Beyond Biomarkers: Machine Learning-Driven Multiomics for Personalized Medicine in Gastric Cancer
Abstract
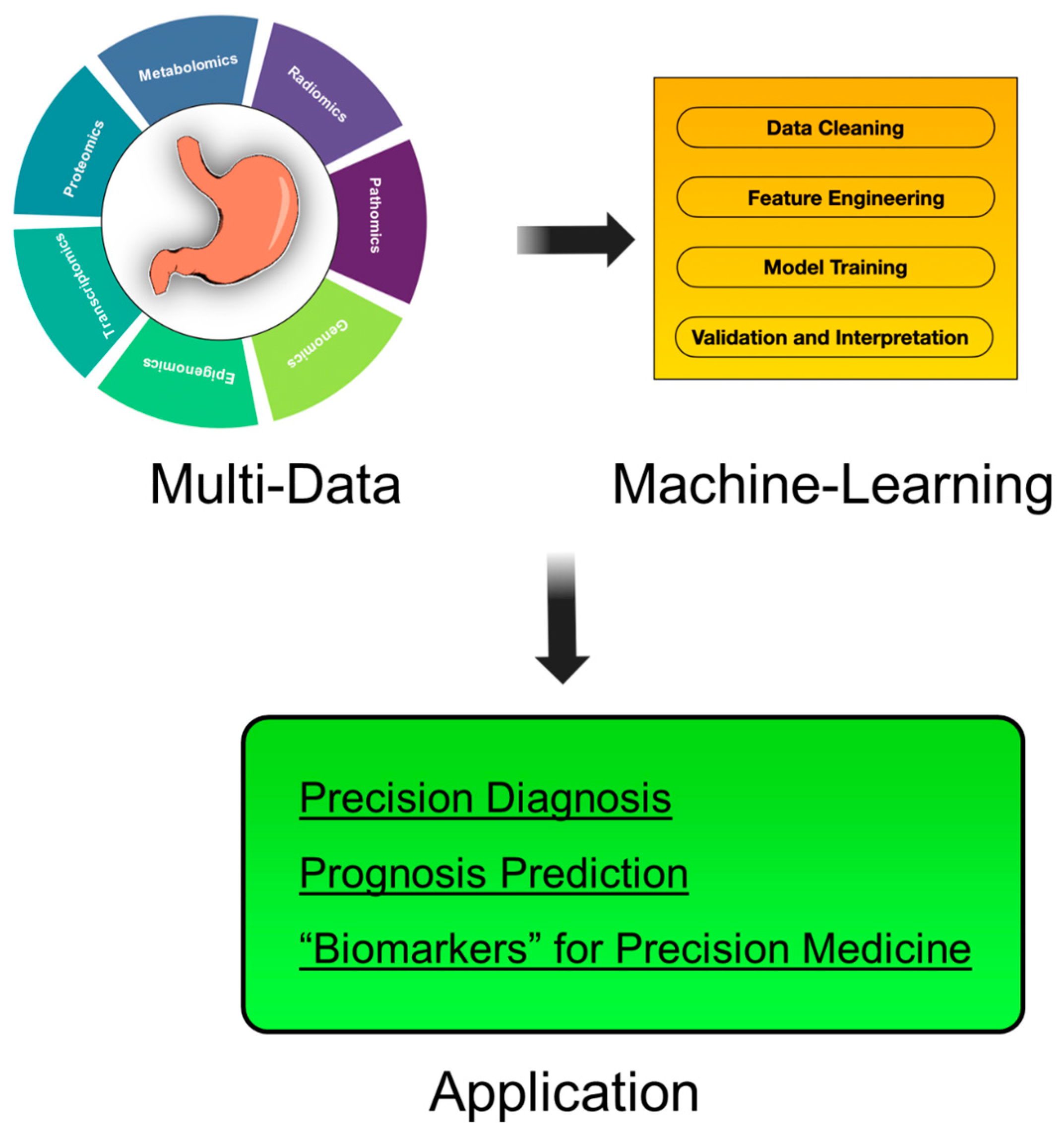
1. Introduction
2. Multiomics Data Types in GC
2.1. Imaging-Based Omics
2.1.1. Radiomics (Computed Tomography)
2.1.2. Radiomics (Endoscopy)
2.1.3. Pathomics
2.2. Molecular Omics
2.2.1. Genomics
2.2.2. Epigenomics
2.2.3. Transcriptomics
2.2.4. Proteomics
2.2.5. Metabolomics
3. ML for GC Research
3.1. Multiomics Integration
3.2. ML Algorithms
3.3. Computational Hardware Requirements (GPU)
4. ML-Driven Multiomics for Personalized Medicine in GC
4.1. Precision Diagnosis
4.1.1. Endoscopy-Driven Diagnosis
4.1.2. Liquid Biopsy and Multiomics Biomarkers
4.1.3. Pathomics for Definitive Diagnosis
4.2. Prognosis Prediction
4.2.1. Molecular Biomarkers
4.2.2. Treatment Response Prediction
4.3. “Biomarkers” for Personalized Medicine
4.3.1. Advancements in Imaging-Based “Biomarkers”
4.3.2. Imaging for Characterization
5. Challenges and Limitations
5.1. Technical Challenges
5.2. Clinical Translation Barriers
5.3. Ethical and Regulatory Issues
6. Future Directions
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Registration Committee of the Japanese GC Association; Katai, H.; Ishikawa, T.; Akazawa, K.; Isobe, Y.; Miyashiro, I.; Oda, I.; Tsujitani, S.; Ono, H.; Tanabe, S.; et al. Five-year survival analysis of surgically resected GC cases in Japan: A retrospective analysis of more than 100,000 patients from the nationwide registry of the Japanese GC Association (2001–2007). Gastric Cancer 2018, 21, 144–154. [Google Scholar] [CrossRef]
- Yashiro, M.; Matsuoka, T.; Ohira, M. The significance of scirrhous GC cell lines: The molecular characterization using cell lines and mouse models. Hum. Cell 2018, 31, 271–281. [Google Scholar] [CrossRef] [PubMed]
- Sohn, B.H.; Hwang, J.-E.; Jang, H.-J.; Lee, H.-S.; Oh, S.C.; Shim, J.-J.; Lee, K.-W.; Kim, E.H.; Yim, S.Y.; Lee, S.H.; et al. Clinical Significance of Four Molecular Subtypes of GC Identified by The Cancer Genome Atlas Project. Clin. Cancer Res. 2017, 23, 4441–4449. [Google Scholar] [CrossRef]
- Farzan, R. Artificial intelligence in Immuno-genetics. Bioinformation 2024, 20, 29–35. [Google Scholar] [CrossRef]
- Matsuoka, T.; Yashiro, M. Bioinformatics Analysis and Validation of Potential Markers Associated with Prediction and Prognosis of GC. Int. J. Mol. Sci. 2024, 25, 5880. [Google Scholar] [CrossRef] [PubMed]
- Cui, C.; Yang, H.; Wang, Y.; Zhao, S.; Asad, Z.; Coburn, L.A.; Wilson, K.T.; Landman, B.A.; Huo, Y. Deep multimodal fusion of image and non-image data in disease diagnosis and prognosis: A review. Prog. Biomed. Eng. 2023, 5, 022001. [Google Scholar] [CrossRef]
- Almeida, M.F.A.; Verza, L.; Bitencourt, A.G.V.; Boaventura, C.S.; Barbosa, P.N.V.P.; Chojniak, R. Computed tomography with a stomach protocol and virtual gastroscopy in the staging of GC: An initial experience. Radiol. Bras. 2018, 51, 211–217. [Google Scholar] [CrossRef]
- Bi, W.L.; Hosny, A.; Schabath, M.B.; Giger, M.L.; Birkbak, N.J.; Mehrtash, A.; Allison, T.; Arnaout, O.; Abbosh, C.; Dunn, I.F.; et al. Artificial intelligence in cancer imaging: Clinical challenges and applications. CA Cancer J. Clin. 2019, 69, 127–157. [Google Scholar] [CrossRef]
- Liu, Y.; Zhao, S.; Wu, Z.; Liang, H.; Chen, X.; Huang, C.; Lu, H.; Yuan, M.; Xue, X.; Luo, C.; et al. Virtual biopsy using CT radiomics for evaluation of disagreement in pathology between endoscopic biopsy and postoperative specimens in patients with GC: A dual-energy CT generalizability study. Insights Imaging 2023, 14, 118. [Google Scholar] [CrossRef]
- Ma, T.; Cui, J.; Wang, L.; Li, H.; Ye, Z.; Gao, X. A multiphase contrast-enhanced CT radiomics model for prediction of human epidermal growth factor receptor 2 status in advanced GC. Front. Genet. 2022, 13, 968027. [Google Scholar] [CrossRef]
- Dong, D.; Fang, M.-J.; Tang, L.; Shan, X.-H.; Gao, J.-B.; Giganti, F.; Wang, R.-P.; Chen, X.; Wang, X.-X.; Palumbo, D.; et al. Deep learning radiomic nomogram can predict the number of lymph node metastasis in locally advanced GC: An international multicenter study. Ann. Oncol. 2020, 31, 912–920. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Deng, J.; Dong, D.; Fang, M.; Ye, Z.; Hu, Y.; Li, H.; Zhong, L.; Cao, R.; Zhao, X.; et al. Deep learning-based radiomics model can predict extranodal soft tissue metastasis in GC. Med. Phys. 2024, 51, 267–277. [Google Scholar] [CrossRef]
- Zhu, Z.-N.; Feng, Q.-X.; Li, Q.; Xu, W.-Y.; Liu, X.-S. Machine learning-based CT radiomics approach for predicting occult peritoneal metastasis in advanced GC preoperatively. Clin. Radiol. 2025, 80, 106727. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Zhou, K.; Sun, Z.; Wang, H.; Xie, J.; Zhang, T.; Sang, S.; Islam, M.T.; Wang, J.-Y.; Chen, C.; et al. Non-invasive tumor microenvironment evaluation and treatment response prediction in GC using deep learning radiomics. Cell Rep. Med. 2023, 4, 101146. [Google Scholar] [CrossRef] [PubMed]
- Hao, D.; Li, Q.; Feng, Q.-X.; Qi, L.; Liu, X.-S.; Arefan, D.; Zhang, Y.-D.; Wu, S. SurvivalCNN: A deep learning-based method for GC survival prediction using radiological imaging data and clinicopathological variables. Artif. Intell. Med. 2022, 134, 102424. [Google Scholar] [CrossRef]
- Zhi, H.; Xiang, Y.; Chen, C.; Zhang, W.; Lin, J.; Gao, Z.; Shen, Q.; Shao, J.; Yang, X.; Yang, Y.; et al. Development and validation of a machine learning-based 18F-fluorodeoxyglucose PET/CT radiomics signature for predicting GC survival. Cancer Imaging 2024, 24, 99. [Google Scholar] [CrossRef]
- Adili, D.; Mohetaer, A.; Zhang, W. Diagnostic accuracy of radiomics-based machine learning for neoadjuvant chemotherapy response and survival prediction in GC patients: A systematic review and meta-analysis. Eur. J. Radiol. 2024, 173, 111249. [Google Scholar] [CrossRef]
- Wu, L.; He, X.; Liu, M.; Xie, H.; An, P.; Zhang, J.; Zhang, H.; Ai, Y.; Tong, Q.; Guo, M.; et al. Evaluation of the effects of an artificial intelligence system on endoscopy quality and preliminary testing of its performance in detecting early GC: A randomized controlled trial. Endoscopy 2021, 53, 1199–1207. [Google Scholar] [CrossRef]
- Klang, E.; Soroush, A.; Nadkarni, G.; Sharif, K.; Lahat, A. Deep Learning and GC: Systematic Review of AI-Assisted Endoscopy. Diagnostics 2023, 13, 3613. [Google Scholar] [CrossRef]
- Islam, M.M.; Poly, T.N.; Walther, B.A.; Lin, M.-C.; Li, Y.-C. (Jack) Artificial Intelligence in GC: Identifying GC Using Endoscopic Images with Convolutional Neural Network. Cancers 2021, 13, 5253. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Wang, Q.-C.; Xu, M.-D.; Zhang, Z.; Cheng, J.; Zhong, Y.-S.; Zhang, Y.-Q.; Chen, W.-F.; Yao, L.-Q.; Zhou, P.-H.; et al. Application of convolutional neural network in the diagnosis of the invasion depth of GC based on conventional endoscopy. Gastrointest. Endosc. 2019, 89, 806–815.e1. [Google Scholar] [CrossRef]
- Goto, A.; Kubota, N.; Nishikawa, J.; Ogawa, R.; Hamabe, K.; Hashimoto, S.; Ogihara, H.; Hamamoto, Y.; Yanai, H.; Miura, O.; et al. Cooperation between artificial intelligence and endoscopists for diagnosing invasion depth of early GC. Gastric Cancer 2023, 26, 116–122. [Google Scholar] [CrossRef] [PubMed]
- Gong, Y.; Gong, L.; Luo, F. Deep-learning-based clinical decision support system for gastric neoplasms in real-time endoscopy: Development and validation study. SSRN J. 2022, 58, 701–708. [Google Scholar] [CrossRef]
- Pornvoraphat, P.; Tiankanon, K.; Pittayanon, R.; Sunthornwetchapong, P.; Vateekul, P.; Rerknimitr, R. Real-time gastric intestinal metaplasia diagnosis tailored for bias and noisy-labeled data with multiple endoscopic imaging. Comput. Biol. Med. 2023, 154, 106582. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Dong, Z.; Wu, L.; Li, Y.; Liu, J.; Luo, C.; Zeng, X.; Deng, Y.; Cheng, D.; Diao, W.; et al. A deep-learning based system using multi-modal data for diagnosing gastric neoplasms in real-time (with video). Gastric Cancer 2023, 26, 275–285. [Google Scholar] [CrossRef]
- Choi, S.; Kim, S. Artificial Intelligence in the Pathology of GC. J. Gastric Cancer 2023, 23, 410. [Google Scholar] [CrossRef] [PubMed]
- Jenkinson, E.; Arandjelović, O. Whole Slide Image Understanding in Pathology: What Is the Salient Scale of Analysis? BioMedInformatics 2024, 4, 489–518. [Google Scholar] [CrossRef]
- Jang, H.-J.; Lee, A.; Kang, J.; Song, I.H.; Lee, S.H. Prediction of genetic alterations from GC histopathology images using a fully automated deep learning approach. World J. Gastroenterol. 2021, 27, 7687–7704. [Google Scholar] [CrossRef]
- Veldhuizen, G.P.; Röcken, C.; Behrens, H.-M.; Cifci, D.; Muti, H.S.; Yoshikawa, T.; Arai, T.; Oshima, T.; Tan, P.; Ebert, M.P.; et al. Deep learning-based subtyping of GC histology predicts clinical outcome: A multi-institutional retrospective study. Gastric Cancer 2023, 26, 708–720. [Google Scholar] [CrossRef]
- Lee, S.H.; Lee, Y.; Jang, H. Deep learning captures selective features for discrimination of microsatellite instability from pathologic tissue slides of GC. Int. J. Cancer 2023, 152, 298–307. [Google Scholar] [CrossRef] [PubMed]
- Matsushima, J.; Sato, T.; Ohnishi, T.; Yoshimura, Y.; Mizutani, H.; Koto, S.; Ikeda, J.; Kano, M.; Matsubara, H.; Hayashi, H. The Use of Deep Learning-Based Computer Diagnostic Algorithm for Detection of Lymph Node Metastases of Gastric Adenocarcinoma. Int. J. Surg. Pathol. 2023, 31, 975–981. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, W.; Ruan, R.; Zhang, Z.; Wang, Z.; Guan, T.; Lin, Q.; Tang, W.; Deng, J.; Wang, Z.; et al. Deep learning based digital pathology for predicting treatment response to first-line PD-1 blockade in advanced GC. J. Transl. Med. 2024, 22, 438. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhao, J.; Marostica, E.; Yuan, W.; Jin, J.; Zhang, J.; Li, R.; Tang, H.; Wang, K.; Li, Y.; et al. A pathology foundation model for cancer diagnosis and prognosis prediction. Nature 2024, 634, 970–978. [Google Scholar] [CrossRef] [PubMed]
- Dongare, D.B.; Nishad, S.S.; Mastoli, S.Y.; Saraf, S.A.; Srivastava, N.; Dey, A. High-throughput sequencing: A breakthrough in molecular diagnosis for precision medicine. Funct. Integr. Genom. 2025, 25, 22. [Google Scholar] [CrossRef]
- Zhang, S.; Li, Z.; Hu, G.; Chen, H. Integrative single-cell and multi-omics analyses reveal ferroptosis-associated gene expression and immune microenvironment heterogeneity in GC. Discov. Oncol. 2025, 16, 57. [Google Scholar] [CrossRef]
- Reddavid, R.; Dagatti, S.; Franco, C.; Puca, L.; Tomatis, M.; Corso, S.; Giordano, S.; Degiuli, M. Molecularly Targeted Therapies for GC. State of the Art. Cancers 2021, 13, 4094. [Google Scholar] [CrossRef]
- Fan, P.; Zhang, Z.; Lu, L.; Guo, X.; Hao, Z.; Wang, X.; Ye, Y. Association of single nucleotide polymorphisms (SNPs) with GC susceptibility and prognosis in population in Wuwei, Gansu, China. World J. Surg. Oncol. 2022, 20, 194. [Google Scholar] [CrossRef]
- Chen, Q.; Wang, Y.; Liu, Y.; Xi, B. ESRRG, ATP4A, and ATP4B as Diagnostic Biomarkers for GC: A Bioinformatic Analysis Based on Machine Learning. Front. Physiol. 2022, 13, 905523. [Google Scholar] [CrossRef]
- Cristiano, S.; Leal, A.; Phallen, J.; Fiksel, J.; Adleff, V.; Bruhm, D.C.; Jensen, S.Ø.; Medina, J.E.; Hruban, C.; White, J.R.; et al. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature 2019, 570, 385–389. [Google Scholar] [CrossRef]
- Canale, M.; Casadei-Gardini, A.; Ulivi, P.; Arechederra, M.; Berasain, C.; Lollini, P.-L.; Fernández-Barrena, M.G.; Avila, M.A. Epigenetic Mechanisms in GC: Potential New Therapeutic Opportunities. Int. J. Mol. Sci. 2020, 21, 5500. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zeng, W.; Ni, X.; Liu, Q.; Li, W.; Stackpole, M.L.; Zhou, Y.; Gower, A.; Krysan, K.; Ahuja, P.; et al. Comprehensive tissue deconvolution of cell-free DNA by deep learning for disease diagnosis and monitoring. Proc. Natl. Acad. Sci. USA 2023, 120, e2305236120. [Google Scholar] [CrossRef] [PubMed]
- Su, C.; Lin, Z.; Ye, Z.; Liang, J.; Yu, R.; Wan, Z.; Hou, J. Development of a prognostic model for early-stage GC-related DNA methylation-driven genes and analysis of immune landscape. Front. Mol. Biosci. 2024, 11, 1455890. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.; Hong, B.; Wang, S.; Wang, J.; Fang, J.; Sun, R.; Nie, J.; Wang, H. Plasma cell-free DNA methylome-based liquid biopsy for accurate GC detection. Cancer Sci. 2024, 115, 3426–3438. [Google Scholar] [CrossRef]
- Kandimalla, R.; Xu, J.; Link, A.; Matsuyama, T.; Yamamura, K.; Parker, M.I.; Uetake, H.; Balaguer, F.; Borazanci, E.; Tsai, S.; et al. EpiPanGI Dx: A Cell-free DNA Methylation Fingerprint for the Early Detection of Gastrointestinal Cancers. Clin. Cancer Res. 2021, 27, 6135–6144. [Google Scholar] [CrossRef]
- Sasaki, A.; Takeshima, H.; Yamashita, S.; Ichita, C.; Kawachi, J.; Naito, W.; Ohashi, Y.; Takeuchi, C.; Fukuda, M.; Furuichi, Y.; et al. Severe induction of aberrant DNA methylation by nodular gastritis in adults. J. Gastroenterol. 2024, 59, 442–456. [Google Scholar] [CrossRef]
- Wu, Z.; Wang, W.; Zhang, K.; Fan, M.; Lin, R. Epigenetic and Tumor Microenvironment for Prognosis of Patients with GC. Biomolecules 2023, 13, 736. [Google Scholar] [CrossRef]
- Ho, S.W.T.; Sheng, T.; Xing, M.; Ooi, W.F.; Xu, C.; Sundar, R.; Huang, K.K.; Li, Z.; Kumar, V.; Ramnarayanan, K.; et al. Regulatory enhancer profiling of mesenchymal-type GC reveals subtype-specific epigenomic landscapes and targetable vulnerabilities. Gut 2023, 72, 226–241. [Google Scholar] [CrossRef]
- Nie, C.; Zhai, J.; Wang, Q.; Zhu, X.; Xiang, G.; Liu, C.; Liu, T.; Wang, W.; Wang, Y.; Zhao, Y.; et al. Comprehensive Analysis of an Individualized Immune-Related lncRNA Pair Signature in GC. Front. Cell Dev. Biol. 2022, 10, 805623. [Google Scholar] [CrossRef]
- Peng, Q.; Zhang, P.; Liu, G.; Lu, L. Integrated single-cell and bulk RNA sequencing analyses identify an immunotherapy nonresponse-related fibroblast signature in GC. Anti-Cancer Drugs 2024, 35, 952–968. [Google Scholar] [CrossRef]
- Liu, J.; Cheng, Z. Identification of Hub Genes Associated with Tumor-Infiltrating Immune Cellsand ECM Dynamics as the Potential Therapeutic Targets in GCthrough an Integrated Bioinformatic Analysis and Machine Learning Methods. Comb. Chem. High Throughput Screen. 2023, 26, 653–667. [Google Scholar] [CrossRef] [PubMed]
- Xie, R.; Liu, L.; Lu, X.; He, C.; Li, G. Identification of the diagnostic genes and immune cell infiltration characteristics of GC using bioinformatics analysis and machine learning. Front. Genet. 2023, 13, 1067524. [Google Scholar] [CrossRef]
- Sundar, R.; Barr Kumarakulasinghe, N.; Huak Chan, Y.; Yoshida, K.; Yoshikawa, T.; Miyagi, Y.; Rino, Y.; Masuda, M.; Guan, J.; Sakamoto, J.; et al. Machine-learning model derived gene signature predictive of paclitaxel survival benefit in GC: Results from the randomised phase III SAMIT trial. Gut 2022, 71, 676–685. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, Z.; Zhang, C. Integrated Analysis of Serum and Tissue microRNA Transcriptome for Biomarker Discovery in GC. Environ. Toxicol. 2025, 40, 281–290. [Google Scholar] [CrossRef]
- Lee, S.; Park, J.; Oh, S.; Kwack, K. Downregulation of LOC441461 Promotes Cell Growth and Motility in Human GC. Cancers 2022, 14, 1149. [Google Scholar] [CrossRef]
- Zhong, M.; Yu, Z.; Wu, Q.; Lu, B.; Sun, P.; Zhang, X.; Yang, L.; Wu, H. PCDHGA10 as a potential prognostic biomarker and correlated with immune infiltration in GC. Front. Immunol. 2024, 15, 1500478. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Li, X.; Wang, Q.; Chen, P.; Zhao, L.; Gao, Y. Proteomic profiling and biomarker discovery for predicting the response to PD-1 inhibitor immunotherapy in GC patients. Front. Pharmacol. 2024, 15, 1349459. [Google Scholar] [CrossRef]
- Zhou, B.; Zhou, Z.; Chen, Y.; Deng, H.; Cai, Y.; Rao, X.; Yin, Y.; Rong, L. Plasma proteomics-based identification of novel biomarkers in early GC. Clin. Biochem. 2020, 76, 5–10. [Google Scholar] [CrossRef]
- Gu, J.; Xie, S.; Li, X.; Wu, Z.; Xue, L.; Wang, S.; Wei, W. Identification of plasma proteomic signatures associated with the progression of cardia GC and precancerous lesions. J. Natl. Cancer Cent. 2023, 3, 286–294. [Google Scholar] [CrossRef]
- Li, J.; Zhao, W.; Yang, J.; Lu, P.; Sun, H.; Zhang, Z.; Gu, J. Proteomic and serological markers for diagnosing cardia GC and precursor lesions in a Chinese population. Sci. Rep. 2024, 14, 25309. [Google Scholar] [CrossRef]
- Vizeacoumar, F.S.; Guo, H.; Dwernychuk, L.; Zaidi, A.; Freywald, A.; Wu, F.-X.; Vizeacoumar, F.J.; Ahmed, S. Mining the plasma-proteome associated genes in patients with gastro-esophageal cancers for biomarker discovery. Sci. Rep. 2021, 11, 7590. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Guo, T.; Wu, X.; Gan, X.; Wang, Y.; Jia, F.; Zhang, Y.; Xing, X.; Gao, X.; Li, Z. Clinical Significance and Immune Infiltration Analyses of the Cuproptosis-Related Human Copper Proteome in GC. Biomolecules 2022, 12, 1459. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Cai, G.; Jiang, J.; He, C.; Chen, Y.; Ding, Y.; Lu, J.; Zhao, W.; Yang, Y.; Zhang, Y.; et al. Proteomic profiling of GC with peritoneal metastasis identifies a protein signature associated with immune microenvironment and patient outcome. Gastric Cancer 2023, 26, 504–516. [Google Scholar] [CrossRef]
- Lee, J.; Lee, S.W.; Kang, S.H.; Seol, D.; Yoo, M.; Hwang, D.; Lee, E.; Park, Y.S.; Ahn, S.-H.; Suh, Y.-S.; et al. MUC16 as a serum-based prognostic indicator of prometastatic GC. Sci. Rep. 2024, 14, 15173. [Google Scholar] [CrossRef]
- Sabit, H.; Arneth, B.; Pawlik, T.M.; Abdel-Ghany, S.; Ghazy, A.; Abdelazeem, R.M.; Alqosaibi, A.; Al-Dhuayan, I.S.; Almulhim, J.; Alrabiah, N.A.; et al. Leveraging Single-Cell Multi-Omics to Decode Tumor Microenvironment Diversity and Therapeutic Resistance. Pharmaceuticals 2025, 18, 75. [Google Scholar] [CrossRef] [PubMed]
- Barberis, E.; Khoso, S.; Sica, A.; Falasca, M.; Gennari, A.; Dondero, F.; Afantitis, A.; Manfredi, M. Precision Medicine Approaches with Metabolomics and Artificial Intelligence. Int. J. Mol. Sci. 2022, 23, 11269. [Google Scholar] [CrossRef]
- Han, Y.; Yoo, H.J.; Jee, S.H.; Lee, J.H. High serum levels of l-carnitine and citric acid negatively correlated with alkaline phosphatase are detectable in Koreans before GC onset. Metabolomics 2022, 18, 62. [Google Scholar] [CrossRef]
- Liu, Z.-C.; Wu, W.-H.; Huang, S.; Li, Z.-W.; Li, X.; Shui, G.-H.; Lam, S.M.; Li, B.-W.; Li, Z.-X.; Zhang, Y.; et al. Plasma lipids signify the progression of precancerous gastric lesions to GC: A prospective targeted lipidomics study. Theranostics 2022, 12, 4671–4683. [Google Scholar] [CrossRef]
- Wei, M.X.; Yang, Z.; Wang, P.P.; Zhao, X.K.; Song, X.; Xu, R.H.; Hu, J.F.; Zhong, K.; Lei, L.L.; Han, W.L.; et al. Novel metabolic biomarker for early detection and diagnosis to the patients with gastric cardia adenocarcinoma. Cancer Med. 2024, 13, e7015. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, B.; Zhao, Y.; Shao, X.; Wang, M.; Ma, F.; Yang, L.; Nie, M.; Jin, P.; Yao, K.; et al. Metabolomic machine learning predictor for diagnosis and prognosis of GC. Nat. Commun. 2024, 15, 1657. [Google Scholar] [CrossRef]
- Kaji, S.; Irino, T.; Kusuhara, M.; Makuuchi, R.; Yamakawa, Y.; Tokunaga, M.; Tanizawa, Y.; Bando, E.; Kawamura, T.; Kami, K.; et al. Metabolomic profiling of GC tissues identified potential biomarkers for predicting peritoneal recurrence. Gastric Cancer 2020, 23, 874–883. [Google Scholar] [CrossRef] [PubMed]
- Qu, T.; Zhang, S.; Yang, S.; Li, S.; Wang, D. Utilizing serum metabolomics for assessing postoperative efficacy and monitoring recurrence in GC patients. BMC Cancer 2024, 24, 27. [Google Scholar] [CrossRef]
- Li, J.; Liu, Y.; Zhou, P.; Fan, Q.; Liu, H. Mass spectrometry-based pseudotargeted metabolomics reveals metabolic variations in a2-induced GC cell. Eur. J. Mass Spectrom. 2024, 30, 199–206. [Google Scholar] [CrossRef]
- Wang, F.; Pang, R.; Zhao, X.; Zhou, B.; Tian, Y.; Ma, Y.; Rong, L. Plasma metabolomics and lipidomics reveal potential novel biomarkers in early GC: An explorative study. Int. J. Biol. Markers 2024, 39, 226–238. [Google Scholar] [CrossRef]
- Fu, L.; Song, L.; Zhou, X.; Chen, L.; Zheng, L.; Hu, D.; Zhu, S.; Hu, Y.; Gong, D.; Chen, C.-L.; et al. Serum metabolomics analysis of malnutrition in patients with GC: A cross sectional study. BMC Cancer 2024, 24, 1195. [Google Scholar] [CrossRef] [PubMed]
- Tao, J.; Liu, D.; Hu, F.-B.; Zhang, X.; Yin, H.; Zhang, H.; Zhang, K.; Huang, Z.; Yang, K. Development and Validation of a Computed Tomography–Based Model for Noninvasive Prediction of the T Stage in GC: Multicenter Retrospective Study. J. Med. Internet Res. 2024, 26, e56851. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Dong, Z.; Cheng, J.; Bu, X.; Qiu, K.; Yang, C.; Wang, J.; Niu, W.; Wu, X.; Xu, J.; et al. Diagnosis and segmentation effect of the ME-NBI-based deep learning model on gastric neoplasms in patients with suspected superficial lesions—A multicenter study. Front. Oncol. 2023, 12, 1075578. [Google Scholar] [CrossRef]
- Saldanha, O.L.; Muti, H.S.; Grabsch, H.I.; Langer, R.; Dislich, B.; Kohlruss, M.; Keller, G.; Van Treeck, M.; Hewitt, K.J.; Kolbinger, F.R.; et al. Direct prediction of genetic aberrations from pathology images in GC with swarm learning. Gastric Cancer 2023, 26, 264–274. [Google Scholar] [CrossRef]
- Cheong, J.-H.; Wang, S.C.; Park, S.; Porembka, M.R.; Christie, A.L.; Kim, H.; Kim, H.S.; Zhu, H.; Hyung, W.J.; Noh, S.H.; et al. Development and validation of a prognostic and predictive 32-gene signature for GC. Nat. Commun. 2022, 13, 774. [Google Scholar] [CrossRef]
- Li, Y.; Sun, R.; Zhang, Y.; Yuan, Y.; Miao, Y. A methylation-based mRNA signature predicts survival in patients with GC. Cancer Cell Int. 2020, 20, 284. [Google Scholar] [CrossRef]
- Kong, J.; Ha, D.; Lee, J.; Kim, I.; Park, M.; Im, S.-H.; Shin, K.; Kim, S. Network-based machine learning approach to predict immunotherapy response in cancer patients. Nat. Commun. 2022, 13, 3703. [Google Scholar] [CrossRef]
- Momeni, Z.; Hassanzadeh, E.; Saniee Abadeh, M.; Bellazzi, R. A survey on single and multi omics data mining methods in cancer data classification. J. Biomed. Inform. 2020, 107, 103466. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, F.; Yang, Y.; Yu, N.; Weng, X.; Yang, Y.; Gong, Z.; Huang, S.; Gan, L.; Sun, S.; et al. Integrated DNA and RNA sequencing reveals early drivers involved in metastasis of GC. Cell Death Dis. 2022, 13, 392. [Google Scholar] [CrossRef] [PubMed]
- Pereira, B.S.; Wisnieski, F.; Calcagno, D.Q.; Santos, L.C.; Gigek, C.O.; Chen, E.S.; Rasmussen, L.T.; Payão, S.L.M.; Almeida, R.S.; Pinto, C.A.; et al. Genetic and Transcriptional Analysis of 8q24.21 Cluster in GC. Anticancer Res. 2022, 42, 4381–4394. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Peng, J.; Xie, J.; Xie, Y. Comprehensive analysis of the function of helicobacter-associated ferroptosis gene YWHAE in GC through multi-omics integration, molecular docking, and machine learning. Apoptosis 2024, 29, 439–456. [Google Scholar] [CrossRef] [PubMed]
- Park, C.H.; Hong, C.; Lee, A.-R.; Sung, J.; Hwang, T.H. Multi-omics reveals microbiome, host gene expression, and immune landscape in gastric carcinogenesis. iScience 2022, 25, 103956. [Google Scholar] [CrossRef]
- Wang, J.; Feng, J.; Chen, X.; Weng, Y.; Wang, T.; Wei, J.; Zhan, Y.; Peng, M. Integrated multi-omics analysis and machine learning identify hub genes and potential mechanisms of resistance to immunotherapy in GC. Aging 2024, 16, 7331–7356. [Google Scholar] [CrossRef]
- Li, J.; Xu, S.; Zhu, F.; Shen, F.; Zhang, T.; Wan, X.; Gong, S.; Liang, G.; Zhou, Y. Multi-omics Combined with Machine Learning Facilitating the Diagnosis of GC. Curr. Med. Chem. 2024, 31, 6692–6712. [Google Scholar] [CrossRef]
- Theodorakis, N.; Feretzakis, G.; Tzelves, L.; Paxinou, E.; Hitas, C.; Vamvakou, G.; Verykios, V.S.; Nikolaou, M. Integrating Machine Learning with Multi-Omics Technologies in Geroscience: Towards Personalized Medicine. J. Pers. Med. 2024, 14, 931. [Google Scholar] [CrossRef]
- Cavicchioli, R.; Hu, J.C.; Loli Piccolomini, E.; Morotti, E.; Zanni, L. GPU acceleration of a model-based iterative method for Digital Breast Tomosynthesis. Sci. Rep. 2020, 10, 43. [Google Scholar] [CrossRef]
- Lee, S.; Amgad, M.; Mobadersany, P.; McCormick, M.; Pollack, B.P.; Elfandy, H.; Hussein, H.; Gutman, D.A.; Cooper, L.A.D. Interactive Classification of Whole-Slide Imaging Data for Cancer Researchers. Cancer Res. 2021, 81, 1171–1177. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.Y.; Chen, B.; Williamson, D.F.K.; Chen, R.J.; Liang, I.; Ding, T.; Jaume, G.; Odintsov, I.; Le, L.P.; Gerber, G.; et al. A visual-language foundation model for computational pathology. Nat. Med. 2024, 30, 863–874. [Google Scholar] [CrossRef] [PubMed]
- White, C.D.; Chetty, R.; Weldon, J.; Morrissey, M.E.; Sykes, R.; Gîrleanu, C.; Colleuori, M.; Fitzgerald, J.; Power, A.; Ahmad, A.; et al. A deep learning approach to case prioritisation of colorectal biopsies. Histopathology 2025, 86, 373–384. [Google Scholar] [CrossRef]
- Gustav, M.; Reitsam, N.G.; Carrero, Z.I.; Loeffler, C.M.L.; Van Treeck, M.; Yuan, T.; West, N.P.; Quirke, P.; Brinker, T.J.; Brenner, H.; et al. Deep learning for dual detection of microsatellite instability and POLE mutations in colorectal cancer histopathology. NPJ Precis. Oncol. 2024, 8, 115. [Google Scholar] [CrossRef]
- Hilgers, L.; Ghaffari Laleh, N.; West, N.P.; Westwood, A.; Hewitt, K.J.; Quirke, P.; Grabsch, H.I.; Carrero, Z.I.; Matthaei, E.; Loeffler, C.M.L.; et al. Automated curation of large-scale cancer histopathology image datasets using deep learning. Histopathology 2024, 84, 1139–1153. [Google Scholar] [CrossRef]
- Yang, Z.; Zhang, Y.; Zhuo, L.; Sun, K.; Meng, F.; Zhou, M.; Sun, J. Prediction of prognosis and treatment response in ovarian cancer patients from histopathology images using graph deep learning: A multicenter retrospective study. Eur. J. Cancer 2024, 199, 113532. [Google Scholar] [CrossRef]
- Huang, K.; Li, M.; Li, Q.; Chen, Z.; Zhang, Y.; Gu, Z. Image-based profiling and deep learning reveal morphological heterogeneity of colorectal cancer organoids. Comput. Biol. Med. 2024, 173, 108322. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Shi, Z.; Zhang, Y.; Wei, L.; Duan, M.; Xiao, M.; Wang, J.; Chen, S.; Wang, Q.; Huang, J.; et al. Development and validation of a deep learning model for morphological assessment of myeloproliferative neoplasms using clinical data and digital pathology. Br. J. Haematol. 2024, 206, 596–606. [Google Scholar] [CrossRef]
- Liao, J.; Li, X.; Gan, Y.; Han, S.; Rong, P.; Wang, W.; Li, W.; Zhou, L. Artificial intelligence assists precision medicine in cancer treatment. Front. Oncol. 2023, 12, 998222. [Google Scholar] [CrossRef]
- Díaz Del Arco, C.; Fernández Aceñero, M.J.; Ortega Medina, L. Molecular Classifications in GC: A Call for Interdisciplinary Collaboration. Int. J. Mol. Sci. 2024, 25, 2649. [Google Scholar] [CrossRef]
- Tao, Y.; Xing, S.; Zuo, S.; Bao, P.; Jin, Y.; Li, Y.; Li, M.; Wu, Y.; Chen, S.; Wang, X.; et al. Cell-free multi-omics analysis reveals potential biomarkers in gastrointestinal cancer patients’ blood. Cell Rep. Med. 2023, 4, 101281. [Google Scholar] [CrossRef]
- Yuan, Q.; Deng, D.; Pan, C.; Ren, J.; Wei, T.; Wu, Z.; Zhang, B.; Li, S.; Yin, P.; Shang, D. Integration of transcriptomics, proteomics, and metabolomics data to reveal HER2-associated metabolic heterogeneity in GC with response to immunotherapy and neoadjuvant chemotherapy. Front. Immunol. 2022, 13, 951137. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y.; Okamoto, K.; Kawano, Y.; Kasai, A.; Kawaguchi, T.; Sagawa, T.; Sogabe, M.; Miyamoto, H.; Takayama, T. Novel Biomarkers of GC: Current Research and Future Perspectives. J. Clin. Med. 2023, 12, 4646. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Feng, L.; Huang, Y.; Xue, J.; Feng, Z.-B.; Long, L. Development and validation of a Radiopathomics model based on CT scans and whole slide images for discriminating between Stage I-II and Stage III GC. BMC Cancer 2024, 24, 368. [Google Scholar] [CrossRef] [PubMed]
- Tian, M.; Yao, Z.; Zhou, Y.; Gan, Q.; Wang, L.; Lu, H.; Wang, S.; Zhou, P.; Dai, Z.; Zhang, S.; et al. DeepRisk network: An AI-based tool for digital pathology signature and treatment responsiveness of GC using whole-slide images. J. Transl. Med. 2024, 22, 182. [Google Scholar] [CrossRef]
- Huang, W.; Zhou, K.; Jiang, Y.; Chen, C.; Yuan, Q.; Han, Z.; Xie, J.; Yu, S.; Sun, Z.; Hu, Y.; et al. Radiomics Nomogram for Prediction of Peritoneal Metastasis in Patients With GC. Front. Oncol. 2020, 10, 1416. [Google Scholar] [CrossRef]
- Zhao, H.; Gao, J.; Bai, B.; Wang, R.; Yu, J.; Lu, H.; Cheng, M.; Liang, P. Development and external validation of a non-invasive imaging biomarker to estimate the microsatellite instability status of GC and its prognostic value: The combination of clinical and quantitative CT-imaging features. Eur. J. Radiol. 2023, 162, 110719. [Google Scholar] [CrossRef]
- Kim, K.W.; Huh, J.; Urooj, B.; Lee, J.; Lee, J.; Lee, I.-S.; Park, H.; Na, S.; Ko, Y. Artificial Intelligence in GC Imaging With Emphasis on Diagnostic Imaging and Body Morphometry. J. Gastric Cancer 2023, 23, 388. [Google Scholar] [CrossRef]
- Martínez-García, M.; Hernández-Lemus, E. Data Integration Challenges for Machine Learning in Precision Medicine. Front. Med. 2022, 8, 784455. [Google Scholar] [CrossRef]
- Hayes, C.N.; Nakahara, H.; Ono, A.; Tsuge, M.; Oka, S. From Omics to Multi-Omics: A Review of Advantages and Tradeoffs. Genes 2024, 15, 1551. [Google Scholar] [CrossRef]
- Goecks, J.; Jalili, V.; Heiser, L.M.; Gray, J.W. How Machine Learning Will Transform Biomedicine. Cell 2020, 181, 92–101. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Ye, Q.; Xia, J. Unbox the black-box for the medical explainable AI via multi-modal and multi-centre data fusion: A mini-review, two showcases and beyond. Inf. Fusion 2022, 77, 29–52. [Google Scholar] [CrossRef] [PubMed]
- Norgeot, B.; Muenzen, K.; Peterson, T.A.; Fan, X.; Glicksberg, B.S.; Schenk, G.; Rutenberg, E.; Oskotsky, B.; Sirota, M.; Yazdany, J.; et al. Protected Health Information filter (Philter): Accurately and securely de-identifying free-text clinical notes. NPJ Digit. Med. 2020, 3, 57. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Galal, G.; Etemadi, M.; Vaidyanathan, M. Evaluation and Mitigation of Racial Bias in Clinical Machine Learning Models: Scoping Review. JMIR Med. Inf. 2022, 10, e36388. [Google Scholar] [CrossRef]
- Chen, R.J.; Wang, J.J.; Williamson, D.F.K.; Chen, T.Y.; Lipkova, J.; Lu, M.Y.; Sahai, S.; Mahmood, F. Algorithmic fairness in artificial intelligence for medicine and healthcare. Nat. Biomed. Eng. 2023, 7, 719–742. [Google Scholar] [CrossRef]
- Wei, L.; Niraula, D.; Gates, E.D.H.; Fu, J.; Luo, Y.; Nyflot, M.J.; Bowen, S.R.; El Naqa, I.M.; Cui, S. Artificial intelligence (AI) and machine learning (ML) in precision oncology: A review on enhancing discoverability through multiomics integration. Br. J. Radiol. 2023, 96, 20230211. [Google Scholar] [CrossRef]
| Omics | Author (Year) | Data Source | Sample Type and Size | Method | Task | Biomarkers | Performance Metrics |
|---|---|---|---|---|---|---|---|
| Radiomics | Jiang et al. (2023) [15] | Multicenter study (Southern Medical University, Stanford University, Sun Yat-sen University Cancer Center, Guangdong Provincial Hospital of Chinese Medicine) | CT imaging (GC patients, N = 2686) | Deep learning (CNN-based) | Treatment response prediction | Deep learning-based image features | AUC = 0.722 |
| Radiomics | Tao et al. (2024) [76] | West China Hospital, Sichuan University, The First Affiliated Hospital of Chengdu Medical College, People’s Hospital of Leshan | CT imaging (GC patients, N = 771) | Deep learning (vision transformer-based) | Diagnosis prediction (T stage: T1–T2 vs. T3–T4) | Deep learning features (1280 features) combined with radiomics features (512 features) | AUC = 0.972 |
| Radiomics (Endoscopic) | Zhu et al. (2019) [22] | Endoscopy Center, Zhongshan Hospital, Fudan University | Endoscopic images (N = 790) | Deep learning (CNN-based) | Diagnosis prediction (invasion depth) | Deep learning-based image features | AUC = 0.94 |
| Radiomics (Endoscopic) | Liu et al. (2022) [77] | Shanghai General Hospital-South and Shanghai Jiao Tong University Affiliated Sixth People Hospital | Endoscopic images (N = 6177) | Deep learning (CNN-based) | Diagnosis prediction (gastric neoplastic lesions) | Deep learning-based image features | AUC = 0.928 |
| Pathomics | Veldhuizen et al. (2023) [30] | TCGA | WSIs (N = 166) | Deep learning | Diagnosis prediction (diffuse vs. intestinal) | Deep learning-based histopathology features | AUROC: 0.93 |
| Pathomics | Saldanha et al. (2023) [78] | Four patient cohorts from Switzerland, Germany, the UK, and the USA | WSIs (N = 60,530) | swarm learning | Diagnosis prediction (MSI, EBV status) | Deep learning-based histopathology features | AUROC: 0.8092, 0.8372 (MSI, EBV prediction, respectively) |
| Genomics | Cheong et al. (2022) [79] | TCGA, GEO, ACRG, Yonsei cohort | Tissue (N = 567) | NTriPath, SVM | Prognosis prediction | 32-gene signature (including TP53, BRCA1, MSH6, PARP1, ACTA2) | AUC = 0.981 |
| Genomics | Wu et al. (2023) [47] | TCGA-STAD, GEO (GSE84437, GSE54129, GSE65801) | Tissue (N = 443) | NMF, SVM, neural networks, LASSO | Prognosis prediction (OS) | SRMS, MET, OLFML2B, KIF24, CLDN9, RNF43, NETO2, PRSS21 | AUC > 0.7 |
| Epigenomics | Kandimalla et al. (2021) [45] | TCGA, GSE72872 | Plasma (N = 300) | Random forest | Diagnosis prediction | 3 DMR panels | AUC = 0.90 |
| Epigenomics | Li et al. (2020) [80] | GEO, TCGA | Tissue (N = 368) | LASSO | Prognosis prediction (OS) | TREM2, RAI14, NRP1, YAP1, MATN3, PCSK5, INHBA, MICAL2 | AUC = 0.74 |
| Transcriptomics | Kong et al. (2022) [81] | TCGA, STRING database | Tissue (N ≥ 700) | Network-based machine learning | Treatment response prediction (ICI) | Network-derived transcriptomic biomarkers | AUC = 0.72 |
| Transcriptomics | Lee et al. (2022) [55] | TCGA-STAD, UCSC Xena | Tissue (N = 379) | Hierarchical clustering | Prognosis prediction | LOC441461 | other |
| Proteomics | Li et al. (2024) [60] | Multicenter study (China) | Serum (N = 60) | XGBoost | Diagnosis prediction (CGC vs. healthy control) | CDHR2, ICAM4, PTPRM, CDC27, FLT1 | AUC = 0.931 |
| Proteomics | Sun et al. (2024) [57] | First Affiliated Hospital of Zhengzhou University | Tissue (N = 28) | SVM, Boruta | Treatment response prediction (ICIs) | COL15A1, SAMHD1, DHX15, PTDSS1, CFI, ORM2, VWF, APOA1, EMC2, COL6A2 | AUC = 0.96 |
| Metabolomics | Liu et al. (2022) [68] | National Upper Gastrointestinal Cancer Early Detection Program (China) | Plasma (N = 200) | OPLS-DA | Diagnosis or prognosis prediction | PC38:6(20:4), PC38:5(20:4), PC34:3, LysoPC18:3, LysoPC20:4, LPI18:0, LPI20:4, FFA20:4 (arachidonic acid), FFA18:3 (α-linolenic acid), FFA18:0 (stearic acid), PA32:1 | AUC = 0.97(for diagnosis) 0.82(for prognosis) |
| Metabolomics | Chen et al. (2024) [70] | Multicenter plasma metabolomics dataset (China) | Plasma (N = 702) | LASSO, random forest, SVM | Diagnosis prediction (GC vs. NGC) | Succinate, Uridine, Lactate, SAM, Pyroglutamate, 2-Aminooctanoate, Neopterin, GlcNAc6p, Serotonin, NMN | AUC = 0.967 |
| Author (Year) | University, Country | Task | Dataset Size | Model | Patch Size | Batch Size | GPU Type (Memory) | Training Epochs | Performance Metrics |
|---|---|---|---|---|---|---|---|---|---|
| Lu et al. (2024) [92] | Harvard Medical School, USA | Zero-shot visual-language pathology AI | 21,442 WSIs | CONCH | 448 × 448 px | 1536 patches | 8 × NVIDIA A100 (80 GB each) | 40 epochs | Zero-shot accuracy: 91.3% |
| Wang et al. (2024) [34] | Harvard Medical School, USA | Cancer diagnosis and prognosis prediction | 60,530 WSIs | CHIEF | 256 × 256 px | 1 WSI | 8 × NVIDIA V100 (32 GB each) | 50 epochs | C-index: 0.74 |
| White et al. (2024) [93] | Mater Misericordiae University Hospital, Ireland | Biopsy prioritization | 24,983 WSIs | MIL | 512 × 512 px | Not specified | 8 × NVIDIA V100 (32 GB) | 200 epochs | F1 Score: 0.949 |
| Gustav et al. (2024) [94] | Technical University Dresden, Germany | Predicting MSI and POLE mutations in colorectal cancer | 2039 WSIs | Vision Transformer | Not specified | Not specified | NVIDIA RTX A6000 (48 GB) | Not specified | AUROC: 0.94 |
| Hilgers et al. (2024) [95] | Technical University Dresden, Germany | Automated curation of WSIs | 32,975 WSIs | ResNet18 | 224 × 224 px | 128 patches | NVIDIA RTX A6000 (48 GB) | 500 epochs | AUROC: 0.995 |
| Liu et al. (2024) [94] | Sun Yat-sen University, China | Predicting response to PD-1 blockade in advanced GC | 313 WSIs | DenseNet121, EfficientNet-B4, Swin V2 | 1024 × 1024 px | 32 patches | 2 × NVIDIA RTX 3090 (24 GB each) | 100 epochs | AUROC: 0.92–1.00 |
| Yang et al. (2024) [96] | Wenzhou Medical University, China | Prognosis and treatment response prediction | 1481 WSIs | OCDPI | 224 × 224 px | 8 patches | NVIDIA RTX 4090 (24 GB) | 40 epochs | Not specified |
| Huang et al. (2024) [97] | Southeast University, China | Morphological profiling of CRC organoids | 31,360 bright field images + 17,000 fluorescent images | Generative | 1360 × 1024 px | Not specified | NVIDIA RTX 3090 (24 GB) | 200 epochs | Not specified |
| Choudhury et al. (2024) [98] | University of Chicago, USA | HPV status prediction | 941 WSIs | Xception-based CNN | 299 × 299 px | Not specified | NVIDIA Titan RTX (24 GB) | 1 epoch | AUROC: 0.84 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, D.; Fan, C.; Sano, T.; Kawabata, K.; Nishikubo, H.; Imanishi, D.; Sakuma, T.; Maruo, K.; Yamamoto, Y.; Matsuoka, T.; et al. Beyond Biomarkers: Machine Learning-Driven Multiomics for Personalized Medicine in Gastric Cancer. J. Pers. Med. 2025, 15, 166. https://doi.org/10.3390/jpm15050166
Ma D, Fan C, Sano T, Kawabata K, Nishikubo H, Imanishi D, Sakuma T, Maruo K, Yamamoto Y, Matsuoka T, et al. Beyond Biomarkers: Machine Learning-Driven Multiomics for Personalized Medicine in Gastric Cancer. Journal of Personalized Medicine. 2025; 15(5):166. https://doi.org/10.3390/jpm15050166
Chicago/Turabian StyleMa, Dongheng, Canfeng Fan, Tomoya Sano, Kyoka Kawabata, Hinano Nishikubo, Daiki Imanishi, Takashi Sakuma, Koji Maruo, Yurie Yamamoto, Tasuku Matsuoka, and et al. 2025. "Beyond Biomarkers: Machine Learning-Driven Multiomics for Personalized Medicine in Gastric Cancer" Journal of Personalized Medicine 15, no. 5: 166. https://doi.org/10.3390/jpm15050166
APA StyleMa, D., Fan, C., Sano, T., Kawabata, K., Nishikubo, H., Imanishi, D., Sakuma, T., Maruo, K., Yamamoto, Y., Matsuoka, T., & Yashiro, M. (2025). Beyond Biomarkers: Machine Learning-Driven Multiomics for Personalized Medicine in Gastric Cancer. Journal of Personalized Medicine, 15(5), 166. https://doi.org/10.3390/jpm15050166






