021, 11, 764912. [Google Scholar] [CrossRef]
- Puccini, A.; Seeber, A.; Berger, M.D. Biomarkers in Metastatic Colorectal Cancer: Status Quo and Future Perspective. Cancers 2022, 14, 4828. [Google Scholar] [CrossRef]
- Van Cutsem, E.; Lenz, H.J.; Kohne, C.H.; Heinemann, V.; Tejpar, S.; Melezinek, I.; Beier, F.; Stroh, C.; Rougier, P.; van Krieken, J.H.; et al. Fluorouracil, leucovorin, and irinotecan plus cetuximab treatment and RAS mutations in colorectal cancer. J. Clin. Oncol. 2015, 33, 692–700. [Google Scholar] [CrossRef] [PubMed]
- Tougeron, D.; Lecomte, T.; Pages, J.C.; Villalva, C.; Collin, C.; Ferru, A.; Tourani, J.M.; Silvain, C.; Levillain, P.; Karayan-Tapon, L. Effect of low-frequency KRAS mutations on the response to anti-EGFR therapy in metastatic colorectal cancer. Ann. Oncol. 2013, 24, 1267–1273. [Google Scholar] [CrossRef] [PubMed]
- Koulouridi, A.; Karagianni, M.; Messaritakis, I.; Sfakianaki, M.; Voutsina, A.; Trypaki, M.; Bachlitzanaki, M.; Koustas, E.; Karamouzis, M.V.; Ntavatzikos, A.; et al. Prognostic Value of KRAS Mutations in Colorectal Cancer Patients. Cancers 2022, 14, 3320. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Pei, L.; Xia, H.; Tang, Q.; Bi, F. Role of oncogenic KRAS in the prognosis, diagnosis and treatment of colorectal cancer. Mol. Cancer 2021, 20, 143. [Google Scholar] [CrossRef]
- Knebel, F.H.; Bettoni, F.; da Fonseca, L.G.; Camargo, A.A.; Sabbaga, J.; Jardim, D.L. Circulating Tumor DNA Detection in the Management of Anti-EGFR Therapy for Advanced Colorectal Cancer. Front. Oncol. 2019, 9, 170. [Google Scholar] [CrossRef]
- Guttlein, L.; Luca, M.R.; Esteso, F.; Fresno, C.; Mariani, J.; Otero Pizarro, M.; Brest, E.; Starapoli, S.; Kreimberg, K.; Teves, P.; et al. Liquid biopsy for KRAS, NRAS and BRAF mutation testing in advanced colorectal cancer patients: The Argentinean experience. Future Oncol. 2022, 18, 3277–3287. [Google Scholar] [CrossRef] [PubMed]
- van ‘t Erve, I.; Greuter, M.J.E.; Bolhuis, K.; Vessies, D.C.L.; Leal, A.; Vink, G.R.; van den Broek, D.; Velculescu, V.E.; Punt, C.J.A.; Meijer, G.A.; et al. Diagnostic Strategies toward Clinical Implementation of Liquid Biopsy RAS/BRAF Circulating Tumor DNA Analyses in Patients with Metastatic Colorectal Cancer. J. Mol. Diagn. 2020, 22, 1430–1437. [Google Scholar] [CrossRef]
- Holm, M.; Andersson, E.; Osterlund, E.; Ovissi, A.; Soveri, L.M.; Anttonen, A.K.; Kytola, S.; Aittomaki, K.; Osterlund, P.; Ristimaki, A. Detection of KRAS mutations in liquid biopsies from metastatic colorectal cancer patients using droplet digital PCR, Idylla, and next generation sequencing. PLoS ONE 2020, 15, e0239819. [Google Scholar] [CrossRef]
- Keller, L.; Belloum, Y.; Wikman, H.; Pantel, K. Clinical relevance of blood-based ctDNA analysis: Mutation detection and beyond. Br. J. Cancer 2021, 124, 345–358. [Google Scholar] [CrossRef]
- Heitzer, E.; Haque, I.S.; Roberts, C.E.S.; Speicher, M.R. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat. Rev. Genet. 2019, 20, 71–88. [Google Scholar] [CrossRef] [PubMed]
- Li, M.M.; Datto, M.; Duncavage, E.J.; Kulkarni, S.; Lindeman, N.I.; Roy, S.; Tsimberidou, A.M.; Vnencak-Jones, C.L.; Wolff, D.J.; Younes, A.; et al. Standards and Guidelines for the Interpretation and Reporting of Sequence Variants in Cancer: A Joint Consensus Recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J. Mol. Diagn. 2017, 19, 4–23. [Google Scholar] [CrossRef]
- Yang, W.; Zou, J.; Li, Y.; Liu, R.; Yan, Z.; Chen, S.; Zhao, X.; Guo, W.; Huang, M.; Li, W.; et al. Longitudinal Circulating Tumor DNA Profiling in Metastatic Colorectal Cancer During Anti-EGFR Therapy. Front. Oncol. 2022, 12, 830816. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Zang, W.; Ge, Y.; Weygant, N.; Yu, P.; Li, L.; Rao, G.; Jiang, Z.; Yan, R.; He, L.; et al. RAS/BRAF Circulating Tumor DNA Mutations as a Predictor of Response to First-Line Chemotherapy in Metastatic Colorectal Cancer Patients. Can J. Gastroenterol. Hepatol. 2018, 2018, 4248971. [Google Scholar] [CrossRef] [PubMed]
- Pietrantonio, F.; Vernieri, C.; Siravegna, G.; Mennitto, A.; Berenato, R.; Perrone, F.; Gloghini, A.; Tamborini, E.; Lonardi, S.; Morano, F.; et al. Heterogeneity of Acquired Resistance to Anti-EGFR Monoclonal Antibodies in Patients with Metastatic Colorectal Cancer. Clin. Cancer Res. 2017, 23, 2414–2422. [Google Scholar] [CrossRef] [PubMed]
- Topham, J.T.; O’Callaghan, C.J.; Feilotter, H.; Kennecke, H.F.; Lee, Y.S.; Li, W.; Banks, K.C.; Quinn, K.; Renouf, D.J.; Jonker, D.J.; et al. Circulating Tumor DNA Identifies Diverse Landscape of Acquired Resistance to Anti-Epidermal Growth Factor Receptor Therapy in Metastatic Colorectal Cancer. J. Clin. Oncol. 2023, 41, 485–496. [Google Scholar] [CrossRef]
- Cremolini, C.; Rossini, D.; Dell’Aquila, E.; Lonardi, S.; Conca, E.; Del Re, M.; Busico, A.; Pietrantonio, F.; Danesi, R.; Aprile, G.; et al. Rechallenge for Patients With RAS and BRAF Wild-Type Metastatic Colorectal Cancer With Acquired Resistance to First-line Cetuximab and Irinotecan: A Phase 2 Single-Arm Clinical Trial. JAMA Oncol. 2019, 5, 343–350. [Google Scholar] [CrossRef]
- Kim, S.T.; Park, K.H.; Shin, S.W.; Kim, Y.H. Dose KRAS Mutation Status Affect on the Effect of VEGF Therapy in Metastatic Colon Cancer Patients? Cancer Res. Treat. 2014, 46, 48–54. [Google Scholar] [CrossRef]
- Fiala, O.; Buchler, T.; Mohelnikova-Duchonova, B.; Melichar, B.; Matejka, V.M.; Holubec, L.; Kulhankova, J.; Bortlicek, Z.; Bartouskova, M.; Liska, V.; et al. G12V and G12A KRAS mutations are associated with poor outcome in patients with metastatic colorectal cancer treated with bevacizumab. Tumour. Biol. 2016, 37, 6823–6830. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Jin, Q.; Xin, Y. Recent clinical advances in PI3K inhibitors on colorectal cancer. Pharmazie 2021, 76, 568–573. [Google Scholar]
- Gong, S.; Xu, D.; Zhu, J.; Zou, F.; Peng, R. Efficacy of the MEK Inhibitor Cobimetinib and its Potential Application to Colorectal Cancer Cells. Cell. Physiol. Biochem. 2018, 47, 680–693. [Google Scholar] [CrossRef] [PubMed]
- Taghizadeh, H.; Mader, R.M.; Mullauer, L.; Erhart, F.; Kautzky-Willer, A.; Prager, G.W. Precision Medicine for the Management of Therapy Refractory Colorectal Cancer. J. Pers. Med. 2020, 10, 272. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.Y.; Zhong, W.Z.; Zhang, X.C.; Su, J.; Xie, Z.; Liu, S.Y.; Tu, H.Y.; Chen, H.J.; Sun, Y.L.; Zhou, Q.; et al. Potential Predictive Value of TP53 and KRAS Mutation Status for Response to PD-1 Blockade Immunotherapy in Lung Adenocarcinoma. Clin. Cancer Res. 2017, 23, 3012–3024. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Huang, X.; Liu, H.; Wei, C.; Ru, H.; Qin, H.; Lai, H.; Meng, Y.; Wu, G.; Xie, W.; et al. Immune landscape and prognostic immune-related genes in KRAS-mutant colorectal cancer patients. J. Transl. Med. 2021, 19, 27. [Google Scholar] [CrossRef]
- Yaeger, R.; Weiss, J.; Pelster, M.S.; Spira, A.I.; Barve, M.; Ou, S.I.; Leal, T.A.; Bekaii-Saab, T.S.; Paweletz, C.P.; Heavey, G.A.; et al. Adagrasib with or without Cetuximab in Colorectal Cancer with Mutated KRAS G12C. N. Engl. J. Med. 2023, 388, 44–54. [Google Scholar] [CrossRef]
- Pfeiffer, P.; Qvortrup, C. KRAS(G12C) inhibition in colorectal cancer. Lancet Oncol. 2022, 23, 10–11. [Google Scholar] [CrossRef]
- Feng, J.; Hu, Z.; Xia, X.; Liu, X.; Lian, Z.; Wang, H.; Wang, L.; Wang, C.; Zhang, X.; Pang, X. Feedback activation of EGFR/wild-type RAS signaling axis limits KRAS(G12D) inhibitor efficacy in KRAS(G12D)-mutated colorectal cancer. Oncogene 2023, 42, 1620–1633. [Google Scholar] [CrossRef]
- Tran, E.; Robbins, P.F.; Lu, Y.C.; Prickett, T.D.; Gartner, J.J.; Jia, L.; Pasetto, A.; Zheng, Z.; Ray, S.; Groh, E.M.; et al. T-Cell Transfer Therapy Targeting Mutant KRAS in Cancer. N. Engl. J. Med. 2016, 375, 2255–2262. [Google Scholar] [CrossRef]
| Patient | Age (Years) | Gender | Primary Location | Metastatic Site | Months since Diagnosis | Surgical Resection before ctDNA Test | Tumor Tissue RAS Mutation | MSS Tumor | Treatment History before ctDNA Test |
|---|---|---|---|---|---|---|---|---|---|
| P1 | 47 | F | Left | Liver | 4.5 | No | N/A | N/A | Anti-EGFR (panitumumab) + FOLFOX |
| P2 | 67 | F | Left | Liver, lung, bone | 71 | Yes (resection of liver metastatic site) | RAS wt | Yes | Anti-VEGF (bevacizumab) + FOLFIRI; Anti-EGFR (cetuximab) + FOLFIRI |
| P3 | 69 | F | Right | Liver, lung | 36 | Yes (resection of liver metastatic site) | N/A | Yes | Anti-VEGF (bevacizumab) + FOLFOX; Nivolumab; Anti-VEGFR2 (ramucirumab) |
| P4 | 73 | F | Rectum | Liver, lung | 1 | Yes (resection of primary and liver metastatic site) | KRAS p.G12D | Yes | FOLFIRI |
| P5 | 62 | M | Rectum | Liver, lung | 1 | Yes (resection of primary site) | KRAS p.G12S | Yes | FOLFIRI + Oxaliplatin + Anti-VEGF (bevacizumab) |
| P6 | 44 | F | Rectum | lung | 1 | Yes (resection of primary site) | KRAS p.G12V † | Yes | FOLFOX |
| Patient | KRAS (VAF%) | APC (VAF%) | TP53 (VAF%) | MAP2K1 (VAF%) | KEAP1 (VAF%) | CTNNB1 (VAF%) |
|---|---|---|---|---|---|---|
| P1 | p.G12V (1.04%) † | p.E1397* (6.05%) | p.R342* (8.31%) | p.K57T (0.37%), p.L177M (0.38%) | ||
| P2 | p.G12D (0.38%) ‡ | p.R499* (1.24%), p.E1309Dfs*4 (1.60%) | ||||
| P3 | p.G13D (0.46%) † | |||||
| P4 | p.G12D (1.23%) | p.R1463X (1.29%) | p.N387K (0.72%) | |||
| P5 | p.G12S (1.02%) | p.R175H (0.54%), p.C135W (0.38%) | p.T142M (0.39%) | |||
| P6 | p.G12V (0.26%) † |
| Genetic Mutation | Anti-EGFR | Anti-VEGF | Anti-VEGFR | PI3K Inhibitor + MEK Inhibitor | Anti-VEGFR + HDAC Inhibitor | Anti-PD1 | CDK4/6 Inhibitor | Anti-EGFR + MEK Inhibitor |
|---|---|---|---|---|---|---|---|---|
| KRAS G12V | R (Tier-IA) | R | S | |||||
| KRAS G12D/G12S/G13D | R (Tier-IA) | C | ||||||
| TP53 mutation | S | S | ||||||
| KRAS + TP53 mutation | R | S | S | |||||
| APC mutation | ||||||||
| KRAS +APC mutation | S | |||||||
| MAP2K1 K57T | R | S | ||||||
| KEAP1 mutation | ||||||||
| CTNNB1 mutation |
| Patient | Change in Clinical Treatment within 1 Month after ctDNA Test | Time to Tumor Size Progression on CT/PET after ctDNA Test (Months) |
|---|---|---|
| P1 | Add anti-VEGF (bevacizumab) | 1.5 |
| P2 | Shift to anti-VEGF (bevacizumab) + anti-EGFR (cetuximab) + FOLFIRI | 3 |
| P3 | Add anti-VEGF (bevacizumab) | 6 |
| P4 | Shift to FOLFIRI + oxaliplatin | 3 |
| P5 | Dose titration in FOLFIRI + oxaliplatin | 5 |
| P6 | Shift to FOLFIRI + anti-EGFR (cetuximab) | 7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, C.-Y.; Shen, M.-Y.; Chen, W.T.-L.; Yang, C.-A. Evaluation of the Prognostic Value of Low-Frequency KRAS Mutation Detection in Circulating Tumor DNA of Patients with Metastatic Colorectal Cancer. J. Pers. Med. 2023, 13, 1051. https://doi.org/10.3390/jpm13071051
Lin C-Y, Shen M-Y, Chen WT-L, Yang C-A. Evaluation of the Prognostic Value of Low-Frequency KRAS Mutation Detection in Circulating Tumor DNA of Patients with Metastatic Colorectal Cancer. Journal of Personalized Medicine. 2023; 13(7):1051. https://doi.org/10.3390/jpm13071051
Chicago/Turabian StyleLin, Chien-Yu, Ming-Yin Shen, William Tzu-Liang Chen, and Chin-An Yang. 2023. "Evaluation of the Prognostic Value of Low-Frequency KRAS Mutation Detection in Circulating Tumor DNA of Patients with Metastatic Colorectal Cancer" Journal of Personalized Medicine 13, no. 7: 1051. https://doi.org/10.3390/jpm13071051
APA StyleLin, C.-Y., Shen, M.-Y., Chen, W. T.-L., & Yang, C.-A. (2023). Evaluation of the Prognostic Value of Low-Frequency KRAS Mutation Detection in Circulating Tumor DNA of Patients with Metastatic Colorectal Cancer. Journal of Personalized Medicine, 13(7), 1051. https://doi.org/10.3390/jpm13071051






