The Case for Pre-Emptive Pharmacogenetic Screening in South Africa
Abstract
1. Introduction
2. Materials and Methods
2.1. Processing and Analysis of Hospital Dispensary Files
2.2. Extraction of Allele Frequency Data
2.3. Analysis of the South African Health Products Regulatory Authority VigiLyze Dataset
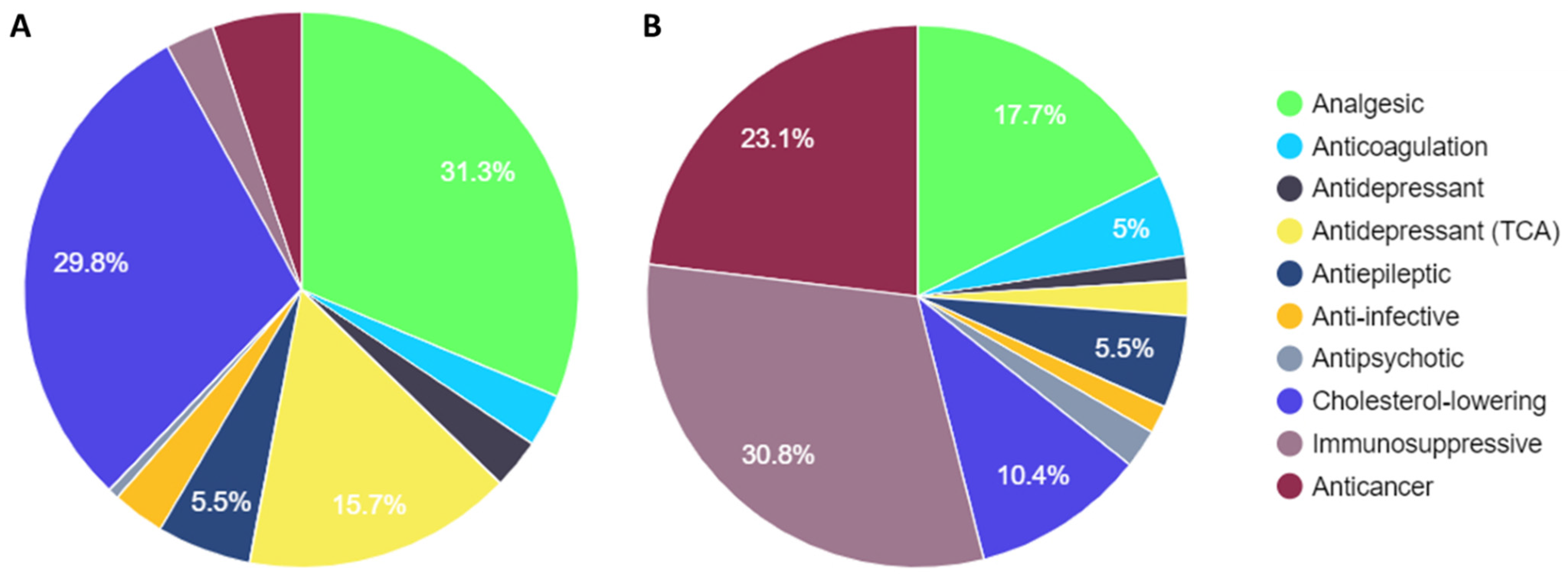
3. Results
3.1. Comparison of PREPARE Study Drugs with Steve Biko Academic Hospital Dispensary Data
3.2. Population-Based Allele Frequency Comparisons
3.3. Analysis of the South African Health Products Regulatory Authority VigiLyze Dataset
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kalow, W. Pharmacogenetics and Pharmacogenomics: Origin, Status, and the Hope for Personalized Medicine. Pharmacogenomics J. 2006, 6, 162–165. [Google Scholar] [CrossRef] [PubMed]
- Sirugo, G.; Williams, S.M.; Tishkoff, S.A. The Missing Diversity in Human Genetic Studies. Cell 2019, 177, 26–31. [Google Scholar] [CrossRef] [PubMed]
- Stark, Z.; Dolman, L.; Manolio, T.A.; Ozenberger, B.; Hill, S.L.; Caulfied, M.J.; Levy, Y.; Glazer, D.; Wilson, J.; Lawler, M.; et al. Integrating Genomics into Healthcare: A Global Responsibility. Am. J. Hum. Genet. 2019, 104, 13–20. [Google Scholar] [CrossRef]
- Visscher, H.; Sistonen, J.; Brunham, L.R.; Pussegoda, K.; Loo, T.T.; Rieder, M.J.; Koren, G.; Carleton, B.C.; Hayden, M.R.; Maggo, S.D.S.; et al. The Canadian Pharmacogenomics Network for Drug Safety: A Model for Safety Pharmacology. Thyroid® 2010, 20, 681–687. [Google Scholar] [CrossRef]
- Swen, J.J.; Nijenhuis, M.; de Boer, A.; Grandia, L.; Maitland-van der Zee, A.H.; Mulder, H.; Rongen, G.A.P.J.M.; Van Schaik, R.H.N.; Schalekamp, T.; Touw, D.J.; et al. Pharmacogenetics: From Bench to Byte—An Update of Guidelines. Clin. Pharmacol. Ther. 2011, 89, 662–673. [Google Scholar] [CrossRef]
- Relling, M.V.; Klein, T.E. CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin. Pharmacol. Ther. 2011, 89, 464–467. [Google Scholar] [CrossRef]
- Relling, M.V.; Klein, T.E.; Gammal, R.S.; Whirl-Carrillo, M.; Hoffman, J.M.; Caudle, K.E. The Clinical Pharmacogenetics Implementation Consortium: 10 Years Later. Clin. Pharmacol. Ther. 2020, 107, 171–175. [Google Scholar] [CrossRef]
- Whirl-Carrillo, M.; Huddart, R.; Gong, L.; Sangkuhl, K.; Thorn, C.F.; Whaley, R.; Klein, T.E. An Evidence-Based Framework for Evaluating Pharmacogenomics Knowledge for Personalized Medicine. Clin. Pharmacol. Ther. 2021, 110, 563–572. [Google Scholar] [CrossRef]
- Abdullah-Koolmees, H.; van Keulen, A.M.; Nijenhuis, M.; Deneer, V.H.M. Pharmacogenetics Guidelines: Overview and Comparison of the DPWG, CPIC, CPNDS, and RNPGx Guidelines. Front. Pharmacol. 2021, 11, 595219. [Google Scholar] [CrossRef]
- Yoon, D.Y.; Lee, S.; Ban, M.S.; Jang, I.-J.; Lee, S. Pharmacogenomic Information from CPIC and DPWG Guidelines and Its Application on Drug Labels. Transl. Clin. Pharmacol. 2020, 28, 189–198. [Google Scholar] [CrossRef]
- Swen, J.J.; van der Wouden, C.H.; Manson, L.E.; Abdullah-Koolmees, H.; Blagec, K.; Blagus, T.; Böhringer, S.; Cambon-Thomsen, A.; Cecchin, E.; Cheung, K.-C.; et al. A 12-Gene Pharmacogenetic Panel to Prevent Adverse Drug Reactions: An Open-Label, Multicentre, Controlled, Cluster-Randomised Crossover Implementation Study. Lancet 2023, 401, 347–356. [Google Scholar] [CrossRef] [PubMed]
- Sherman, R.M.; Forman, J.; Antonescu, V.; Puiu, D.; Daya, M.; Rafaels, N.; Boorgula, M.P.; Chavan, S.; Vergara, C.; Ortega, V.E.; et al. Assembly of a Pan-Genome from Deep Sequencing of 910 Humans of African Descent. Nat. Genet. 2019, 51, 30–35. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, A.; Aron, S.; Botigué, L.R.; Sengupta, D.; Botha, G.; Bensellak, T.; Wells, G.; Kumuthini, J.; Shriner, D.; Fakim, Y.J.; et al. High-depth African Genomes Inform Human Migration and Health. Nature 2020, 586, 741–748. [Google Scholar] [CrossRef]
- Wonkam, A. Sequence Three Million Genomes across Africa. Nature 2021, 590, 209–211. [Google Scholar] [CrossRef] [PubMed]
- Liao, W.-W.; Asri, M.; Ebler, J.; Doerr, D.; Haukness, M.; Hickey, G.; Lu, S.; Lucas, J.K.; Monlong, J.; Abel, H.J.; et al. A Draft Human Pangenome Reference. Nature 2023, 617, 312–324. [Google Scholar] [CrossRef] [PubMed]
- Rajman, I.; Knapp, L.; Morgan, T.; Masimirembwa, C. African Genetic Diversity: Implications for Cytochrome P450-Mediated Drug Metabolism and Drug Development. EBioMedicine 2017, 17, 67–74. [Google Scholar] [CrossRef] [PubMed]
- da Rocha, J.E.B.; Othman, H.; Botha, G.; Cottino, L.; Twesigomwe, D.; Ahmed, S.; Drögemöller, B.I.; Fadlelmola, F.M.; Machanick, P.; Mbiyavanga, M.; et al. The Extent and Impact of Variation in ADME Genes in Sub-Saharan African Populations. Front. Pharmacol. 2021, 12, 634016. [Google Scholar] [CrossRef]
- Mpye, K.L.; Matimba, A.; Dzobo, K.; Chirikure, S.; Wonkam, A.; Dandara, C. Disease Burden and the Role of Pharmacogenomics in African Populations. Glob. Heal. Epidemiol. Genom. 2017, 2, e1. [Google Scholar] [CrossRef]
- Radouani, F.; Zass, L.; Hamdi, Y.; da Rocha, J.; Sallam, R.; Abdelhak, S.; Ahmed, S.; Azzouzi, M.; Benamri, I.; Benkahla, A.; et al. A Review of Clinical Pharmacogenetics Studies in African Populations. Pers. Med. 2020, 17, 155–170. [Google Scholar] [CrossRef]
- Mulder, N.; Abimiku, A.; Adebamowo, S.N.; de Vries, J.; Matimba, A.; Olowoyo, P.; Ramsay, M.; Skelton, M.; Stein, D.J. H3Africa: Current Perspectives. Pharmacogenomics Pers. Med. 2018, 11, 59–66. [Google Scholar] [CrossRef]
- Dandara, C.; Masimirembwa, C.; Haffani, Y.Z.; Ogutu, B.; Mabuka, J.; Aklillu, E.; Bolaji, O. African Pharmacogenomics Consortium: Consolidating Pharmacogenomics Knowledge, Capacity Development and Translation in Africa. AAS Open Res. 2019, 2, 19. [Google Scholar] [CrossRef] [PubMed]
- Katsukunya, J.N.; Soko, N.D.; Naidoo, J.; Rayner, B.; Blom, D.; Sinxadi, P.; Chimusa, E.R.; Dandara, M.; Dzobo, K.; Jones, E.; et al. Pharmacogenomics of Hypertension in Africa: Paving the Way for a Pharmacogenetic-Based Approach for the Treatment of Hypertension in Africans. Int. J. Hypertens. 2023, 2023, e9919677. [Google Scholar]
- Adiukwu, F.; Essien, E.A.; Adesokun, O.; Ofori, S. Psychiatry Pharmacogenomics: Africans Are Not at the Table. Lancet Psychiatry 2023, 10, 80. [Google Scholar] [CrossRef] [PubMed]
- Sitabule, B.R.; Othman, H.; Choudhury, A.; Twesigomwe, D.; Hanchard, N.A. Promoting Pharmacogenomics in Africa: Perspectives From Variation in G6PD and Other Pharmacogenes. Clin. Pharmacol. Ther. 2023, 113, 476–479. [Google Scholar] [CrossRef] [PubMed]
- Jiménez, C.M.; Ramírez, C.P.; Martín, A.S.; Maroun, S.V.; Santiago, S.A.; Tortosa, M.C.R.; Morales, A.J. Clinical Application of Pharmacogenetic Markers in the Treatment of Dermatologic Pathologies. Pharmaceuticals 2021, 14, 905. [Google Scholar] [CrossRef]
- ART Clinical Guidelines for the Management of HIV in Adults, Pregnancy, Adolescents, Children, Infants and Neonates|Department of Health Knowledge Hub. Available online: https://www.knowledgehub.org.za/elibrary/2019-art-clinical-guidelines-management-hiv-adults-pregnancy-adolescents-children-infants (accessed on 31 December 2021).
- Cooper-DeHoff, R.M.; Niemi, M.; Ramsey, L.B.; Luzum, J.A.; Tarkiainen, E.K.; Straka, R.J.; Gong, L.; Tuteja, S.; Wilke, R.A.; Wadelius, M.; et al. The Clinical Pharmacogenetics Implementation Consortium Guideline for SLCO1B1, ABCG2, and CYP2C9 Genotypes and Statin-Associated Musculoskeletal Symptoms. Clin. Pharmacol. Ther. 2022, 111, 1007–1021. [Google Scholar] [CrossRef]
- Hicks, J.K.; Sangkuhl, K.; Swen, J.J.; Ellingrod, V.L.; Müller, D.J.; Shimoda, K.; Bishop, J.R.; Kharasch, E.D.; Skaar, T.C.; Gaedigk, A.; et al. Clinical Pharmacogenetics Implementation Consortium Guideline (CPIC) for CYP2D6 and CYP2C19 Genotypes and Dosing of Tricyclic Antidepressants: 2016 Update. Clin. Pharmacol. Ther. 2017, 102, 37–44. [Google Scholar] [CrossRef]
- Crews, K.R.; Monte, A.A.; Huddart, R.; Caudle, K.E.; Kharasch, E.D.; Gaedigk, A.; Dunnenberger, H.M.; Leeder, J.S.; Callaghan, J.T.; Samer, C.F.; et al. Clinical Pharmacogenetics Implementation Consortium Guideline for CYP2D6, OPRM1, and COMT Genotypes and Select Opioid Therapy. Clin. Pharmacol. Ther. 2021, 110, 888–896. [Google Scholar] [CrossRef]
- Birdwell, K.; Decker, B.; Barbarino, J.; Peterson, J.; Stein, C.; Sadee, W.; Wang, D.; Vinks, A.; He, Y.; Swen, J.; et al. Clinical Pharmacogenetics Implementation Consortium (CPIC®) Guidelines for CYP3A5 Genotype and Tacrolimus Dosing. Clin. Pharmacol. Ther. 2015, 98, 19–24. [Google Scholar] [CrossRef]
- Steyn, H. Profile of Adverse Drug Reaction Reports in South Africa: An Analysis of VigiBase for the Year 2017. S. Afr. Med. J. 2023, 113, 1205–1212. [Google Scholar] [CrossRef]
- Matlala, M.F.; Lubbe, M.S.; Steyn, H. The completeness of adverse drug reaction reports in South Africa: An analysis in VigiBase®. Afr. J. Prim. Heal. Care Fam. Med. 2023, 15, 3659. [Google Scholar] [CrossRef] [PubMed]
- Ampadu, H.H.; Hoekman, J.; de Bruin, M.L.; Pal, S.N.; Olsson, S.; Sartori, D.; Leufkens, H.G.M.; Dodoo, A.N.O. Adverse Drug Reaction Reporting in Africa and a Comparison of Individual Case Safety Report Characteristics Between Africa and the Rest of the World: Analyses of Spontaneous Reports in VigiBase®. Drug Saf. 2016, 39, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Semakula, J.R.; Mouton, J.P.; Jorgensen, A.; Hutchinson, C.; Allie, S.; Semakula, L.; French, N.; Lamorde, M.; Toh, C.-H.; Blockman, M.; et al. A Cross-Sectional Evaluation of Five Warfarin Anticoagulation Services in Uganda and South Africa. PLoS ONE 2020, 15, e0227458. [Google Scholar] [CrossRef] [PubMed]
- Ndadza, A.; Muyambo, S.; Mntla, P.; Wonkam, A.; Chimusa, E.; Kengne, A.P.; Ntsekhe, M.; Dandara, C. Profiling of Warfarin Pharmacokinetics-Associated Genetic Variants: Black Africans Portray Unique Genetic Markers Important for an African Specific Warfarin Pharmacogenetics-Dosing Algorithm. J. Thromb. Haemost. 2021, 19, 2957–2973. [Google Scholar] [CrossRef] [PubMed]
- Masimirembwa, C.; Hasler, J. Pharmacogenetics in Africa, an Opportunity for Appropriate Drug Dosage Regimens: On the Road to Personalized Healthcare. CPT Pharmacomet. Syst. Pharmacol. 2013, 2, 45. [Google Scholar] [CrossRef]
- Munsami, L.; Schutte, C.M.; de Villiers, M.; Hiesgen, J. Late-Onset Efavirenz Toxicity: A Descriptive Study from Pretoria, South Africa. S. Afr. J. HIV Med. 2023, 24, 1439. [Google Scholar] [CrossRef] [PubMed]
- Global Tuberculosis Report 2023. Available online: https://www.who.int/publications-detail-redirect/9789240083851 (accessed on 20 November 2023).
- Mthiyane, T.; Millard, J.; Adamson, J.; Balakrishna, Y.; Connolly, C.; Owen, A.; Rustomjee, R.; Dheda, K.; McIlleron, H.; Pym, A.S. N-Acetyltransferase 2 Genotypes among Zulu-Speaking South Africans and Isoniazid and N. -Acetyl-Isoniazid Pharmacokinetics during Antituberculosis Treatment. Antimicrob. Agents Chemother. 2020, 64, e02376-19. [Google Scholar] [CrossRef]
- Metushi, I.G.; Uetrecht, J.; Phillips, E. Mechanism of Isoniazid-Induced Hepatotoxicity: Then and now. Br. J. Clin. Pharmacol. 2016, 81, 1030–1036. [Google Scholar] [CrossRef]
- Recht, J.; Ashley, E.A.; White, N.J. Use of Primaquine and Glucose-6-Phosphate Dehydrogenase Deficiency Testing: Divergent policies and Practices in Malaria Endemic Countries. PLoS Neglected Trop. Dis. 2018, 12, e0006230. [Google Scholar] [CrossRef]
- Gammal, R.S.; Pirmohamed, M.; Somogyi, A.A.; Morris, S.A.; Formea, C.M.; Elchynski, A.L.; Oshikoya, K.A.; McLeod, H.L.; Haidar, C.E.; Whirl-Carrillo, M.; et al. Expanded Clinical Pharmacogenetics Implementation Consortium Guideline for Medication Use in the Context of G6PD Genotype. Clin. Pharmacol. Ther. 2023, 113, 973–985. [Google Scholar] [CrossRef]
- Mbavha, B.T.; Kanji, C.R.; Stadler, N.; Stingl, J.; Stanglmair, A.; Scholl, C.; Wekwete, W.; Masimirembwa, C. Population Genetic Polymorphisms of Pharmacogenes in Zimbabwe, a Potential Guide for the Safe and Efficacious Use of Medicines in People of African Ancestry. Pharmacogenetics Genom. 2022, 32, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Raga, S.V.; Wilmshurst, J.M.; Smuts, I.; Meldau, S.; Bardien, S.; Schoonen, M.; van der Westhuizen, F.H. A Case for Genomic Medicine in South African Paediatric Patients with Neuromuscular Disease. Front. Pediatr. 2022, 10, 1033299. [Google Scholar] [CrossRef] [PubMed]
- Soko, N.D.; Muyambo, S.; Dandara, M.T.L.; Kampira, E.; Blom, D.; Jones, E.S.W.; Rayner, B.; Shamley, D.; Sinxadi, P.; Dandara, C. Towards Evidence-Based Implementation of Pharmacogenomics in Southern Africa: Comorbidities and Polypharmacy Profiles across Diseases. J. Pers. Med. 2023, 13, 1185. [Google Scholar] [CrossRef] [PubMed]
- Kanji, C.R.; Mbavha, B.T.; Masimirembwa, C.; Thelingwani, R.S. Analytical Validation of GenoPharm a Clinical Genotyping open Array Panel of 46 Pharmacogenes Inclusive of Variants Unique to people of African Ancestry. PLoS ONE 2023, 18, e0292131. [Google Scholar] [CrossRef] [PubMed]
- Van der Linden, L. The PREPARE Study: Benefits of Pharmacogenetic Testing are Unclear. Lancet 2023, 401, 1851. [Google Scholar] [CrossRef]
- Curtis, D. The PREPARE Study: Benefits of Pharmacogenetic Testing are Unclear. Lancet 2023, 401, 1850–1851. [Google Scholar] [CrossRef] [PubMed]
- Caudle, K.E.; Gammal, R.S.; Whirl-Carrillo, M.; Hoffman, J.M.; Relling, M.V.; Klein, T.E. Evidence and Resources to Implement Pharmacogenetic Knowledge for Precision Medicine. Am. J. Heal. Pharm. 2016, 73, 1977–1985. [Google Scholar] [CrossRef][Green Version]
- Hoffman, J.M.; Dunnenberger, H.M.; Hicks, J.K.; Caudle, K.E.; Carrillo, M.W.; Freimuth, R.R.; Williams, M.S.; Klein, T.E.; Peterson, J.F. Developing Knowledge Resources to Support Precision Medicine: Principles from the Clinical Pharmacogenetics Implementation Consortium (CPIC). J. Am. Med. Inform. Assoc. 2016, 23, 796–801. [Google Scholar] [CrossRef]
- Hansen, J.M.; Nørgaard, J.D.; Sporrong, S.K. A Systematic Review of Pharmacogenetic Testing in Primary Care: Attitudes of Patients, General Practitioners, and Pharmacists. Res. Soc. Adm. Pharm. 2022, 18, 3230–3238. [Google Scholar] [CrossRef]
- Muzoriana, N.; Gavi, S.; Nembaware, V.; Dhoro, M.; Matimba, A. Knowledge, Attitude, and Perceptions of Pharmacists and Pharmacy Students towards Pharmacogenomics in Zimbabwe. Pharmacy 2017, 5, 36. [Google Scholar] [CrossRef]
- Mufwambi, W.; Stingl, J.; Masimirembwa, C.; Manasa, J.; Nhachi, C.; Stadler, N.; Mwila, C.; Kalungia, A.C.; Mukosha, M.; Mutiti, C.S.; et al. Healthcare Professionals’ Knowledge of Pharmacogenetics and Attitudes Towards Antimicrobial Utilization in Zambia: Implications for a Precision Medicine Approach to Reducing Antimicrobial Resistance. Front. Pharmacol. 2020, 11, 551522. [Google Scholar] [CrossRef] [PubMed]
- Kudzi, W.; Addy, B.; Dzudzor, B. Knowledge of Pharmacogenetics among Healthcare Professionals and Faculty Members of Health Training Institutions in Ghana. Ghana. Med. J. 2015, 49, 50–56. [Google Scholar] [CrossRef] [PubMed]
| Pharmacy | Year | Prescription Number | Cumulative Prescriptions | Total Cost (ZAR) | Cumulative Cost (ZAR) |
|---|---|---|---|---|---|
| Inpatient | 2018 | 477,254 | 1,751,386 | 28,900,337.69 | 117,892,639.02 |
| 2019 | 459,999 | 30,319,287.36 | |||
| 2020 | 453,137 | 31,785,757.32 | |||
| 2021 | 360,996 | 26,887,256.65 | |||
| Outpatient | 2018 | 1,169,679 | 4,288,797 | 43,350,476.40 | 186,686,832.97 |
| 2019 | 1,106,479 | 45,458,498.01 | |||
| 2020 | 1,103,182 | 52,555,138.32 | |||
| 2021 | 909,457 | 45,322,720.24 | |||
| Oncology | 2018 | 205,341 | 758,697 | 18,034,013.16 | 64,540,893.00 |
| 2019 | 207,669 | 13,907,246.91 | |||
| 2020 | 198,983 | 24,021,552.54 | |||
| 2021 | 146,704 | 8,578,080.39 | |||
| Total | 2018–2021 | 6,798,880 | 369,120,364.99 |
| Drug Class | Prescription Number | Total Cost (ZAR) | Genes Monitored | dbSNPs Monitored |
|---|---|---|---|---|
| Analgesic | 217,572 | 5,848,133.71 | CYP2D6 | 13 |
| Cholesterol-lowering | 207,018 | 3,444,997.81 | SLCO1B1 | 3 |
| Antidepressant (TCA) | 109,345 | 684,720.60 | CYP2D6/CYP2C19 | 23 |
| Antiepileptic | 38,216 | 1,834,534.52 | HLA B*5701/CYP2C19 | 6 |
| Anticancer | 37,801 | 7,648,288.64 | DPYD/UGT1A1/CYP2D6 | 20 |
| Anticoagulation | 21,370 | 1,642,319.46 | CYP2C9/CYP2C19/VKOR1 | 11 |
| Anti-infective | 21,357 | 553,183.30 | CYP2B6/HLA B*5701/CYP2C9 | 14 |
| Antidepressant | 20,180 | 476,304.87 | CYP2D6/CYP2C19 | 23 |
| Immunosuppressive | 18,123 | 10,206,762.30 | TPMT/CYP3A5 | 6 |
| Antipsychotic | 4274 | 762,684.76 | CYP2D6/CYP1A2 | 15 |
| Total | 695,256 | 33,101,929.97 |
| Drug Class | Description | Total Number of Prescriptions Issued | Cost (ZAR) |
|---|---|---|---|
| Analgesic | Tramadol | 217,546 | 5,836,578.7 |
| Cholesterol-lowering | Simvastatin | 142,450 | 1,205,186.0 |
| Antidepressant (TCA) | Amitriptyline | 109,340 | 684,461.7 |
| Cholesterol-lowering | Atorvastatin | 64,568 | 2,239,811.8 |
| Antipileptic | Carbamazepine | 29,287 | 1,307,022.6 |
| Anticancer | Tamoxifen | 20,048 | 765,132.7 |
| Anti-infective | Flucloxacillin | 17,754 | 241,172.0 |
| Antidepressant | Citalopram | 15,969 | 176,589.1 |
| Anticoagulant | Clopidogrel | 11 157 | 910,518.3 |
| Anticancer | Fluorouracil | 10,971 | 281,973.2 |
| Anticoagulant | Warfarin | 10,213 | 731,801.2 |
| Immunosuppressive | Tacrolimus | 10,156 | 9,413,587.5 |
| Antiepileptic | Phenytoin | 8929 | 527,511.9 |
| Immunosuppressive | Azathioprine | 7967 | 793,174.8 |
| Anticancer | Capecitabine | 4238 | 4,386,219.7 |
| Anti-infective | Efavirenz | 3565 | 174,476.0 |
| Antipsychotic | Haloperidol | 2965 | 88,551.0 |
| Antidepressant | Sertraline | 2108 | 156,890.4 |
| Antidepressant | Venlafaxine | 2059 | 138,826.1 |
| Immunosuppressive | Mercaptopurine | 1508 | 1,600,266.0 |
| Antipsychotic | Aripiprazole | 1032 | 648,508.4 |
| Anticancer | Irinotecan | 857 | 213,280.9 |
| Antipsychotic | Clozapine | 247 | 23,949.7 |
| Immunosuppressive | Thioguanine | 179 | 401,416.1 |
| Antidepressant | Paroxetine | 44 | 3999.3 |
| Anti-infective | Voriconazole | 38 | 137,535.3 |
| Antipsychotic | Zuclopenthixol | 30 | 1675.7 |
| Analgesic | Codeine | 26 | 11,555.0 |
| Antidepressant (TCA) | Imipramine | 5 | 258.9 |
| Genes | Allele | dbSNP rsID Number | Functional Status | Allele Frequency European | Allele Frequency Sub-Saharan Africa | Fold Enrichment |
|---|---|---|---|---|---|---|
| CYP3A5 | *6 | rs10264272 | No function | 0.00151 | 0.19324 | 127.6 |
| CYP2D6 | *29 | rs61736512/ rs59421388 | Decreased function | 0.00105 | 0.10833 | 103.5 |
| CYP2C9 | *5 | rs28371686 | Decreased function | 0.00017 | 0.01033 | 59.2 |
| UGT1A1 | *37 | rs8175347 | Decreased function | 0.00069 | 0.03707 | 54.0 |
| CYP2D6 | *17 | rs28371706 | Decreased function | 0.00392 | 0.19355 | 49.4 |
| CYP2C9 | *8 | rs7900194 | Decreased function | 0.00181 | 0.07585 | 42.0 |
| CYP2C19 | *9 | rs17884712 | Decreased function | 0.00066 | 0.02696 | 40.8 |
| CYP1A2 ^ | *1C | rs2069514 | Decreased function | 0.01298 | 0.27690 | 21.3 |
| CYP2C9 | *11 | rs28371685 | Decreased function | 0.00164 | 0.02569 | 15.6 |
| TPMT | *3C | rs1142345 | No function | 0.00492 | 0.05288 | 10.7 |
| UGT1A1 ^ | *27 | rs35350960 | Decreased function | 0.00001 | 0.00004 | 5.2 |
| CYP2D6 | *10 | rs1065852 | Decreased function | 0.01571 | 0.04869 | 3.1 |
| CYP2D6 | *5 | Deletion | No function | 0.02948 | 0.06209 | 2.1 |
| CYP2C19 | *3 | rs4986893 | No function | 0.00162 | 0.00267 | 1.6 |
| CYP2B6 | *6 | rs3745274 | Decreased function | 0.23298 | 0.37487 | 1.6 |
| UGT1A1 | *28 | rs8175347 | Decreased function | 0.31647 | 0.40004 | 1.3 |
| CYP2C19 | *2 | rs4244285 | No function | 0.14686 | 0.15684 | 1.1 |
| CYP1A2 ^ | *1F | rs762551 | Increased function | 0.71180 | 0.60500 | 0.8 |
| CYP2C19 | *17 | rs12248560 | Increased function | 0.21544 | 0.17334 | 0.8 |
| CYP2D6 | *41 | rs28371725 | Decreased function | 0.09238 | 0.04529 | 0.5 |
| VKORC1 | X | rs9934438 | Decreased function | 0.41326 | 0.10774 | 0.3 |
| CYP3A5 | *3 | rs776746 | No function | 0.92438 | 0.24095 | 0.3 |
| SLCO1B1 | *15 | rs4149056 | No function | 0.15017 | 0.02793 | 0.2 |
| HLA-B*5701 | X | rs2395029 | 0.03604 | 0.00609 | 0.2 | |
| CYP2D6 | *4 | rs3892097 | No function | 0.18485 | 0.02873 | 0.2 |
| CYP2C9 | *3 | rs1057910(C) | No function | 0.07554 | 0.01116 | 0.1 |
| CYP2C9 | *2 | rs1799853 | Decreased function | 0.12730 | 0.01311 | 0.1 |
| CYP3A5 | *7 | rs41303343 | No function | 0.00000 | 0.08641 | n/a |
| CYP2D6 | *3 | rs35742686 | No function | 0.01592 | 0.00098 | n/a |
| CYP2B6 | *18 | rs28399499 | No function | 0.00000 | 0.05768 | n/a |
| CYP2C19 | *4A/B | rs28399504 | No function | 0.00236 | 0.00000 | n/a |
| CYP2C19 | *5 | rs56337013 | No function | 0.00003 | 0.00000 | n/a |
| CYP2C19 | *6 | rs72552267 | No function | 0.00030 | 0.00000 | n/a |
| CYP2C19 | *7 | rs72558186 | No function | 0.00000 | 0.00000 | n/a |
| CYP2C19 | *8 | rs41291556 | No function | 0.00336 | 0.00000 | n/a |
| CYP2C19 | *10 | rs6413438 | Decreased function | 0.00000 | 0.00000 | n/a |
| CYP2D6 | *6 | rs5030655 | No function | 0.01120 | 0.00000 | n/a |
| CYP2D6 | *8 | rs5030865 | No function | 0.00022 | 0.00000 | n/a |
| CYP2D6 | *9 | rs5030656 | Decreased function | 0.02754 | 0.00000 | n/a |
| CYP2D6 | *14A/B | rs5030865 | Decreased function | 0.00000 | 0.00000 | n/a |
| DPYD | *2A | rs3918290 | No function | 0.00792 | 0.00000 | n/a |
| DPYD | *13 | rs55886062 | No function | 0.00056 | 0.00000 | n/a |
| DPYD | c.2846 A>T | rs67376798 | Decreased function | 0.00374 | 0.00000 | n/a |
| DPYD | c.1236G>A | rs56038477 | Decreased function | 0.02374 | 0.00000 | n/a |
| SLCO1B1 | *5 | rs4149056 | No function | 0.02040 | 0.00000 | n/a |
| TPMT | *2 | rs1800462 | No function | 0.00206 | 0.00000 | n/a |
| TPMT | *3B | rs1800460 | No function | 0.00283 | 0.00000 | n/a |
| UGT1A1 | *6 | rs4148323 | Decreased function | 0.00787 | 0.00000 | n/a |
| Drug Class | Drug | Concomitant | Interacting | Suspected |
|---|---|---|---|---|
| Anti-infective | Efavirenz FDC | 71 | 1 | 522 |
| Anti-infective | Efavirenz | 179 | 1 | 384 |
| Cholesterol-lowering | Atorvastatin | 260 | 285 | |
| Anticoagulant | Warfarin | 78 | 1 | 178 |
| Cholesterol-lowering | Simvastatin | 313 | 136 | |
| Antidepressant | Citalopram | 62 | 125 | |
| Antidepressant (TCA) | Amitriptyline | 89 | 95 | |
| Analgesic | Tramadol | 59 | 84 | |
| Antiepileptic | Carbamazepine | 36 | 64 | |
| Antipsychotic | Clozapine | 4 | 59 | |
| Antipsychotic | Haloperidol | 15 | 48 | |
| Anticoagulant | Clopidogrel | 52 | 36 | |
| Antiepileptic | Phenytoin | 14 | 36 | |
| Antidepressant | Sertraline | 26 | 24 | |
| Antipsychotic | Zuclopenthixol | 14 | 22 | |
| Immunosuppressive | Tacrolimus | 9 | 21 | |
| Anticancer | Capecitabine | 3 | 18 | |
| Anti-infective | Voriconazole | 0 | 17 | |
| Immunosuppressive | Azathioprine | 16 | 13 | |
| Antipsychotic | Aripiprazole | 5 | 11 | |
| Analgesic | Codeine | 15 | 6 | |
| Anticancer | Tamoxifen | 1 | 5 | |
| Anti-infective | Flucloxacillin | 1 | 4 | |
| Anticancer | Irinotecan | 1 | 3 | |
| Antidepressant | Paroxetine | 8 | 3 | |
| Anticancer | Fluorouracil | 2 | 2 | |
| Antidepressant (TCA) | Imipramine | 1 | 1 | |
| Antidepressant | Venlafaxine | 20 | 4 | 0 |
| Total | 1354 | 7 | 2202 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hurrell, T.; Naidoo, J.; Masimirembwa, C.; Scholefield, J. The Case for Pre-Emptive Pharmacogenetic Screening in South Africa. J. Pers. Med. 2024, 14, 114. https://doi.org/10.3390/jpm14010114
Hurrell T, Naidoo J, Masimirembwa C, Scholefield J. The Case for Pre-Emptive Pharmacogenetic Screening in South Africa. Journal of Personalized Medicine. 2024; 14(1):114. https://doi.org/10.3390/jpm14010114
Chicago/Turabian StyleHurrell, Tracey, Jerolen Naidoo, Collen Masimirembwa, and Janine Scholefield. 2024. "The Case for Pre-Emptive Pharmacogenetic Screening in South Africa" Journal of Personalized Medicine 14, no. 1: 114. https://doi.org/10.3390/jpm14010114
APA StyleHurrell, T., Naidoo, J., Masimirembwa, C., & Scholefield, J. (2024). The Case for Pre-Emptive Pharmacogenetic Screening in South Africa. Journal of Personalized Medicine, 14(1), 114. https://doi.org/10.3390/jpm14010114





