Therapeutic Effects of Engineered Exosomes from RAW264.7 Cells Overexpressing hsa-let-7i-5p against Sepsis in Mice—A Comparative Study with Human Placenta-Derived Mesenchymal Stem Cell Exosomes
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Cultures and Animals
2.1.1. Cell Cultures
2.1.2. Animals
2.2. Genetic Modification of RAW264.7 Cells for Overexpressing hsa-let-7i-5p
2.3. Isolation of Engineered Exosomes and hpMSC Exosomes
2.4. Confirmation of Isolated Exosomes and miRNA Analysis in Engineered Exosomes
2.4.1. Morphology
2.4.2. Particle Sizing Analysis
2.4.3. Markers of Exosomes
2.4.4. Analysis of hsa-let-7i-5p in Engineered Exosomes
2.5. Electroporation of hsa-let-7i-5p Inhibitor into Exosomes
2.6. Biodistribution and Pharmacokinetics of Exosomes
2.6.1. Biodistribution Assay
2.6.2. Pharmacokinetics
2.7. Therapeutic Effects of Exosomes against Sepsis
2.7.1. Monomicrobial Sepsis Model
2.7.2. Experimental Protocols
2.7.3. Survivorship, Plasma Sample Collection, and Plasma Cytokine Measurements
2.7.4. Functional Assay of the Lungs
2.7.5. Lung Tissue Collection and Bronchoalveolar Lavage Fluid (BALF) Collection
2.7.6. Histological Analysis of Lung Injury and Wet/Dry Weight Ratio Assay
2.7.7. Lung Inflammation Assay
- Activation of upstream regulator nuclear factor-κB (NF-κB)
- b.
- Cytokines in the lungs
- c.
- Macrophage M1 phase polarization
2.7.8. Lung Oxidation Assay
- a.
- Assay of endogenous antioxidant enzymes
- b.
- Assay of lipid peroxidation
2.7.9. Cell Death Process Apoptosis in the Lungs
- a.
- Assay of pro-apoptotic protein
- b.
- Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL)
2.8. Statistical Analysis
3. Results
3.1. Confirmation of Exosomes
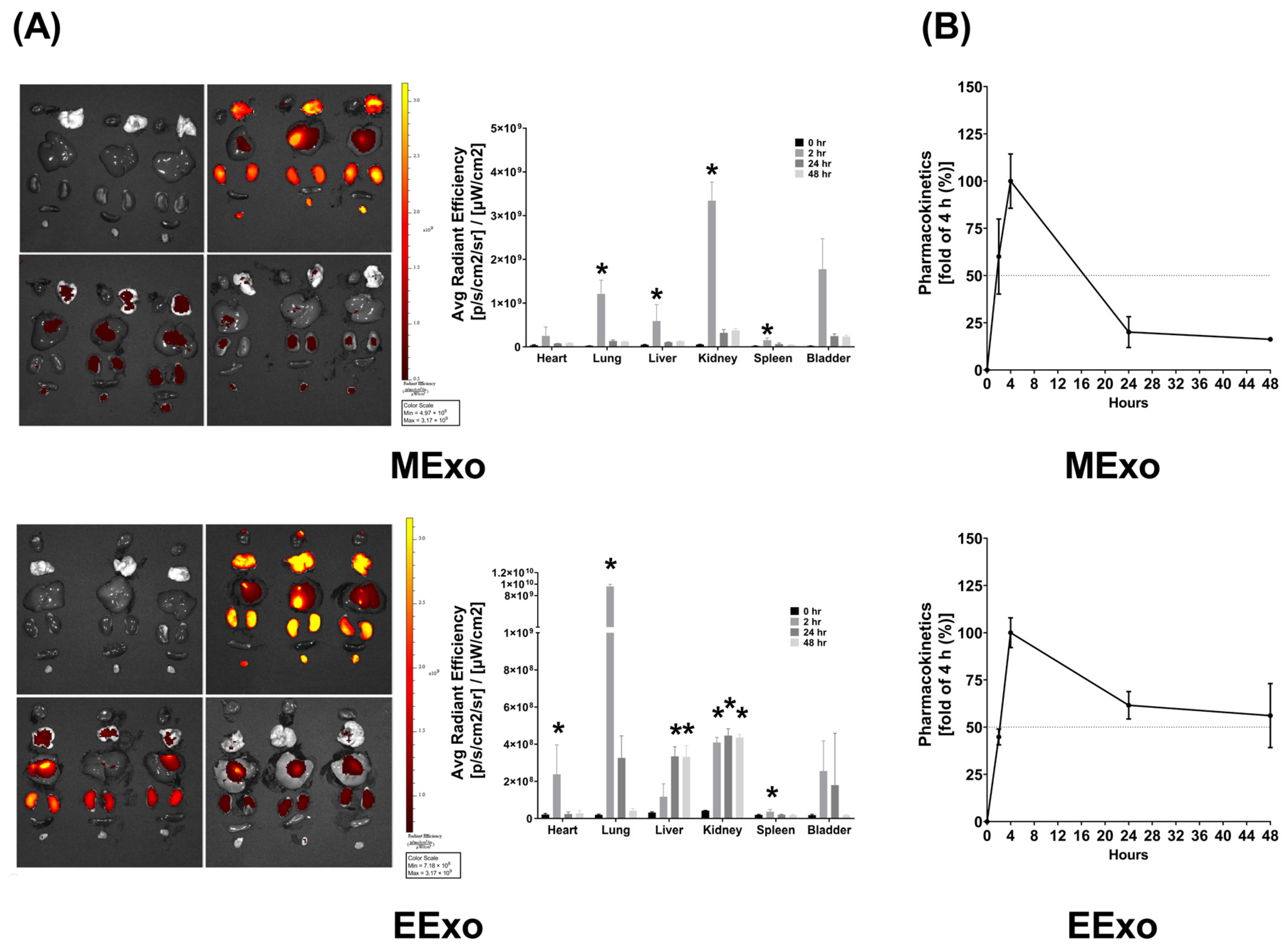
3.2. Biodistribution and Pharmacokinetics of Exosomes
3.3. Survivorship and Plasma Cytokines
3.4. Lung Injury
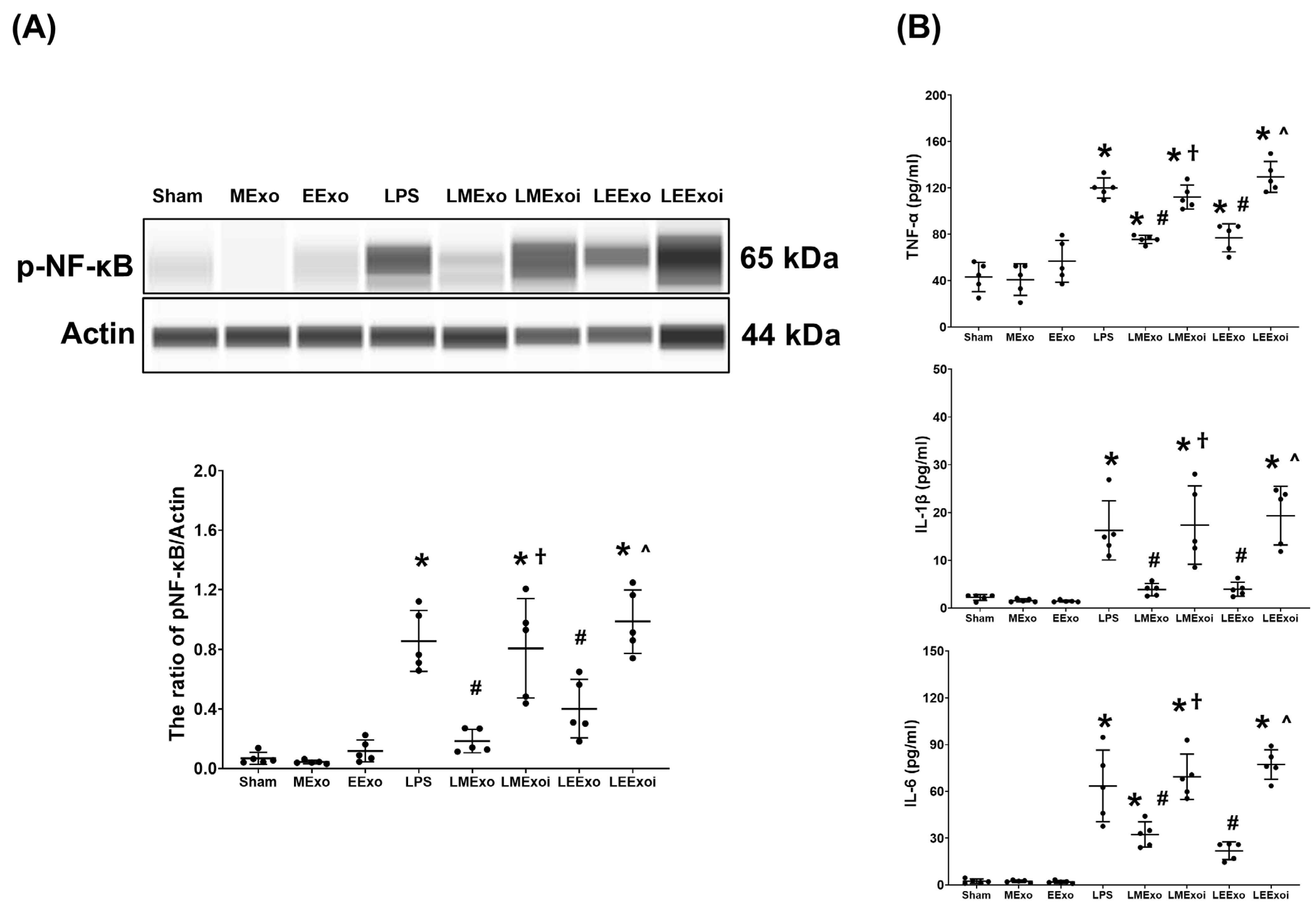
3.5. Lung Inflammation
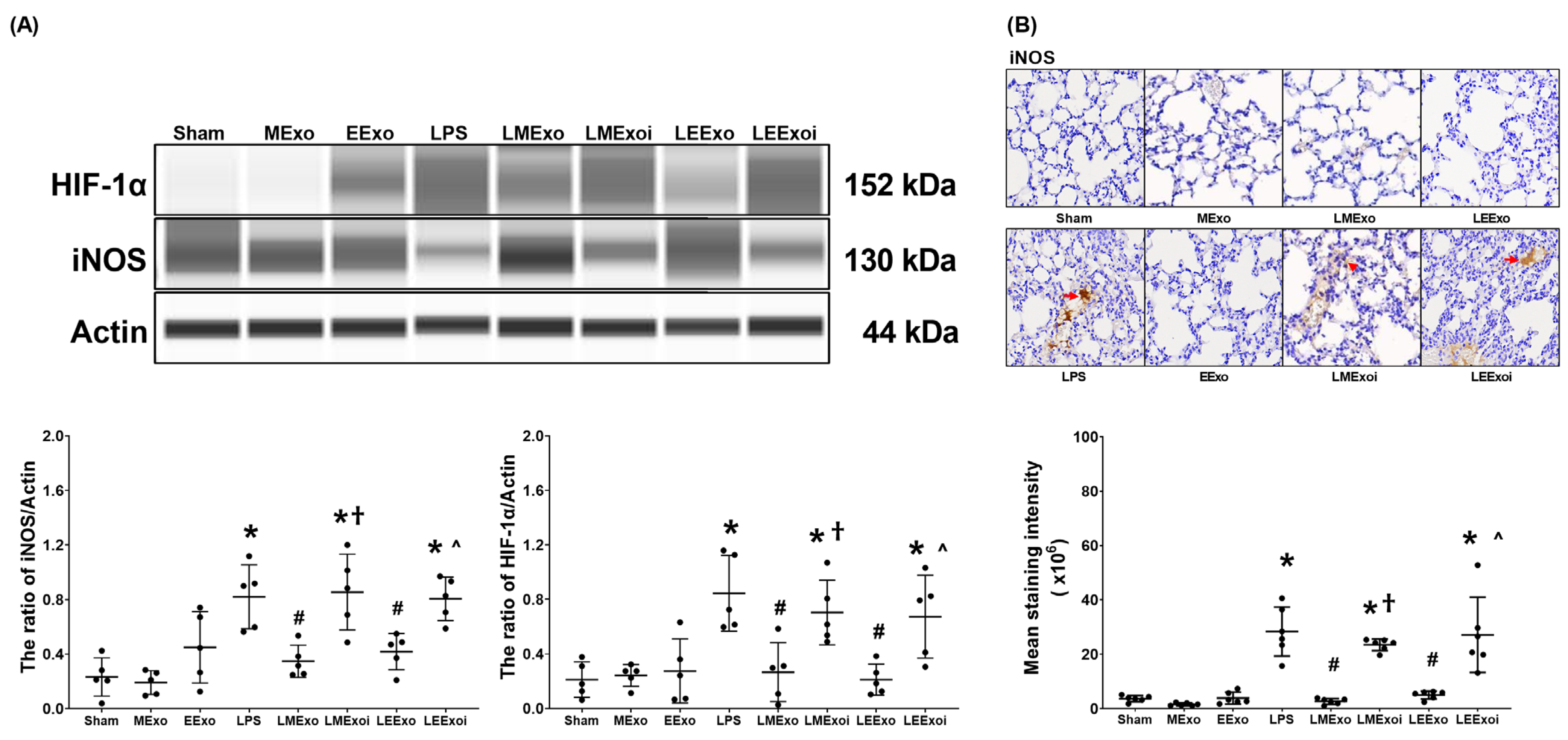
3.6. Macrophage Polarization in Lung Tissues
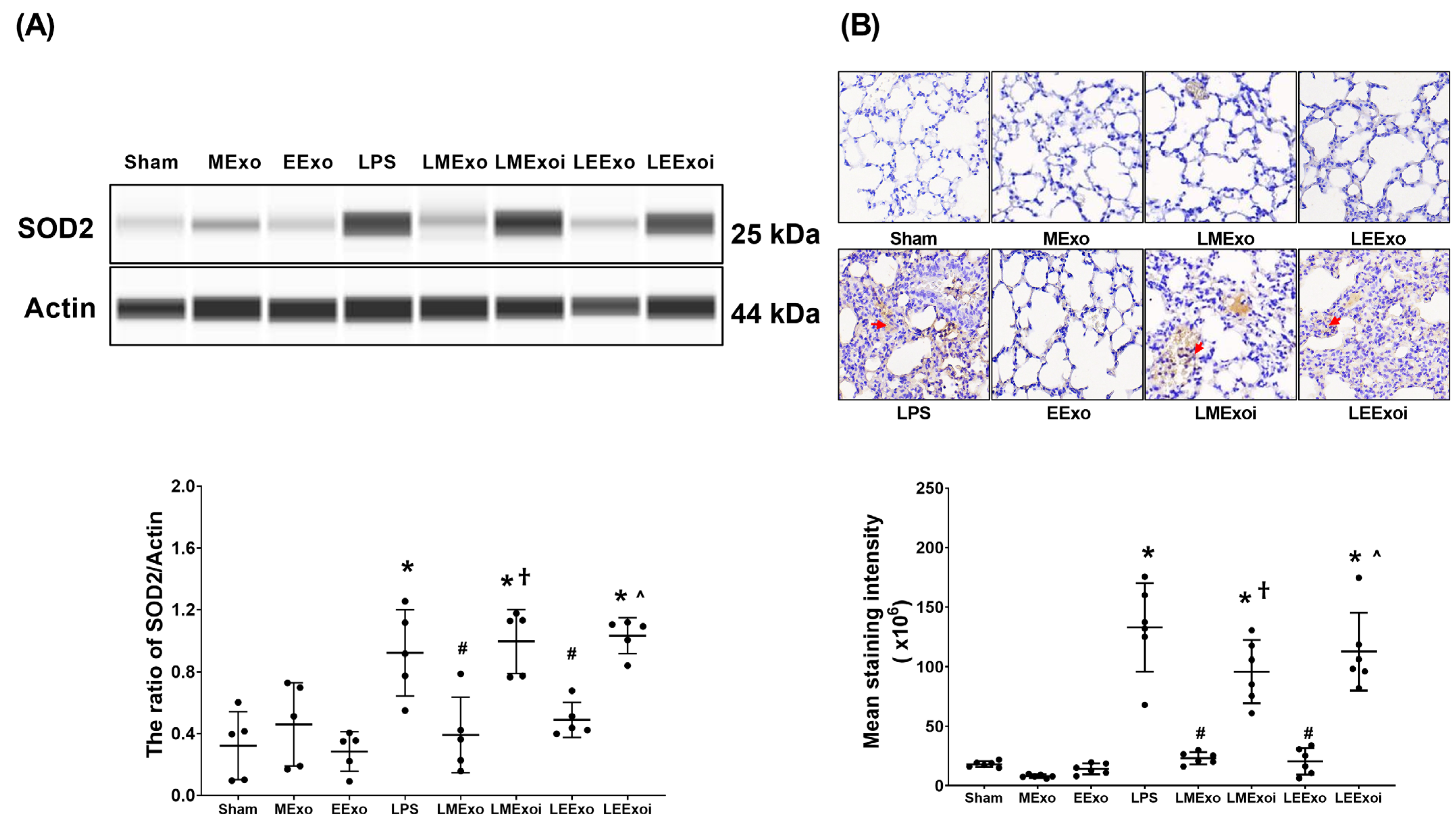
3.7. Lung Oxidation
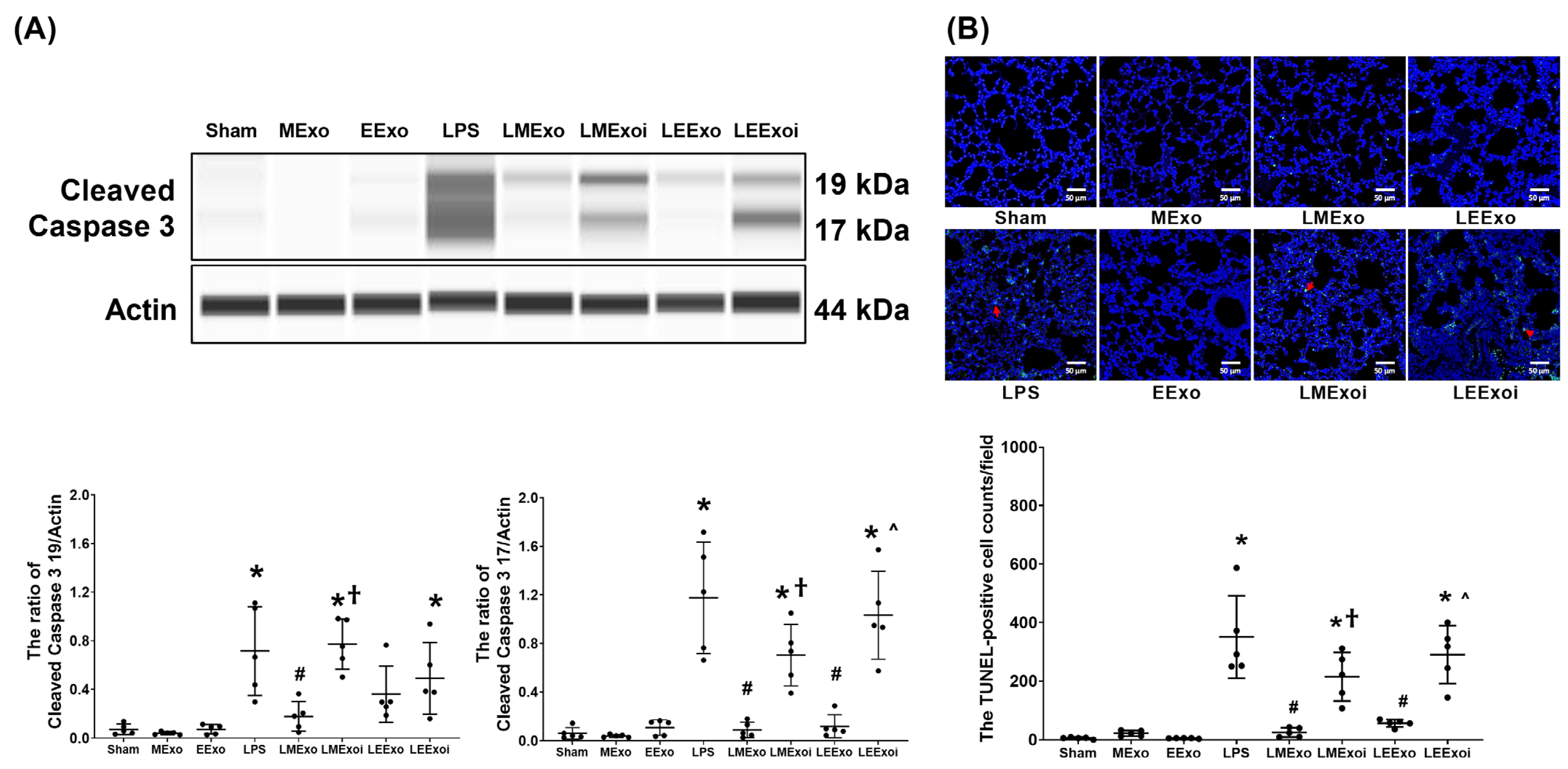
3.8. Lung Apoptosis
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Delano, M.J.; Ward, P.A. The immune system’s role in sepsis progression, resolution, and long-term outcome. Immunol. Rev. 2016, 274, 330–353. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V. Pulmonary Innate Immune Response Determines the Outcome of Inflammation During Pneumonia and Sepsis-Associated Acute Lung Injury. Front. Immunol. 2020, 11, 1722. [Google Scholar] [CrossRef] [PubMed]
- Rudd, K.E.; Johnson, S.C.; Agesa, K.M.; Shackelford, K.A.; Tsoi, D.; Kievlan, D.R.; Colombara, D.V.; Ikuta, K.S.; Kissoon, N.; Finfer, S.; et al. Global, regional, and national sepsis incidence and mortality, 1990–2017: Analysis for the Global Burden of Disease Study. Lancet 2020, 395, 200–211. [Google Scholar] [CrossRef] [PubMed]
- Hotchkiss, R.S.; Nicholson, D.W. Apoptosis and caspases regulate death and inflammation in sepsis. Nat. Rev. Immunol. 2006, 6, 813–822. [Google Scholar] [CrossRef] [PubMed]
- Su, L.-J.; Zhang, J.-H.; Gomez, H.; Murugan, R.; Hong, X.; Xu, D.; Jiang, F.; Peng, Z.-Y. Reactive Oxygen Species-Induced Lipid Peroxidation in Apoptosis, Autophagy, and Ferroptosis. Oxid. Med. Cell Longev. 2019, 2019, 5080843. [Google Scholar] [CrossRef] [PubMed]
- Danielski, L.G.; Giustina, A.D.; Bonfante, S.; Barichello, T.; Petronilho, F. The NLRP3 Inflammasome and Its Role in Sepsis Development. Inflammation 2020, 43, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Joffre, J.; Hellman, J. Oxidative Stress and Endothelial Dysfunction in Sepsis and Acute Inflammation. Antioxid. Redox Signal. 2021, 35, 1291–1307. [Google Scholar] [CrossRef] [PubMed]
- Mushtaq, A.; Kazi, F. Updates in sepsis management. Lancet Infect. Dis. 2022, 22, 24. [Google Scholar] [CrossRef] [PubMed]
- Freeman, B.D.; Natanson, C. Anti-inflammatory therapies in sepsis and septic shock. Expert. Opin. Investig. Drugs. 2000, 9, 1651–1663. [Google Scholar] [CrossRef]
- Mantzarlis, K.; Tsolaki, V.; Zakynthinos, E. Role of Oxidative Stress and Mitochondrial Dysfunction in Sepsis and Potential Therapies. Oxid. Med. Cell Longev. 2017, 2017, 5985209. [Google Scholar] [CrossRef]
- Shi, Y.; Wang, Y.; Li, Q.; Liu, K.; Hou, J.; Shao, C.; Wang, Y. Immunoregulatory mechanisms of mesenchymal stem and stromal cells in inflammatory diseases. Nat. Rev. Nephrol. 2018, 14, 493–507. [Google Scholar] [CrossRef] [PubMed]
- Stavely, R.; Nurgali, K. The emerging antioxidant paradigm of mesenchymal stem cell therapy. Stem Cells Transl. Med. 2020, 9, 985–1006. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Zhou, L.; Qiang, X.; Fang, B. Mesenchymal Stromal Cells Attenuate Infection-Induced Acute Respiratory Distress Syndrome in Animal Experiments: A Meta-Analysis. Cell Transplant. 2020, 29, 963689720969186. [Google Scholar]
- Homma, K.; Bazhanov, N.; Hashimoto, K.; Shimizu, M.; Heathman, T.; Hao, Q.; Nawgiri, R.; Muthukumarana, V.; Lee, J.W.; Prough, D.S.; et al. Mesenchymal stem cell-derived exosomes for treatment of sepsis. Front. Immunol. 2023, 14, 1136964. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chen, T.; Lei, P.; Tang, X.; Huang, P. Exosomes Released by Bone Marrow Mesenchymal Stem Cells Attenuate Lung Injury Induced by Intestinal Ischemia-Reperfusion via the TLR4/NF-κB Pathway. Int. J. Med. Sci. 2019, 16, 1238–1244. [Google Scholar] [CrossRef] [PubMed]
- Giebel, B.; Kordelas, L.; Börger, V. Clinical potential of mesenchymal stem/stromal cell derived extracellular vesicles. Stem Cell Investig. 2017, 4, 84. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Li, P.; Goodwin, A.J.; Cook, J.A.; Halushka, P.V.; Chang, E.; Zingarelli, B.; Fan, H. Exosomes from endothelial progenitor cells improve outcomes of the lipopolysaccharide-induced acute lung injury. Crit. Care 2019, 23, 44. [Google Scholar] [CrossRef] [PubMed]
- Chiang, M.D.; Chang, C.Y.; Shih, H.J.; Le, V.L.; Huang, Y.H.; Huang, C.J. Exosomes from Human Placenta Choriodecidual Membrane-Derived Mesenchymal Stem Cells Mitigate Endoplasmic Reticulum Stress, Inflammation, and Lung Injury in Lipopolysaccharide-Treated Obese Mice. Antioxidants 2022, 11, 615. [Google Scholar] [CrossRef]
- Krylova, S.V.; Feng, D. The Machinery of Exosomes: Biogenesis, Release, and Uptake. Int. J. Mol. Sci. 2023, 24, 1337. [Google Scholar] [CrossRef]
- Cai, J.; Tang, D.; Hao, X.; Liu, E.; Li, W.; Shi, J. Mesenchymal stem cell-derived exosome alleviates sepsis- associated acute liver injury by suppressing MALAT1 through microRNA-26a-5p: An innovative immunopharmacological intervention and therapeutic approach for sepsis. Front. Immunol. 2023, 14, 1157793. [Google Scholar] [CrossRef]
- Chang, C.-Y.; Chen, K.-Y.; Shih, H.-J.; Chiang, M.; Huang, I.-T.; Huang, Y.-H.; Huang, C.-J. Let-7i-5p Mediates the Therapeutic Effects of Exosomes from Human Placenta Choriodecidual Membrane-Derived Mesenchymal Stem Cells on Mitigating Endotoxin-Induced Mortality and Liver Injury in High-Fat Diet-Induced Obese Mice. Pharmaceuticals 2021, 15, 36. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zou, X.; Liu, F. Silencing TTTY15 mitigates hypoxia-induced mitochondrial energy metabolism dysfunction and cardiomyocytes apoptosis via TTTY15/let-7i-5p and TLR3/NF-κB pathways. Cell Signal 2020, 76, 109779. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, Y.; Tsuchiya, A.; Terai, S. The development of mesenchymal stem cell therapy in the present, and the perspective of cell-free therapy in the future. Clin. Mol. Hepatol. 2021, 27, 70–80. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.Y.; Tsai, P.S.; Hung, Y.C.; Huang, C.J. L-type calcium channels are involved in mediating the anti-inflammatory effects of magnesium sulphate. Br. J. Anaesth. 2010, 104, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Fernando, M.R.; Jiang, C.; Krzyzanowski, G.D.; Ryan, W.L. New evidence that a large proportion of human blood plasma cell-free DNA is localized in exosomes. PLoS ONE 2017, 12, e0183915. [Google Scholar] [CrossRef] [PubMed]
- Rumbaugh, G.; Miller, C.A. Epigenetic changes in the brain: Measuring global histone modifications. Methods Mol. Biol. 2011, 670, 263–274. [Google Scholar] [PubMed]
- Chang, C.Y.; Hsu, H.J.; Foo, J.; Shih, H.J.; Huang, C.J. Peptide-Based TNF-α-Binding Decoy Therapy Mitigates Lipopolysaccharide-Induced Liver Injury in Mice. Pharmaceuticals 2020, 13, 280. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.; Wu, L.; Liu, M.; Zhu, L.; Chen, Y.; Zhou, H.; Shi, X.; Wei, J.; Zheng, L.; Hu, X.; et al. Bone Marrow Mesenchymal Stem Cell-Derived Exosomes Attenuate LPS-Induced ARDS by Modulating Macrophage Polarization Through Inhibiting Glycolysis in Macrophages. Shock 2020, 54, 828–843. [Google Scholar] [CrossRef] [PubMed]
- Lo, B.; Parham, L. Ethical issues in stem cell research. Endocr. Rev. 2009, 30, 204–213. [Google Scholar] [CrossRef]
- de Mello, A.H.; Costa, A.B.; Engel, J.D.G.; Rezin, G.T. Mitochondrial dysfunction in obesity. Life Sci. 2018, 192, 26–32. [Google Scholar] [CrossRef]
- Li, Z.-L.; Woollard, J.R.; Ebrahimi, B.; Crane, J.A.; Jordan, K.L.; Lerman, A.; Wang, S.-M.; Lerman, L.O. Transition from obesity to metabolic syndrome is associated with altered myocardial autophagy and apoptosis. Arterioscler. Thromb. Vasc. Biol. 2012, 32, 1132–1141. [Google Scholar] [CrossRef] [PubMed]
- Guha, M.; Mackman, N. LPS induction of gene expression in human monocytes. Cell Signal. 2001, 13, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Nishi, K.; Oda, T.; Takabuchi, S.; Oda, S.; Fukuda, K.; Adachi, T.; Semenza, G.L.; Shingu, K.; Hirota, K. LPS induces hypoxia-inducible factor 1 activation in macrophage-differentiated cells in a reactive oxygen species-dependent manner. Antioxid. Redox Signal. 2008, 10, 983–995. [Google Scholar] [CrossRef] [PubMed]
- Falkenberg, M.; Gaspari, M.; Rantanen, A.; Trifunovic, A.; Larsson, N.-G.; Gustafsson, C.M. Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nat. Genet. 2002, 31, 289–294. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Duan, L.; Lu, J.; Xia, J. Engineering exosomes for targeted drug delivery. Theranostics 2021, 11, 3183–3195. [Google Scholar] [CrossRef] [PubMed]
- Le Blanc, K.; Tammik, C.; Rosendahl, K.; Zetterberg, E.; Ringdén, O. HLA expression and immunologic properties of differentiated and undifferentiated mesenchymal stem cells. Exp. Hematol. 2003, 31, 890–896. [Google Scholar] [CrossRef]
- Li, J.; Ezzelarab, M.B.; Cooper, D.K. Do mesenchymal stem cells function across species barriers? Relevance for xenotransplantation. Xenotransplantation 2012, 19, 273–285. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Le, V.L.; Chang, C.-Y.; Chuang, C.-W.; Syu, S.-H.; Shih, H.-J.; Nguyen Vo, H.-P.; Van, M.N.; Huang, C.-J. Therapeutic Effects of Engineered Exosomes from RAW264.7 Cells Overexpressing hsa-let-7i-5p against Sepsis in Mice—A Comparative Study with Human Placenta-Derived Mesenchymal Stem Cell Exosomes. J. Pers. Med. 2024, 14, 619. https://doi.org/10.3390/jpm14060619
Le VL, Chang C-Y, Chuang C-W, Syu S-H, Shih H-J, Nguyen Vo H-P, Van MN, Huang C-J. Therapeutic Effects of Engineered Exosomes from RAW264.7 Cells Overexpressing hsa-let-7i-5p against Sepsis in Mice—A Comparative Study with Human Placenta-Derived Mesenchymal Stem Cell Exosomes. Journal of Personalized Medicine. 2024; 14(6):619. https://doi.org/10.3390/jpm14060619
Chicago/Turabian StyleLe, Van Long, Chao-Yuan Chang, Ching-Wei Chuang, Syuan-Hao Syu, Hung-Jen Shih, Hong-Phuc Nguyen Vo, Minh Nguyen Van, and Chun-Jen Huang. 2024. "Therapeutic Effects of Engineered Exosomes from RAW264.7 Cells Overexpressing hsa-let-7i-5p against Sepsis in Mice—A Comparative Study with Human Placenta-Derived Mesenchymal Stem Cell Exosomes" Journal of Personalized Medicine 14, no. 6: 619. https://doi.org/10.3390/jpm14060619
APA StyleLe, V. L., Chang, C.-Y., Chuang, C.-W., Syu, S.-H., Shih, H.-J., Nguyen Vo, H.-P., Van, M. N., & Huang, C.-J. (2024). Therapeutic Effects of Engineered Exosomes from RAW264.7 Cells Overexpressing hsa-let-7i-5p against Sepsis in Mice—A Comparative Study with Human Placenta-Derived Mesenchymal Stem Cell Exosomes. Journal of Personalized Medicine, 14(6), 619. https://doi.org/10.3390/jpm14060619






