New Species-Specific Primers for Molecular Diagnosis of Bactrocera minax and Bactrocera tsuneonis (Diptera: Tephritidae) in China Based on DNA Barcodes
Abstract
:1. Introduction
2. Materials and Methods
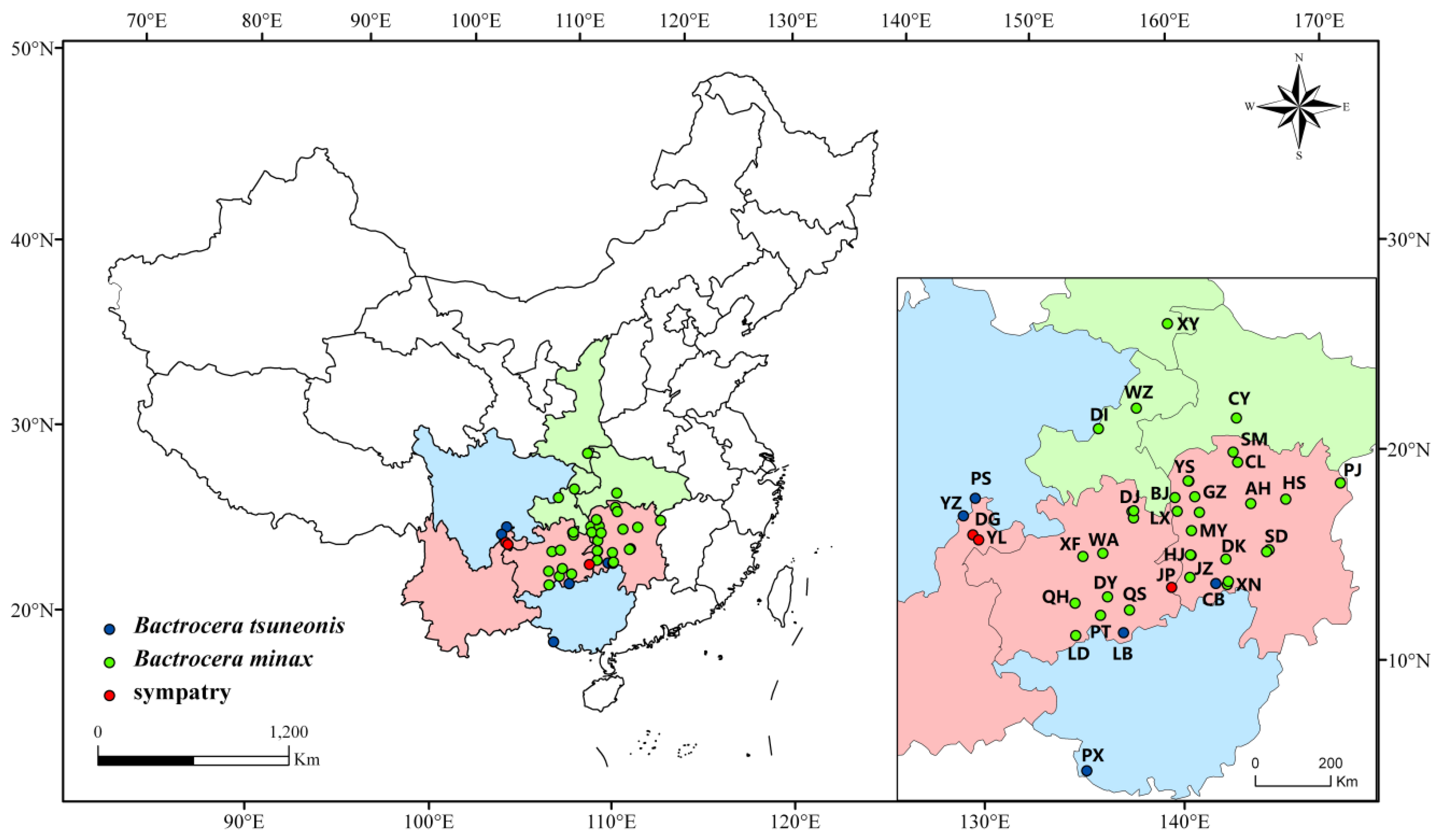
2.1. Sample Collection
2.2. Morphological Diagnostic Method
2.3. DNA Extraction, PCR Amplification and Sequencing
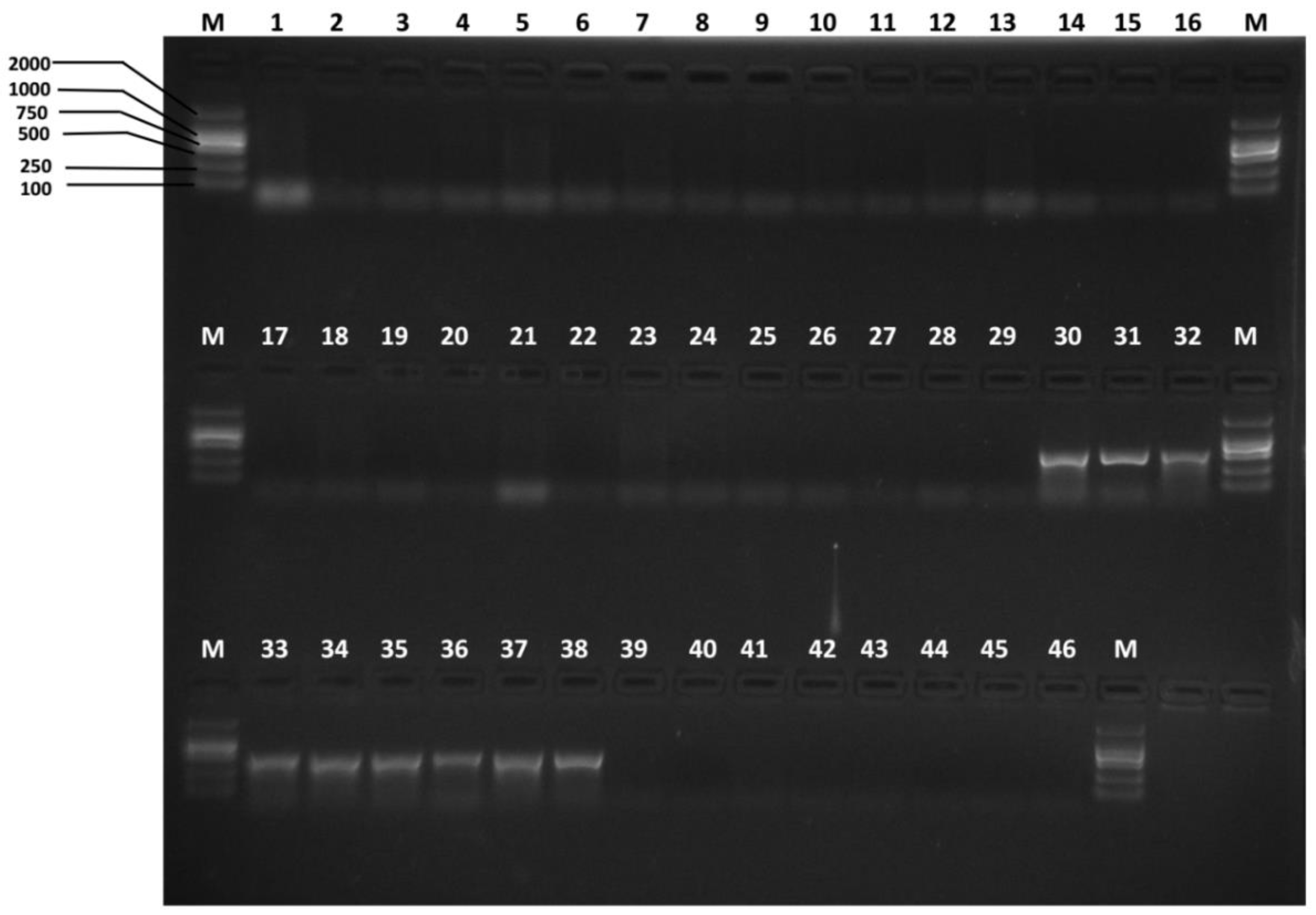
2.4. Specific Primer Design and Specificity Test
2.5. Sensitivity Test
3. Results and Discussion
3.1. DNA Barcode Sequence Analyses
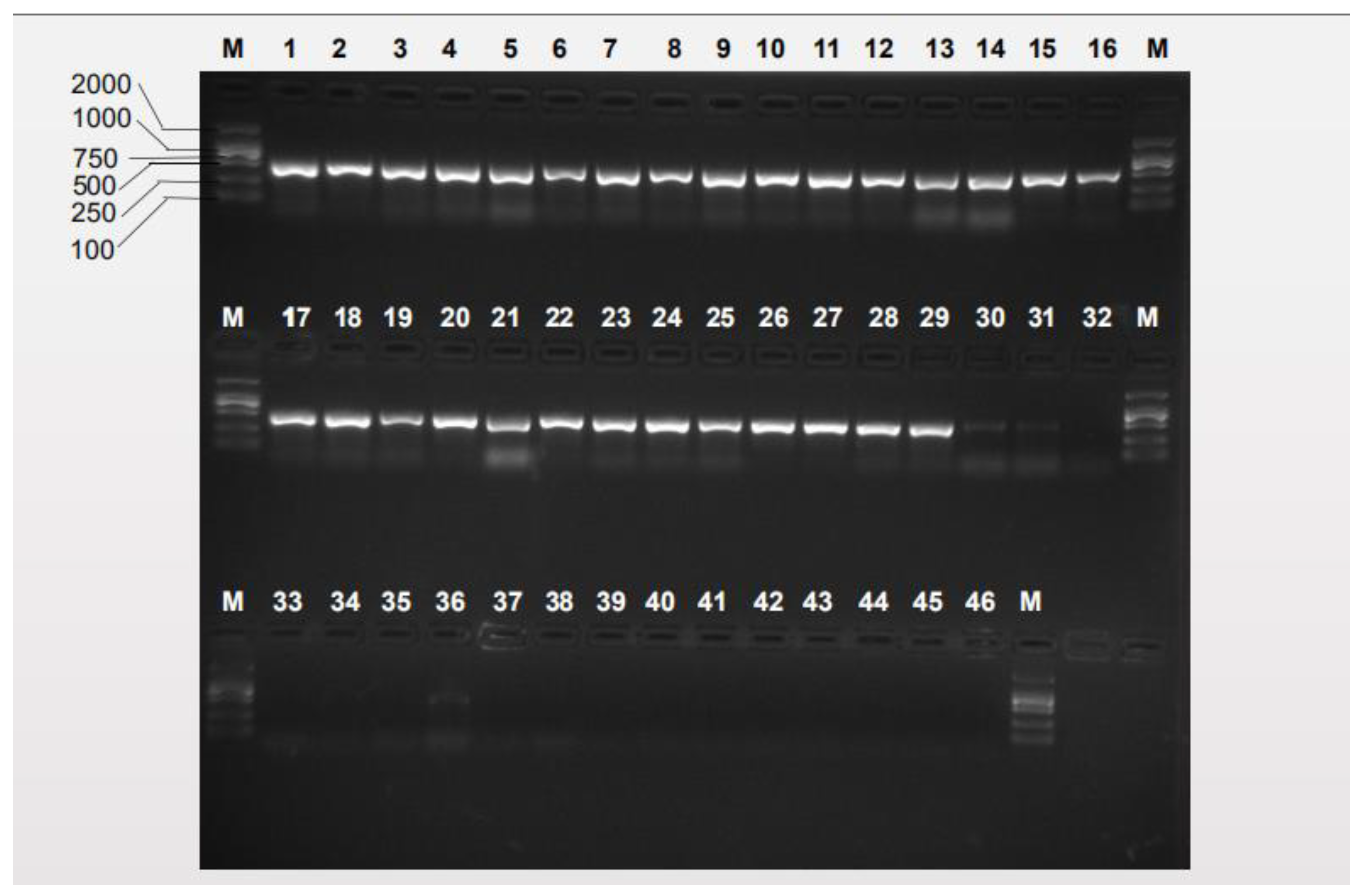
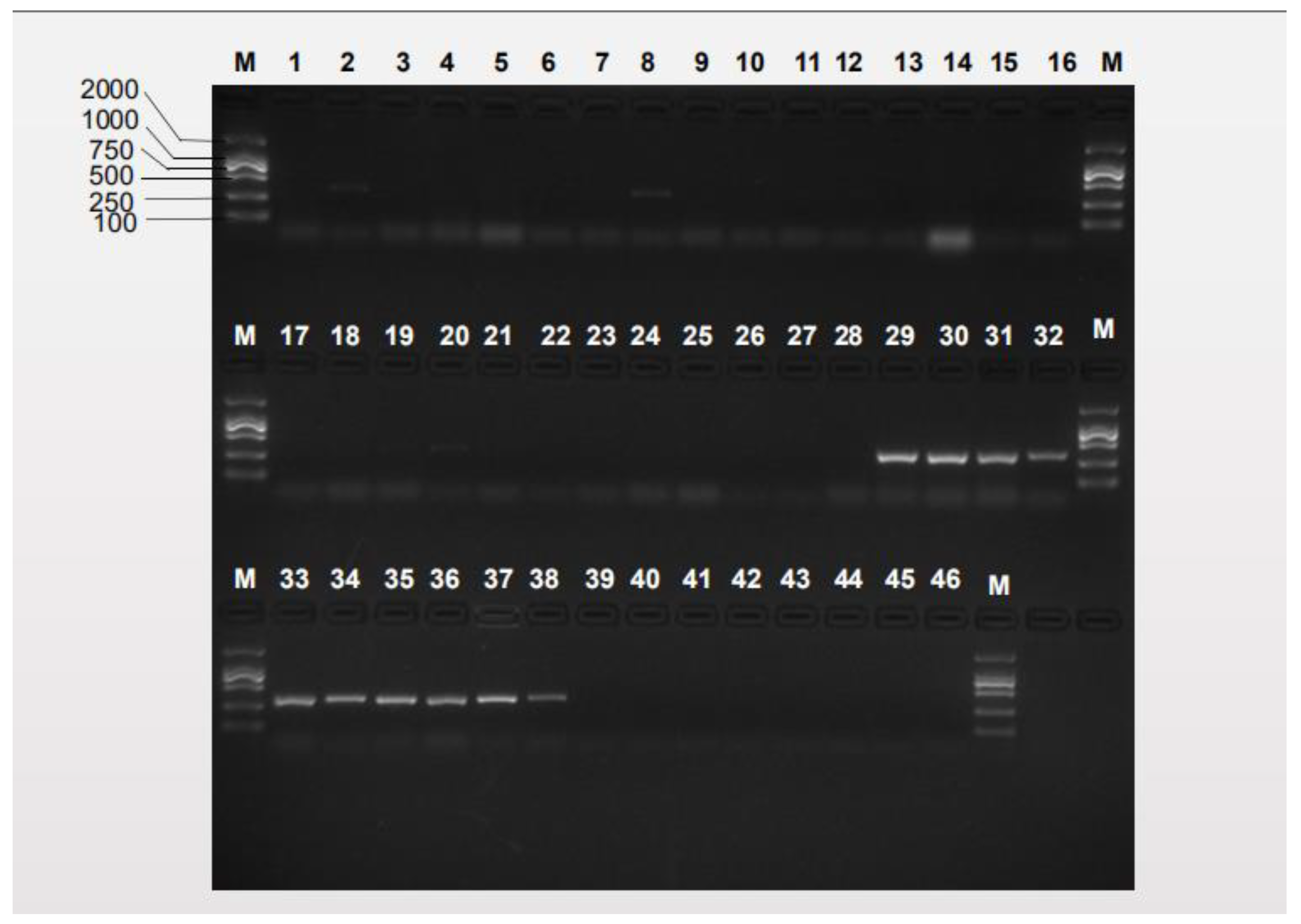
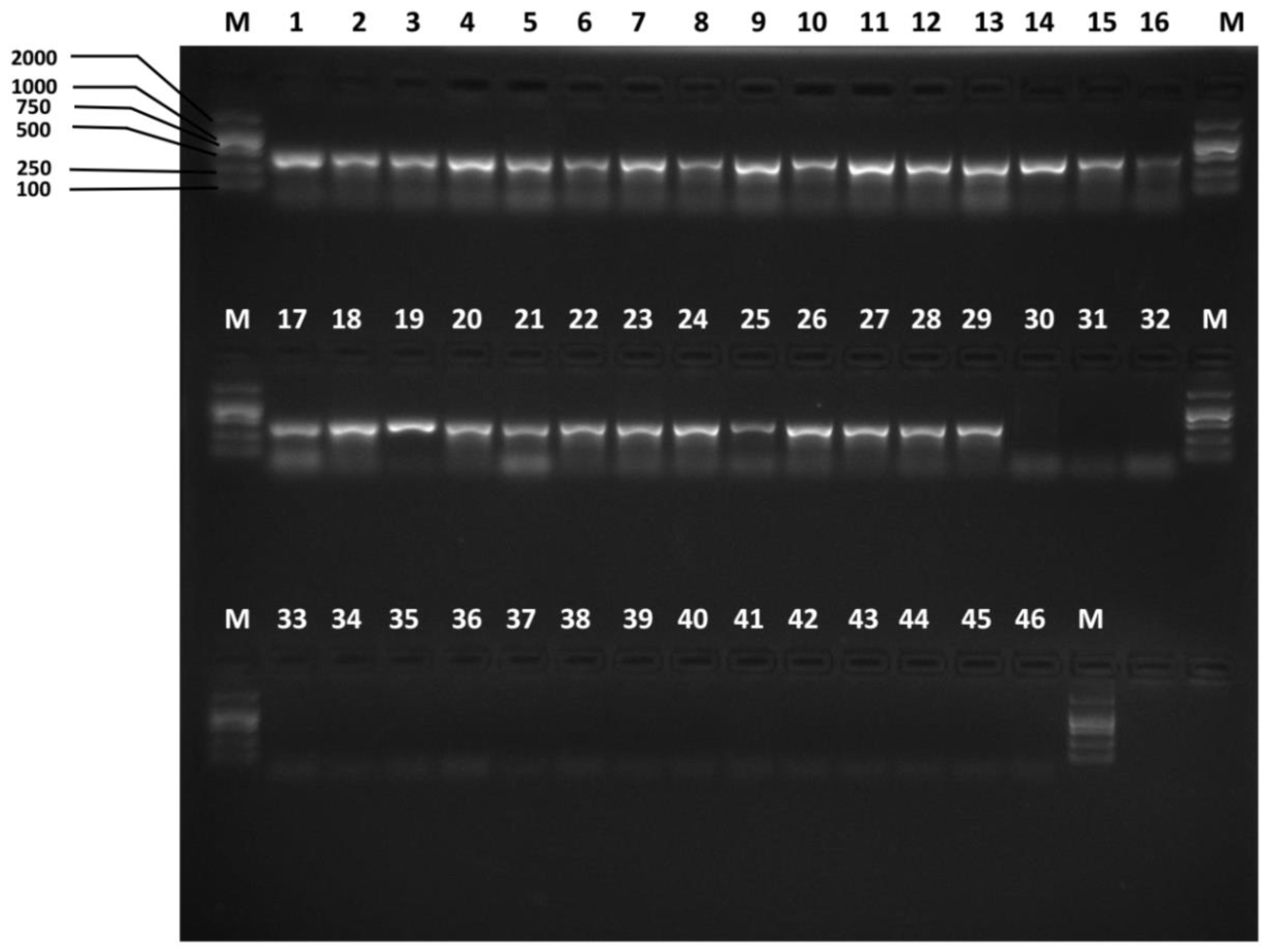
3.2. Species-Specific Primer Selection, Specificity Test and Sensitivity Test
3.3. Advantages of This Diagnosis Method
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- White, I.M.; Elson-Harris, M.M. Fruit Flies of Economic Significance: Their Identification and Bionomics. Entomol. Res. 1992, 82, 433. [Google Scholar]
- Van Houdt, J.K.J.; Breman, F.C.; Virgilio, M.; De Meyer, M. Recovering full DNA barcodes from natural history collections of Tephritid fruit flies (Tephritidae, Diptera) using mini-barcodes. Mol. Ecol. Resour. 2010, 10, 459–465. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.Y.; Xiao, T.G. Studies on the occurrence, biological characteristic and control of Bactrocera (Tetradacus) Minax. Crop Res. 2014, 28, 299–301. [Google Scholar]
- Yasunaga, K.; Nagatomi, A. Studies on the Dacus (Tetradacus) tsuneonis Miyake (Diptera: Trypetidae). 1. Some fundamental and biological investigations essential for its control. Sci. Bull. Fac. Agric. 1959, 17, 129–146. [Google Scholar]
- Wang, X.J. The fruit flies (Diptera: Tephritidae) of the East Asian region. Acta Zootaxon. Sin. 1996, 21, 1–338. [Google Scholar]
- EPPO, PQR Database. Paris, France: European and Mediterranean Plant Protection Organization. Available online: http://www.eppo.int/DATABASES/pqr/pqr.htm/ (accessed on 14 March 2019).
- Zhuo, S.F.; Zhuo, N.J. From “citrus fruit fly incident” to discuss the marketing crisis management caused by the imbalance of consumer psychology. Mark. Mod. 2009, 6, 105–106. [Google Scholar]
- Chen, S.H. Two new Dacinae from Szechwan. Sinensia 1940, 11, 131–135. [Google Scholar]
- Zia, Y. On three new species of tephritid flies from South China and Viet-Nam. Acta Zool. Sin. 1955, 7, 63–68. [Google Scholar]
- White, I.M.; Wang, X.J. Taxonomic notes on some dacine (Diptera: Tephritidae) fruit flies associated with citrus, olives, and cucurbits. Bull. Entomol. Res. 1992, 82, 275–279. [Google Scholar] [CrossRef]
- Shiraki, T. A systematic study of Trypetidae in the Japanese Empire. Mem. Fac. Sci. Agric. Taihoku Imp. Univ. 1933, 8, 1–509. [Google Scholar]
- Ito, S. Die Japanese Borhrfliegen; Maruzen Co., Ltd.: Osaka, Japan, 1983. [Google Scholar]
- Sun, Z.Y.; Du, Y.L. Preliminary observation on the damage symptoms of Bactrocera minax. Entomol. Knowl. 1957, 5, 207–210. [Google Scholar]
- Chen, Y.E.; Shi, Y.L.; Wang, Q.; Xie, L. Occurrence regularity and integrated control technology of Bactrocera minax. Hubei Plant Prot. 2017, 9, 30. [Google Scholar]
- Ma, F.M.; Li, D.S.; Zhang, B.X. Major citrus diseases and insect pests of citrus in China. Chin. J. Biol. Control. 2007, S1, 87–92. [Google Scholar]
- EPPO, The Distribution of Bactrocera minax 2014. Available online: https://www.cabi.org/ (accessed on 14 March 2019).
- EPPO, The Distribution of Bactrocera tsuneonis 2014. Available online: https://www.cabi.org/ (accessed on 14 March 2019).
- Hou, B.H.; Ouyang, G.C.; Lu, H.L.; Ma, J.; Lu, Y.Y.; Xia, Y. First detection of Bactrocera tsuneonis (Diptera: Tephritidae) in Guangdong Province of China. Fla. Entomol. 2018, 101, 533–536. [Google Scholar] [CrossRef]
- Fan, J.A.; Zhao, X.Q.; Zhu, J. Study on cold tolerance and diapause of Tetradacus citri Chen. J. Southwest Agric. Univ. 1994, 16, 530–534. [Google Scholar]
- Blaser, S.; Diem, H.; von Felten, A.; Gueuning, M.; Andreou, M.; Boonham, N.; Tomlinson, J.; Müller, P.; Utzinger, J.; Bühlmann, A. From laboratory to point of entry: Development and implementation of a loop-mediated isothermal amplification (LAMP)-based genetic identification system to prevent introduction of quarantine insect species. Pest Manag. Sci. 2018, 74, 1504–1512. [Google Scholar] [CrossRef] [Green Version]
- Ji, X.Z.; Guo, X.H. Occurrence and damage of Bactrocera minax and comprehensive control measures. Plant Quar. 1992, 6, 289–290. [Google Scholar]
- Mezghani Khemakhem, M.; Ben Lazahr, W.; Bouktila, D.; Ben Slimen, H.; Makni, H.; Makni, M. A rapid diagnostic technique of Bactrocera cucurbitae and Bactrocera zonata (Diptera: Tephritidae) for quarantine application. Pest. Manag. Sci. 2013, 69, 744–746. [Google Scholar] [CrossRef]
- Hebert, P.D.; Cywinska, A.; Ball, S.L.; Dewaard, J.R. Biological identifications through DNA barcodes. Proc. R. Soc. Lond. Ser. B Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef] [Green Version]
- McCulloch, G.A.; Purcell, M.F.; Harms, N.E.; Grodowitz, M.J.; Zhang, J.; Sun-Hee, H.; Walter, G.H. Molecular screening of herbivorous flies collected from Hydrilla verticillata across China and Korea—Setting up hypotheses for further exploratory surveys and tests. Biol. Control 2019, 138, 104051. [Google Scholar] [CrossRef]
- Blattner, L.; Gerecke, R.; Von Fumetti, S. Hidden biodiversity revealed by integrated morphology and genetic species delimitation of spring dwelling water mite species (Acari, Parasitengona: Hydrachnidia). Parasites Vectors 2019, 12, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Renaud, A.K.; Savage, J.; Adamowicz, S.J. DNA barcoding of Northern Nearctic Muscidae (Diptera) reveals high correspondence between morphological and molecular species limits. BMC Ecol. 2012, 12, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Armstrong, K.F.; Ball, S.L. DNA barcodes for biosecurity: Invasive species identification. Philos. Trans. R. Soc. B 2005, 360, 1813–1823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buahom, N.; Li, Z.H.; Wu, J.J.; Liu, J.Q. Molecular identification of fruit fly larvae from Thailand based on DNA barcoding. Plant Quar. 2011, 25, 49–52. [Google Scholar]
- Barr, N.B.; Ruiz-Arce, R.; Farris, R.E.; Silva, J.G.; Lima, K.M.; Dutra, V.S.; Thomas, D.B. Identifying Anastrepha (Diptera; Tephritidae) Species Using DNA Barcodes. J. Econ. Entomol. 2017, 111, 405–421. [Google Scholar] [CrossRef]
- Manger, A.; Behere, G.T.; Firake, D.M.; Sharma, B.; Deshmukh, N.A.; Firake, P.D.; Thakur, N.S.A.; Ngachan, S.V. Genetic characterization of Bactrocera fruit flies (Diptera: Tephritidae) from Northeastern India based on DNA barcodes. Mitochondrial DNA A 2018, 29, 792–799. [Google Scholar] [CrossRef]
- Onah, I.E.; Eyo, J.E.; Taylor, D. Application of PCR-RFLP of COI gene for identification of life stages of Bactrocera dorsalis and Ceratitis anonae infesting citrus in southeastern Nigeria. Afr. Entomol. 2017, 25, 485–493. [Google Scholar] [CrossRef]
- Raquin, V.; Henri, H.; Vallat, M.; Leulier, F.; Gibert, P.; Kremer, N. Development of a PCR-RFLP assay to identify Drosophila melanogaster among field-collected larvae. Ecol. Evol. 2018, 8, 10067–10074. [Google Scholar] [CrossRef] [Green Version]
- Li, D.M.; Nair, S.; Anderson, D.; Doddala, P.; Gunawardana, D.N.; George, S. Real-time PCR assays for rapid detection of Zeugodacus cucumis and Bactrocera jarvisi (Diptera: Tephritidae) for quarantine application. J. Appl. Entomol. 2019, 143, 155–163. [Google Scholar] [CrossRef] [Green Version]
- Jiang, F.; Fu, W.; Clarke, A.R.; Schutze, M.K.; Susanto, A.; Zhu, S.; Li, Z. A high-throughput detection method for invasion fruit fly (Diptera: Tephritidae) species based on microfluidic dynamic array. Mol. Ecol. Resour. 2016, 16, 1378–1388. [Google Scholar] [CrossRef]
- Chua, T.H.; Song, B.K.; Chong, Y.V. Development of allele-specific single-nucleotide polymorphism-based PCR markers in COI for the differentiation of B. papayae and B. carambolae (Diptera: Tephritidae). J. Econ. Entomol. 2010, 103, 1994–1999. [Google Scholar] [CrossRef] [PubMed]
- Asokan, R.; Rebijith, K.B.; Singh, S.K.; Sidhu, A.S.; Siddharthan, S.; Karanth, P.K.; Ellango, R.; Ramamurthy, V.V. Molecular identification and phylogeny of Bactrocera species (Diptera: Tephritidae). Fla. Entomol. 2011, 94, 1026–1035. [Google Scholar] [CrossRef]
- Jiang, F.; Li, Z.H.; Wu, J.J.; Wang, F.X.; Xiong, H.L. A rapid diagnostic tool for two species of Tetradacus (Diptera: Tephritidae: Bactrocera) based on species-specific PCR. J. Appl. Entomol. 2014, 138, 418–422. [Google Scholar] [CrossRef]
- Chua, T.H.; Chong, Y.V.; Lim, S.H. Species determination of Malaysian Bactrocera pests using PCR-RFLP analyses (Diptera: Tephritidae). Pest Manag. Sci. 2010, 66, 379–384. [Google Scholar]
- White, I.M.; Marlene, M.E. Fruit Flies of Economic Significance: Their Identification and Bionomics; International Institute of Entomology: London, UK, 1994; p. 118. [Google Scholar]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. 1994, 3, 294–299. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP v6: DNA Sequence Polymorphism Analysis of Large Datasets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Hall, T.; Biosciences, I.; Carlsbad, C. BioEdit: An important software for molecular biology. GERF Bull. Biosci. 2011, 2, 60–61. [Google Scholar]
- Rychlik, W. OLIGO 7 primer analysis software. In PCR Primer Design; Humana Press: New York, NY, USA, 2007; pp. 35–59. [Google Scholar]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids. Res. 2000, 28, e63. [Google Scholar] [CrossRef] [Green Version]

| Species | Primer | Primers Sequence (5’-3’) | Size (bp) |
|---|---|---|---|
| B. minax | BTmina-F | CTTGTTCGAGCAGAACTAGGC | 499 |
| BTmina-R | GGACTGGGAGGGATAGTAAGAGG | ||
| B. tsuneonis | BTtsun-F | CCATCCCTTACCCTATTGTTACTC | 337 |
| BTtsun-R | AGGATGTATTTAGGTTTCGGTCC |
| Species | Primer | Primers Sequence (5’-3’) | Size (bp) | Tm (°C) |
|---|---|---|---|---|
| B. minax | Bm-F | AATTTATAACGTAATCGTTACAGCC | 422 | 53.9 |
| Bm-R | AAGTATTGTGATAGCTCCGGCTAGG | 60.2 | ||
| B. tsuneonis | Bt-F | TAATGTAATCGTTACTGCTCACGCC | 456 | 59.9 |
| Bt-R | CTGGGTCAAAGAAGGATGTATTTAG | 56.1 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, L.; Zhang, Y.; Yang, W.; Zeng, Y.; Jiang, F.; Qin, Y.; Zhang, J.; Jiang, Z.; Hu, W.; Guo, D.; et al. New Species-Specific Primers for Molecular Diagnosis of Bactrocera minax and Bactrocera tsuneonis (Diptera: Tephritidae) in China Based on DNA Barcodes. Insects 2019, 10, 447. https://doi.org/10.3390/insects10120447
Zheng L, Zhang Y, Yang W, Zeng Y, Jiang F, Qin Y, Zhang J, Jiang Z, Hu W, Guo D, et al. New Species-Specific Primers for Molecular Diagnosis of Bactrocera minax and Bactrocera tsuneonis (Diptera: Tephritidae) in China Based on DNA Barcodes. Insects. 2019; 10(12):447. https://doi.org/10.3390/insects10120447
Chicago/Turabian StyleZheng, Linyu, Yue Zhang, Wenzhao Yang, Yiying Zeng, Fan Jiang, Yujia Qin, Jiafeng Zhang, Zhaochun Jiang, Wenzhao Hu, Dijin Guo, and et al. 2019. "New Species-Specific Primers for Molecular Diagnosis of Bactrocera minax and Bactrocera tsuneonis (Diptera: Tephritidae) in China Based on DNA Barcodes" Insects 10, no. 12: 447. https://doi.org/10.3390/insects10120447
APA StyleZheng, L., Zhang, Y., Yang, W., Zeng, Y., Jiang, F., Qin, Y., Zhang, J., Jiang, Z., Hu, W., Guo, D., Wan, J., Zhao, Z., Liu, L., & Li, Z. (2019). New Species-Specific Primers for Molecular Diagnosis of Bactrocera minax and Bactrocera tsuneonis (Diptera: Tephritidae) in China Based on DNA Barcodes. Insects, 10(12), 447. https://doi.org/10.3390/insects10120447




