Simple Summary
The Neotropical Region harbors a highly diverse and poorly known fauna of leaf miners of the micromoth family Gracillariidae (Lepidoptera). Angelabella is a genus of Gracillariidae whose geographic range is restricted to a few valleys of the arid environments of the Peru-Chile desert. Only one species is currently included in this genus. The aims of this study were to explore the geographic range, determine the spatial distribution of mitochondrial lineages, and test lineage conspecificity hypotheses in Angelabella. The spatial distribution of genetic diversity indicated four spatial clusters, three of which are north of the previously known geographic range. These groups were defined as different species by four species delimitation methods. These results suggest that Angelabella harbors at least four morphologically cryptic species with restricted, not overlapping geographic ranges. This study shows that adequate single locus sequence analysis can be useful to discover surprising biodiversity patterns in underexplored environments, providing the base to plan further studies involving little-known organisms.
Abstract
Angelabella (Lepidoptera: Gracillariidae: Oecophyllembiinae) is considered a monotypic Neotropical genus of leaf miner micromoths known only from a few valleys of the arid environments of the Peru-Chile desert, particularly the southernmost part of Peru and northernmost part of Chile (type locality), where natural populations of its primary host plant occur. The geographic distribution of potential host plants provides a scenario for a wider range for this micromoth genus. The aims of this study were to explore the geographic range of Angelabella, determine the spatial distribution of mitochondrial lineages, and test lineage conspecificity hypotheses. The spatial distribution of genetic diversity indicated the presence of four spatial clusters, three of which are north of the previously known geographic range. Genetic distances were 0.2–0.8% and 3.6–8.3% (K2P) between haplotypes of the same and different spatial clusters, respectively. Phylogenetic relationships indicated reciprocal monophyly among the four spatial clusters, suggesting that allopatric differentiation processes have governed the recent history of Angelabella in these arid environments. These groups were defined as different species by four species delimitation methods, suggesting that Angelabella is not a monotypic genus, but harbors at least four morphologically cryptic allopatric species with restricted geographic ranges, including the type species and three candidate species.
1. Introduction
The Neotropical micromoth genus Angelabella Vargas & Parra (Lepidoptera: Gracillariidae: Oecophyllembiinae) includes only the type species, A. tecomae Vargas & Parra, a leaf miner described from the Azapa Valley, Atacama Desert of northern Chile, whose primary host plant is the shrub Tecoma fulva (Cav.) D. Don (Bignoniaceae) [1] (Figure 1). The small females of A. tecomae, forewing length about 3 mm, select actively growing leaflets for egg laying on this shrub [2]. This behavior ensures food availability to complete larval and pupal development inside a single leaflet, generating an aggregated spatial pattern of the immature stages along the shoot with eggs at the apex, larvae at intermediate positions and pupae at the base [2]. Despite the highly specialized egg laying site selection by the females, the host range of A. tecomae is not restricted to T. f. fulva, as this micromoth is also able to use the exotic ornamental tree Tecoma stans (L.) Juss. Ex Kunth as a host plant [3]. T. stans is a widespread Neotropical tree mainly associated with the Andes in South America [4]. It is restricted to the urban area in the range of A. tecomae [3]. The geographic range of A. tecomae appears to be narrow based on the available records, as besides the type locality, this micromoth has been collected only in a second valley in northern Chile and one valley in southern Peru, with about 50 km between the most distant records [3,5].
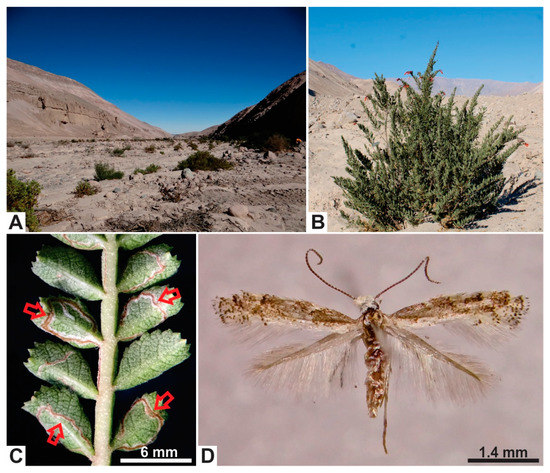
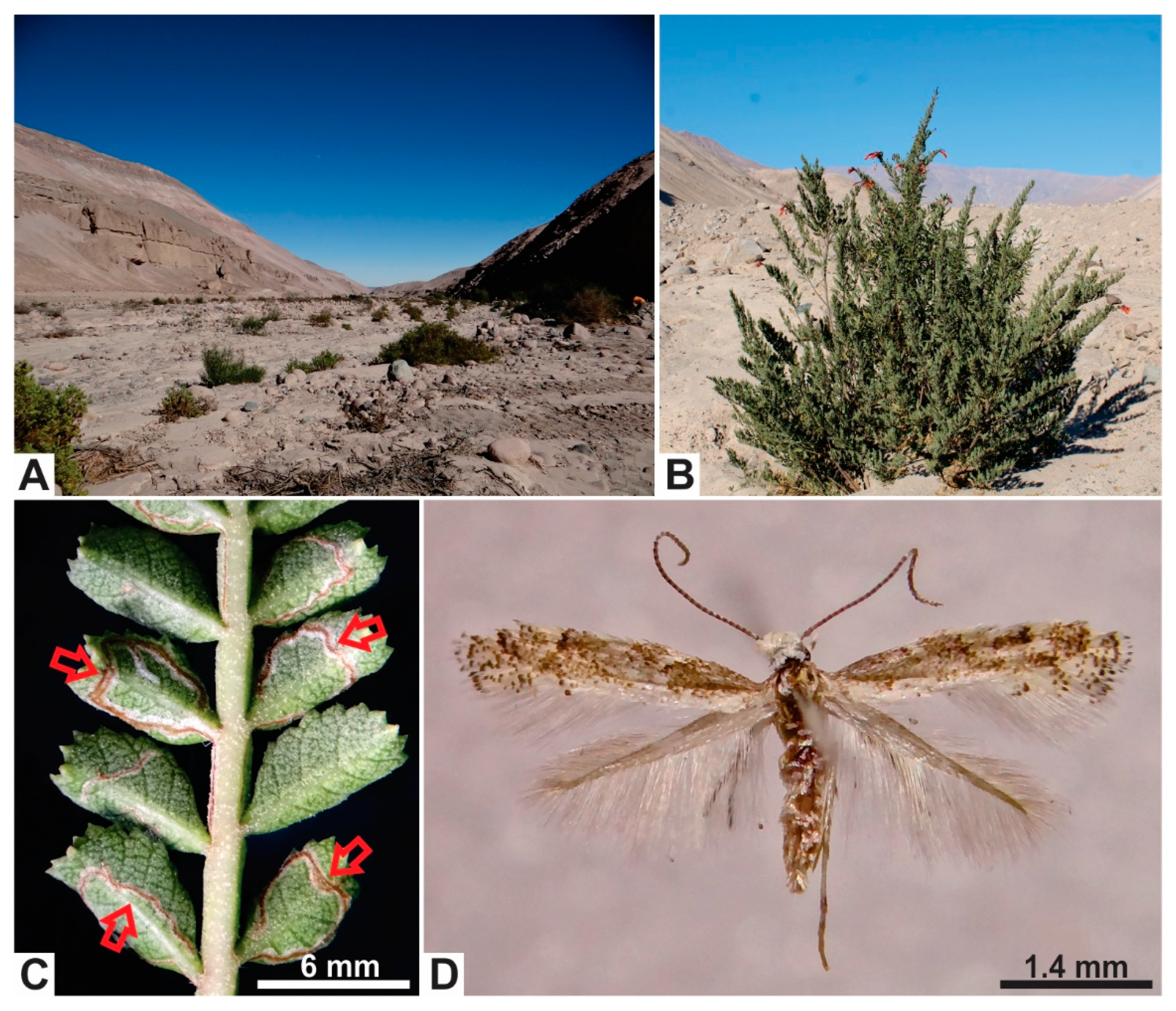
Figure 1.
Habitat and host plant of Angelabella tecomae (Gracillariidae). (A) Habitat of A. tecomae in the type locality, the Azapa Valley, northern Chile. (B) The main host plant in the type locality, Tecoma fulva fulva (Bignoniaceae). (C) Detail of a leaf of T. f. fulva showing leaflets and winged rachis mined by larvae of A. tecomae. (D) Female adult of A. tecomae in dorsal view.
Taxonomic identification of Lepidoptera is mainly based on adult morphology, with special reference to genitalia structures. However, morphological differentiation can be extremely subtle between adults of different species in some genera of Gracillariidae, especially in those of small size such as A. tecomae, hindering identification based exclusively on the morphology of this life stage [6,7]. DNA barcodes [8] are widely recognized as helpful complementary tools in taxonomic identification of Lepidoptera [9,10,11], including Gracillariidae [12,13,14,15]. Their use is especially adequate to identify immature stages [16,17] and to detect putative new species [6,18].
The geographic range of the host plant Tecoma fulva extends from the lowlands of the coastal valleys of south-central Peru and northern Chile to the Andes highlands of northern Argentina, Bolivia, and south-central Peru [4]. Six subspecies are currently recognized in this extensive range, four of which are found between near sea level and at about 2800 m elevation on the western slopes of the Andes of south-central Peru [4]: the nominotypical T. f. fulva, shared with the coastal valleys of northern Chile, T. f. arequipensis (Sprague) J. R. I. Wood., T. f. tanaeciiflora (Kränzlin) J. R. I. Wood and T. f. guarume (A. DC.) J. R. I. Wood.
As the host range of A. tecomae is not restricted to T. f. fulva, the presence of additional subspecies of T. fulva between the Pacific coast and the Andes of south-central Peru provides potential for a wider geographic range for this micromoth. Accordingly, to verify if A. tecomae is more widespread than currently known, it should be surveyed for in habitats where other subspecies of T. fulva occur. Considering the patchy distribution of potential host plants between extensive arid areas in the desert landscape of south-central Peru, together with the expected low vagility of this micromoth suggested by its small size, highly specialized egg lying site selection of the females and the endophytic habit of the larvae, the differentiation of mitochondrial lineages is expected to be associated with localities (patches), allowing us to hypothesize that such lineages are reciprocally monophyletic as reported in another micromoth, Bucculatrix mirnae Vargas & Moreira (Bucculatricidae), in similar environments [19]. If this hypothesis is validated, we can determine if such levels of differentiation have led to the formation of independent lineages, that is, if they can be considered as species.
Molecular approaches to species delimitation allow testing conspecificity hypotheses through a variety of different methods [20], some based on intraspecific genetic distance thresholds (ABGD [21]), a mixture of speciation models and coalescent theory (GMYC [22,23]) or speciation modeling in terms of number of nucleotide substitutions (PTP, [24]), all allowing single locus delimitation [25,26]. Since a given method makes a number of simplifying assumptions that could be violated in a particular system, it is important to test hypotheses by applying a wide range of species delimitation analyses and to rely on delimitations that are congruent across methods [27].
The aims of this study were to explore the geographic range of Angelabella, determine the spatial distribution of mitochondrial lineages and test lineage conspecificity hypotheses using DNA barcodes of individuals collected on potential host plants from the Peru-Chile desert.
2. Materials and Methods
2.1. Sample Collection
Mined leaflets were collected from individuals of three subspecies of T. fulva in four localities of Peru. Identification of the subspecies of T. fulva was based on the study of Wood [4]. From north to south, the sampling sites and their respective plants were as follows: (1) Lima Department (n = 4 pupae), Zuñiga, Lunahuana-Yauyos highway (12°50′ S, 75°57′ W), 950 m, June, 2019, T. f. guarume; (2) Arequipa Department (n = 5 pupae; n = 2 male adults), Pocsi (16°30′ S, 71°25′ W), 2900 m, August, 2018, T. f. arequipensis; (3) Moquegua Department (n = 10 pupae), Yacango (17°05′ S, 70°52′ W), 1900 m, January, 2020, T. f. fulva; and (4) Tacna Department (n = 10 pupae), Tacna (18°03′ S, 70°17′ W), 460 m, January, 2020, T. f. fulva. The maximum distance between localities was nearly 1000 km. The southernmost locality (Tacna Department) corresponds to the only previous Peruvian record of A. tecomae based on morphology [3]. The collected leaflets were dissected under a stereomicroscope to extract pupae, which were placed in ethanol 95% and kept at −20 °C until DNA extraction. Differences in number of individuals collected in each site reflect the availability of samples at the time of field work. The identification of the pupae was undertaken by comparison with pupae from the type locality of A. tecomae. In addition, two male adults were obtained from pupae from Arequipa; their genitalia were dissected and compared with males of A. tecomae from the type locality.
2.2. DNA Extraction and Sequencing
Genomic DNA was extracted from 29 pupae following the procedures described in [28]. A fragment of the COI gene was amplified by polymerase chain reaction (PCR) with the primers LEP-F1 and LEP R1 [29]. PCR reactions were performed in a final volume of 20 μL. Each reaction contained 1 μL of DNA extract, 10 ρmoles of each primer, 2.5 mM of each dNTP, 2 mM MgCl2, 1X PCR buffer (KCl), 1 unit of Taq DNA polymerase (Thermo Scientific) and sterile distilled water. The amplification program was 5 min at 94 °C, 35 cycles of 30 s at 94 °C, 30 s at 47 °C, 1 min at 72 °C, and a final elongation step of 10 min at 72 °C. Three μL of each PCR product was visualized on 1.5% agarose gels stained with gel-red (Biotium). Reactions containing fragments of the expected size were purified and sequenced directly by a commercial facility (Macrogen, South Korea).
2.3. Sequence Analysis
Fifteen sequences of A. tecomae from northern Chile (Arica) [4], including five from the type locality (Azapa Valley, red triangle in Figure 2B), were used in the analyses to evaluate the conspecificity of the 29 new Peruvian samples (Table 1). The software MEGA X [30] was used to perform the 657 bp sequence alignment with ClustalW to determine the nucleotide substitution model (under BIC criteria) and to assess the genetic distance between haplotypes with the Kimura 2-parameter (K2P) model. Genetic diversity indices were calculated in DnaSP 5.10 [31]. To assess whether the dataset represents neutral evolutionary processes, deviations from neutrality were assessed using Tajima’s D test [32] for population genetic data (Angelabella data only), and the Mcdonald-Kreitman (MK) test [33] for comparative data (Angelabella and outgroups), both analyses performed in DnaSP 5.10. The Xia test [34] was used to assess the presence of phylogenetic signal with a substitution saturation analysis in the software Dambe 7.2.1 [35].
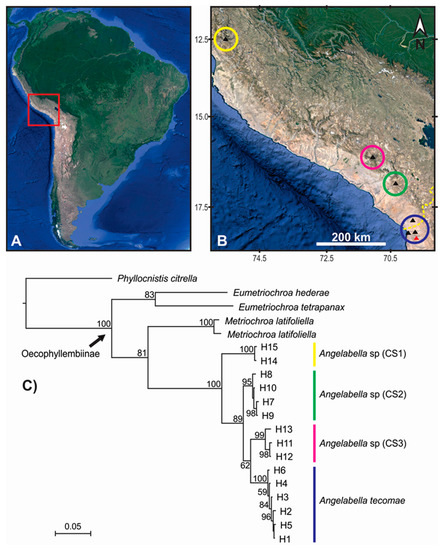
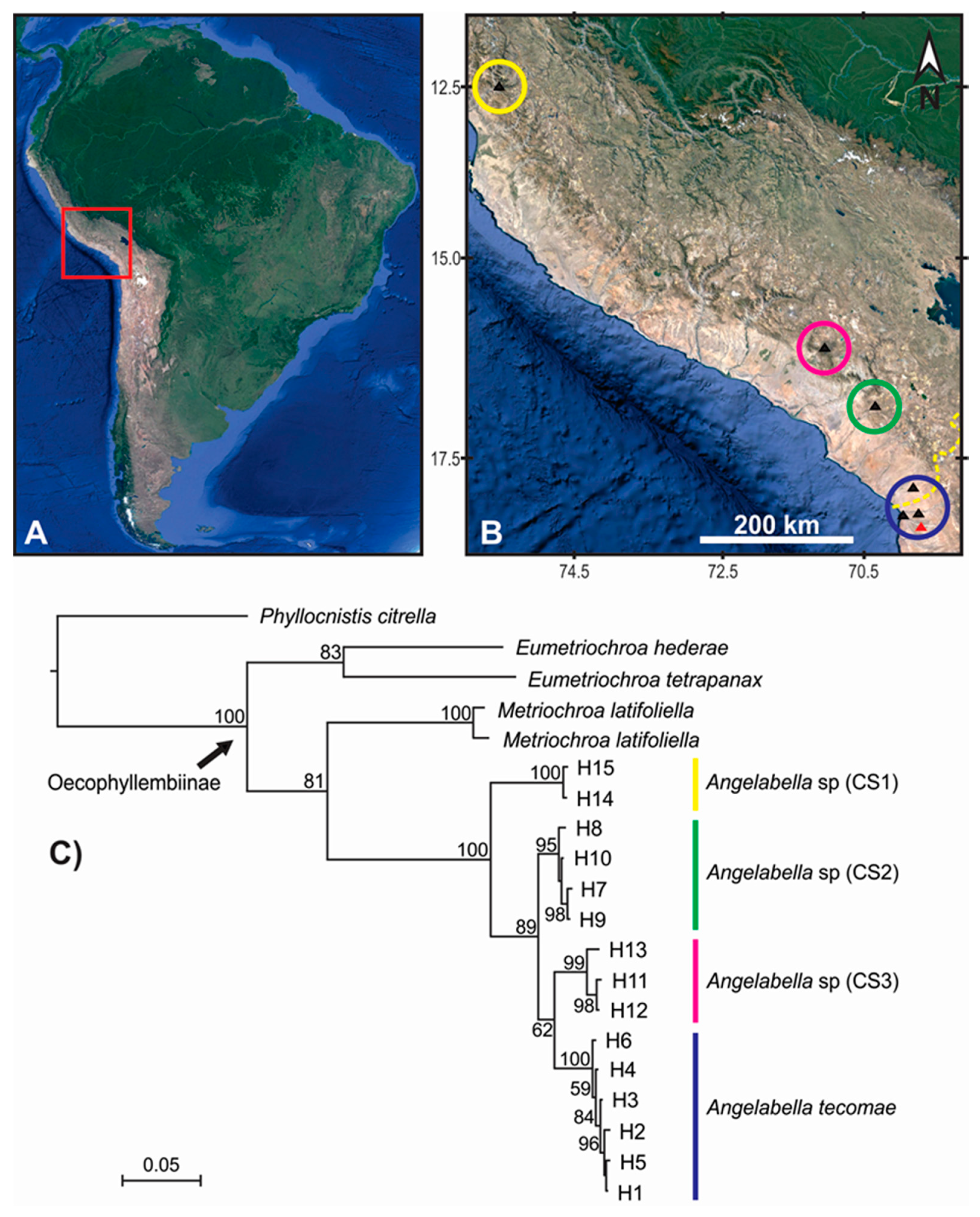
Figure 2.
Spatial clustering and phylogenetic relationships of the COI haplotypes of Angelabella. (A) The study area (red square) in South America. (B) Spatial clustering of the haplotypes suggested by Geneland analysis (circles), with sampling sites marked by triangles (type locality in red). Yellow dashed line in the lower right corner of Figure 2B indicates Peru-Chile geopolitical boundary. (C) Phylogenetic consensus tree of Angelabella tecomae and candidate species (CS); color of the longitudinal stripes corresponds to that of the spatial clusters in B; numbers near branches indicate node support (posterior probability). Yellow = Lima; pink = Arequipa; green = Moquegua; blue = Tacna-Arica.

Table 1.
Sequences Used in the Analyses of this Study. Haplotypes H4 to H15 Are New.
Geneland 4.9.2 [36,37] was used to determine the spatial location of genetic discontinuities of Angelabella. Analyses were carried out with K = 7, with 100,000,000 MCMC iterations, thinning each 10,000 iterations and a burn-in of 10%. The number of genetic clusters was assessed through probability density distribution graphs. In parallel, the geographic barrier isolation hypothesis suggested by Geneland was evaluated by means of an analysis of molecular variance in Arlequin 3.5 [38].
To determine evolutionary relationships between haplotypes and to evaluate the reciprocal monophyly hypothesis, a phylogenetic analysis was performed by Bayesian inference using a mixture model in BayesPhylogenies [39] with a GTR+G as substitution model. To assess the monophyly of Angelabella, sequences of Eumetriochroa Kumata, Metriochroa Busck (Oecophyllembiinae), and Phyllocnistis Zeller (Phyllocnistinae) were included in the phylogenetic analysis as outgroups, following the most recent phylogeny of Gracillariidae [40]. Trees were obtained after running 200,000,000 MCMC. The parameters of the runs were observed in Tracer 1.5. The visualization of the phylogenetic tree was done in BayesTrees 1.3 [41].
To determine if the cryptic diversity suggested by Geneland and the phylogenetic analysis corresponds to different Angelabella species, species discovery delimitation analyses were carried out using a distance-based method, the Automatic Barcode Gap Discovery (ABGD [21]), and three tree-based methods, the single-threshold General Mixed Yule Coalescent (st-GMYC [22]), a Bayesian implementation of the General Mixed Yule Coalescent (bGMYC [42]) and a Bayesian implementation of the PTP model (bPTP [24]). The reason for using Bayesian implementations of GMYC (bGMYC) and PTP (bPTP) methods is because with the determination of evolutionary relationships under single locus approaches (usually DNA barcode sequences), the estimation of phylogenies may be associated with large amounts of uncertainty [43,44]. bGMYC incorporates uncertainty in phylogenetic relationships and allows obtaining marginal probabilities of species identities [42], and bPTP adds Bayesian support values to species delimited on the input tree, so higher Bayesian support value of a node indicates that all descendants from this node are more likely to be conspecific (https://species.h-its.org/). The ABGD analysis was performed through the web server of ABGD (http://wwwabi.snv.jussieu.fr/public/abgd/abgdweb.html) using the K2P evolution model. Outgroups were removed from alignment. A range of prior intraspecific divergence values from 0.001 to 0.3 was assayed, applying a relative gap width of 1.5. For the st-GMYC analysis, an ultrametric phylogenetic tree was generated by Bayesian inference, using the Yule process as the prior method of speciation model under a lognormal relaxed clock model in BEAST 2.5 [45] using a clock rate of 0.023 substitutions per million years [46,47]. The ultrametric tree was used as input for the st-GMYC analysis conducted in R 4.0 using the splits package (http://r-forge.r-project.org/projects/splits/). We initially included only haplotypes of Angelabella. However, st-GMYC results were not significantly different from the null model of coalescence. Thus we added singletons of Metriochroa latifoliela, Eumetriochroa tetrapanax and Eumetriochroa hederae, species from the same subfamily as Angelabella (Oecophillembiinae), to increase the Yule portion of the tree and fit the model to the data better (e.g., [48,49]). The bGMYC model was implemented in R 4.0 using the bGMYC package, using 100 randomly selected trees from BEAST analysis (same priors as for st-GMYC). 50,000 MCMC generations were simulated, sampling every 100 trees and discarding 10% as burn-in. Finally, to implement the bPTP method, an ML tree was determined using RAxML-NG [50] on the website https://raxml-ng.vital-it.ch/, using a GTR +G+I as the substitution model and a bootstrapping cutoff of 0.5. The best resulting RAxML tree was used as input for the bPTP analysis on the website https://species.h-its.org/, using 100,000 MCMC generations, thinning every 100 trees and discarding 10% as burn-in.
3. Results
No morphological differences were found between pupae of A. tecomae from the type locality and those collected in Lima, Arequipa, Moquegua and Tacna, neither between the genitalia of A. tecomae from the type locality and those of adults from Arequipa.
No evidence of stop codons or substitution saturation (ISS < ISS.C, p < 0.05) were detected in the alignments, indicating that datasets were suitable for all the subsequent genetic analyses.
3.1. Genetic Diversity and Spatial Distribution
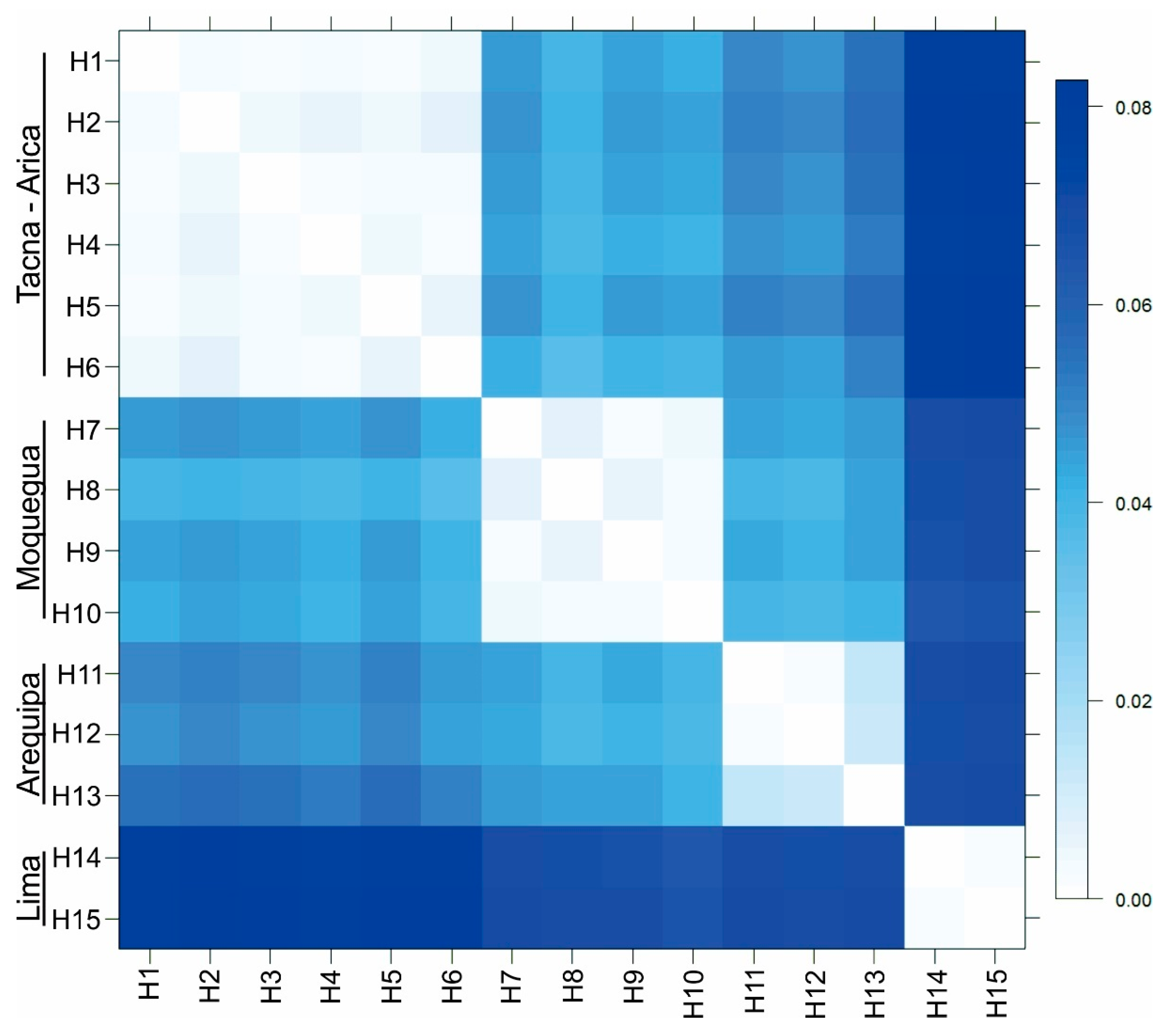
Forty four sequences of Angelabella of 657 base pair (bp) length were analyzed, including 29 provided here and 15 from a previous study [4]. Their alignment showed 73 variable sites, representing fifteen haplotypes (H1-H15): two from Lima, three from Arequipa, four from Moquegua and six from Tacna-Arica. Tajima’s D neutrality test indicated a non-significant deviation from zero (Tajima’s D = 0.458, p > 0.1), thus supporting the null hypothesis that the observed polymorphism has been maintained without selection. The Geneland analysis indicated the maximum a posteriori estimate for four genetic clusters (K = 4) with probabilities greater than 0.4 (Figure S1). The clusters were distributed in Lima (cluster 1), Arequipa (cluster 2), Moquegua (cluster 3) and Tacna-Arica (cluster 4) (Figure 2A,B). Analysis of molecular variance indicated that 94.28% of the variation occurred between these spatial groups (FST = 0.94). Genetic distance was 0.2–0.8 and 3.6–8.3% (K2P) between haplotypes of the same and different spatial clusters, respectively (Figure 3).
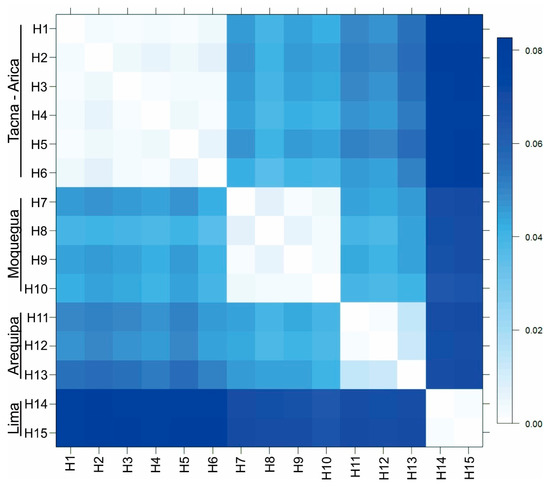
Figure 3.
Genetic distance (K2P) matrix between COI haplotypes (657 bp) of Angelabella ordered according to spatial clusters suggested by Geneland. Tacna-Arica group represents A. tecomae, the type species of the genus.
3.2. Phylogenetic Analysis and Species Delimitation
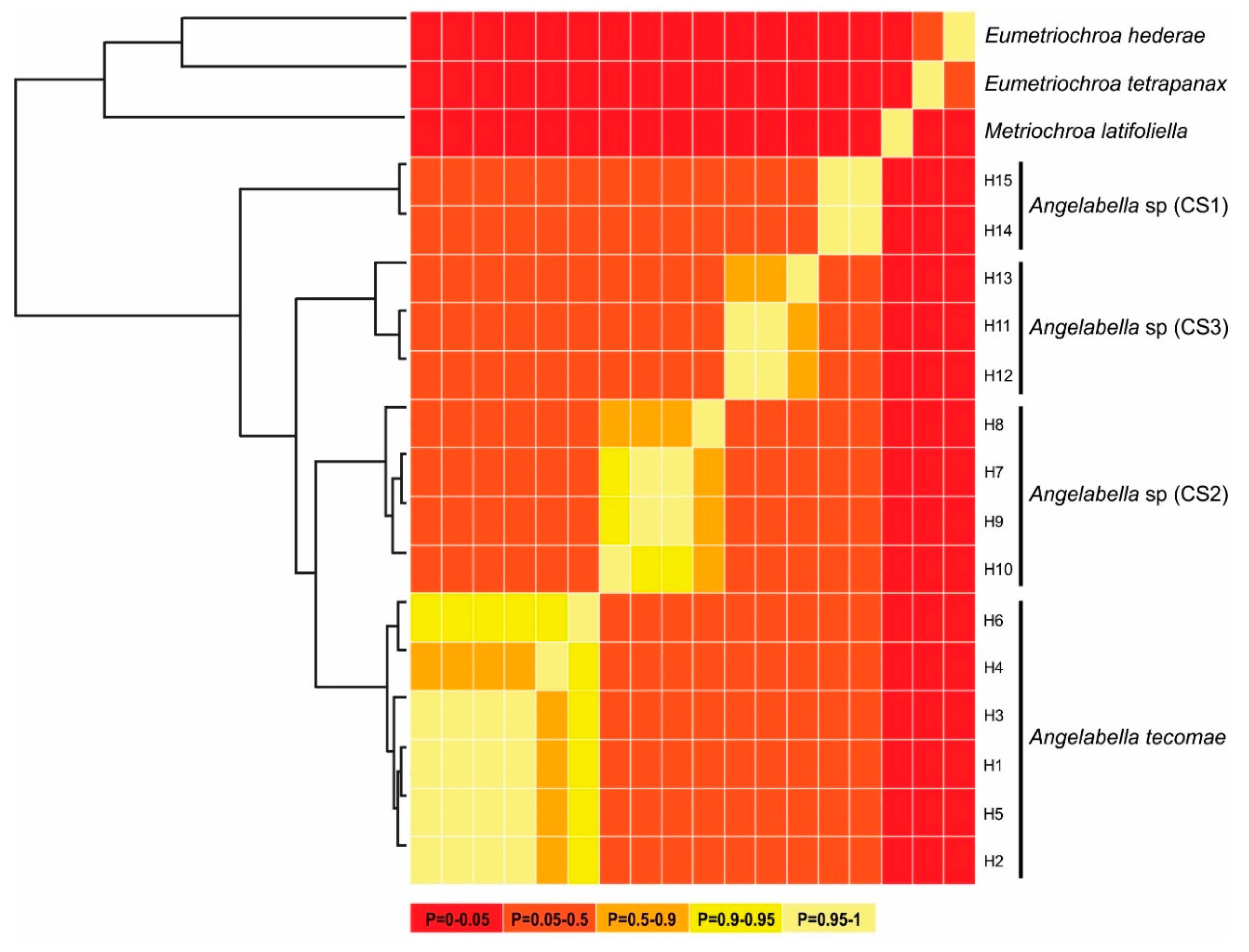
The alignment included 20 sequences of 657 bp, one of each haplotype of Angelabella and five of outgroups. The McDonald-Kreitman (MK) neutrality test indicated that ratio of replacement to synonymous fixed substitutions is the same as the ratio of replacement to synonymous polymorphism (G value = 0.288, p-value = 0.591). Therefore, the evolution represented by the dataset has occurred mainly by neutral processes. The phylogenetic analysis indicated a well-supported clade of the subfamily Oecophyllembiinae, clustering all the sequences of Angelabella in a clade (100% posterior probability) sister to Metriochroa latifoliella (Millière). The haplotypes in the Angelabella clade were clustered in accordance with the spatial groups suggested by Geneland in well-supported reciprocally monophyletic groups (Lima, Arequipa, Moquegua and Tacna-Arica). All species delimitation analyses (ABGD, st-GMYC, bGMYC and bPTP) indicated that Angelabella includes the type species, A. tecomae, represented by lineages of Tacna-Arica, and three candidate species (CSs) represented by the lineages of Lima, Arequipa, and Moquegua (Figure 2C). The ABGD analysis showed three barcode gaps, around 0.02, 0.04, and 0.075 (Figure S2). The st-GMYC analysis indicated that the number of Angelabella species delimited is four (number of ML clusters = 4; confidence interval = 2–4), favoring the st-GMYC model over the null model of coalescence (likelihood ratio = 7.501, p-value < 0.05), with a time threshold between inter- intraspecific branching events of about 350 Kya (Figure S3). The same number of candidate species within Angelabella was suggested by bGMYC analysis using a conspecificity probability threshold of 0.5 (Figure 4), assigning the same haplotypes for each candidate species as the other analyses. The highest Bayesian supported solution of the bPTP analysis indicated that Angelabella included four species, with ranges of support between 0.554–0.971 (Figure S4).
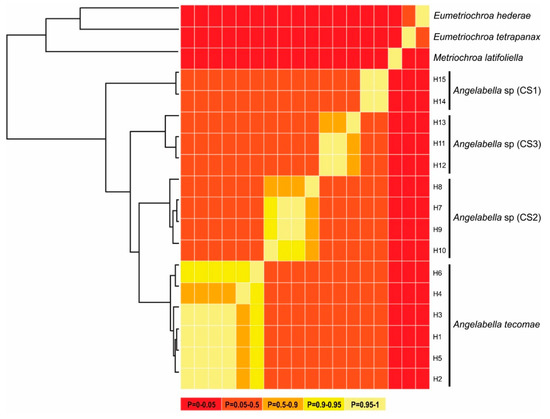
Figure 4.
Summary of Bayesian General Mixed Yule Coalescent (bGMYC) results. Maximum clade credibility tree from BEAST (left) and sequence-by-sequence matrix (right). Colors of cells represent the posterior probability that the sequences are conspecific.
4. Discussion
The taxonomic diversity of the Neotropical Gracillariidae remains poorly researched, mainly due to the scarce collecting efforts and the small number of taxonomists focused on the study of these micromoths in this biogeographic region [13,51]. The geographic distribution is poorly documented for a large number of the currently described Neotropical gracillariids, many of which are recorded only from the type locality [52]. This is the case of the Peruvian fauna of Gracillariidae, with only 28 native species recorded despite the high plant and environmental diversity of this country [52,53,54].
It has been suggested recently that DNA barcodes can be helpful to explore geographic ranges of Lepidoptera, due to the small intraspecific divergence generally found between samples widely separated geographically [55]. In contrast, the divergence between the haplotypes of Angelabella of different spatial groups was relatively high on a small geographic scale. This could be attributed to the particular landscape configuration of the study area, which has relatively small patches of host plants separated by extensive hyperarid lands that appear to represent geographic barriers with low permeability to gene flow of these micromoths between patches. In relation to the geographic distribution of A. tecomae, the results provide molecular evidence in support of the only previous Peruvian record of this species, suggesting a relatively narrow geographic range restricted to a few ravines near the limit between Peru and Chile, as only the samples from Tacna-Arica showed divergence levels within the ranges previously reported as intraspecific for Gracillariidae [56,57,58]. This geographic range may be slightly wider, because T. f. fulva occurs in a few additional ravines in the southernmost part of Peru [18]; this micromoth should be searched for in all of them.
Although morphological comparisons were not an objective of this study, pupae from different sampling sites were compared under stereomicroscope before DNA extraction, and the genitalia of two males obtained from Arequipa were compared under light microscope with those of specimens from the type locality of A. tecomae. However, no clear differences were found in either case. The divergence between haplotypes of Angelabella from different spatial genetic clusters (3.6–8.3% K2P) is either remarkably higher than [5] or near [6,12] those recorded between morphologically cryptic species of two other genera of Gracillariidae. Similar levels of divergence have been interpreted to represent putative heterospecific lineages in the absence of morphological evidence [12,17,58]. Despite the absence of obvious morphological differentiation between samples of Angelabella, the deep divergence between haplotypes of different spatial clusters, their reciprocal monophyly indicated by the phylogenetic analysis and the highly consistent results of the four species delimitation analyses suggest heterospecific status for the geographically isolated lineages analyzed. Patterns of allopatric genetic differentiation similar to those found in this study have been recorded for populations of Bucculatricidae [19] and Tortricidae [59], and pairs of morphologically cryptic allopatric species are known to occur in Cosmopterigidae [60] and Tortricidae [61] near the study area, suggesting the characteristics of these hyperarid landscapes as a causal agent of allopatric diversification processes among populations of micromoths in the Peru-Chile desert.
The only other micromoth associated with T. fulva is the many-plumed moth Alucita danunciae Vargas, 2011 (Lepidoptera: Alucitidae), whose larvae feed on unripe seeds, either on T. f. fulva in the coastal valleys of northern Chile [62] or T. f. arequipensis on the western slopes of the Andes of Arequipa in southern Peru [63]. Divergence of 0.6–0.9% (K2P) was recorded between DNA barcodes of A. danunciae from the two localities [63], contrasting with the 4.7–5.6% (K2P) found in the present study between DNA barcodes of Angelabella from northern Chile and Arequipa, suggesting different lineage diversification scenarios in micromoths associated with the same host plants in the same geographic range, a comparative aspect that deserves further attention.
Although solid taxonomic identifications of Lepidoptera should be based on the analysis of a wide range of data sources, our results show that adequate single locus sequence analysis can be useful to discover surprising biodiversity patterns in underexplored environments, providing the base to plan further studies involving little-known organisms. Angelabella includes only the type species up to now [52]. However, the results provided here suggest that it is not a monotypic genus, but harbors four morphologically cryptic allopatric species, a scenario that should be explored further. As Angelabella has been surveyed only on three of the six subspecies of T. fulva, surveys must be expanded to the remaining three subspecies, two of which are distributed on the eastern slopes of the Andes [18], to characterize better the distribution and species diversity of this micromoth genus. A greater sampling effort is necessary to define lineage diversity at the metapopulation level for the type species and the three candidate species suggested here, which implies more detailed knowledge of their geographic ranges. Additional character sources must be assessed, including morphology of different life stages [64,65] and bi-parental markers [5,6], to understand better the processes and patterns that have governed the evolutionary history of this leaf miner genus.
5. Conclusions
The results obtained in this study suggest that Angelabella is not a monotypic genus, but harbors at least four morphologically cryptic allopatric leaf miner species with restricted geographic ranges, including the type species and three candidate species.
Supplementary Materials
The following are available online at https://www.mdpi.com/2075-4450/11/10/677/s1, Figure S1: Results of Geneland analyses. A. Number of clusters along the chain after burn-in. B. Map of posterior probability to belong to clusters 1-4 (indicated by dark colors), Figure S2: Graphical results of the ABGD analysis. A. Histogram of pairwise Kimura 2-parameter distances for the COI marker, showing three barcode gaps. B. Ranked pairwise distances. C. Number of groups for partitions per value of prior intraspecific divergence, Figure S3: Results of the st-GMYC analysis. A. Number of lineages over time (years) (red vertical line indicates the threshold time between inter- intraspecific branching). B. Likelihood surface through time. C. Tree with species delimited (indicated by red branches), Figure S4: Results of the bPTP analysis. A. Highest Bayesian supported solution. B. Delimitation supports for candidate species (CS) of Angelabella.
Author Contributions
Conceptualization, J.C., J.F., M.V.-O., and H.A.V.; methodology, G.A.-P., A.L.-R., J.C., J.F., W.H.-M., and M.V.-O.; formal analysis, M.V.-O.; writing—original draft preparation, M.V.-O. and H.A.V.; writing—review and editing, G.A.-P., A.L.-R., J.C., J.F., and W.H.-M.; supervision, H.A.V.; funding acquisition, G.A.-P., H.A.V. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Universidad Nacional de San Agustin de Arequipa through the contract number TP IB-08-2020-UNSA to G.A.-P., Universidad de Tarapacá through “UTA-Mayor 9726-20” to H.A.V., and Beca Doctorado Nacional CONICYT-2018 (currently called ANID) to M.V.-O.
Acknowledgments
We thank Steffany Cardenas-Ninasivincha for support in DNA extraction and Lafayette Eaton for checking the English.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Vargas, H.A.; Parra, L.E. Un nuevo género y una nueva especie de oecophyllembiinae (Lepidoptera: Gracillariidae) de Chile. Neotrop. Entomol. 2005, 34, 227–233. [Google Scholar] [CrossRef]
- Storey-Palma, J.; Benítez, H.A.; Mundaca, E.A.; Vargas, H.A. Egg laying site selection by a host plant specialist leaf miner moth at two intra-plant levels in the northern chilean atacama desert. Rev. Bras. Entomol. 2014, 58, 280–284. [Google Scholar] [CrossRef]
- Vargas, H.A. Angelabella tecomae (Lepidoptera: Gracillariidae): An exotic hostplant in northern Chile and first record from Peru. Rev. Colomb. Entomol. 2010, 36, 340–341. [Google Scholar]
- Wood, J.R.I. A revision of tecoma juss. (Bignoniaceae) in Bolivia. Bot. J. Linn. Soc. 2008, 156, 143–172. [Google Scholar] [CrossRef]
- Maita-Maita, J.; Huanca-Mamani, W.; Vargas, H.A. First remarks on genetic variation of the little known leaf miner Angelabella tecomae vargas & parra (Gracillariidae) in the atacama desert of northern chile. J. Lepid. Soc. 2015, 69, 192–196. [Google Scholar]
- Kirichenko, N.; Huemer, P.; Deutsch, H.; Triberti, P.; Rougerie, R.; Lopez-Vaamonde, C. Integrative taxonomy reveals a new species of Callisto (Lepidoptera, Gracillariidae) in the Alps. ZooKeys 2015, 473, 157–176. [Google Scholar] [CrossRef]
- Kirichenko, N.; Triberti, P.; Ohshima, I.; Haran, J.; Byun, B.-K.; Li, H.; Augustin, S.; Roques, A.; Lopez-Vaamonde, C. From east to west across the Palearctic: Phylogeography of the invasive lime leaf miner phyllonorycter issikii (Lepidoptera: Gracillariidae) and discovery of a putative new cryptic species in East Asia. PLoS ONE 2017, 12, e0171104. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L.; deWaard, J.R. Biological identifications through DNA barcodes. Proc. Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef]
- Huemer, P.; Wieser, C.; Stark, W.; Hebert, P.D.N.; Wiesmair, B. DNA barcode library of megadiverse Austrian noctuoidea (Lepidoptera)-a nearly perfect match of linnean taxonomy. Biodivers. Data J. 2019, 7, e37734. [Google Scholar] [CrossRef]
- Huemer, P.; Karsholt, O.; Aarvik, L.; Berggren, K.; Bidzilya, O.; Junnilainen, J.; Landry, J.-F.; Mutanen, M.; Nupponen, K.; Segerer, A.; et al. DNA barcode library for European gelechiidae (Lepidoptera) suggests greatly underestimated species diversity. ZooKeys 2020, 921, 141–157. [Google Scholar] [CrossRef]
- Kim, S.; Lee, Y.; Mutanen, M.; Seung, J.; Lee, S. High functionality of DNA barcodes and revealed cases of cryptic diversity in Korean curved-horn moths (Lepidoptera: Gelechioidea). Sci. Rep. 2020, 10, 6208. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, A.Y.; Jones, M.; Jia, Q.; Lapointe, S.L.; Stansly, P.A. A synthetic pheromone for phyllocnistis citrella (Lepidoptera: Gracillariidae) attracts multiple leafminer species. Fla. Entomol. 2013, 96, 1213–1216. [Google Scholar] [CrossRef]
- Laštůvka, Z.; Laštůvka, A.; Lopez-Vaamonde, C. A revision of the phyllonorycter ulicicolella species group with description of a new species (Lepidoptera: Gracillariidae). SHILAP Revta. Lepid. 2013, 41, 251–265. [Google Scholar]
- Lees, D.C.; Kawahara, A.Y.; Rougerie, R.; Ohshima, I.; Kawakita, A.; Bouteleux, O.; De Prins, J.; Lopez-Vaamonde, C. DNA barcoding reveals a largely unknown fauna of gracillariidae leaf-mining moths in the neotropics. Mol. Ecol. Resour. 2014, 14, 286–296. [Google Scholar] [CrossRef]
- Kirichenko, N.; Triberti, P.; Akulov, E.; Ponomarenko, M.; Gorokhova, S.; Sheiko, V.; Ohshima, I.; Lopez-Vaamonde, C. Exploring species diversity and host plant associations of leaf-mining micromoths (Lepidoptera: Gracillariidae) in the Russian far east using DNA barcoding. Zootaxa 2019, 4652, 1–55. [Google Scholar] [CrossRef]
- Hausmann, A.; Scalercio, S. Host-plant relationships of 29 mediterranean lepidoptera species in forested ecosystems unveiled by DNA Barcoding (Insecta: Lepidoptera). SHILAP. Revta. Lepid. 2016, 44, 463–471. [Google Scholar]
- Hausmann, A.; Diller, J.; Moriniere, J.; Höcherl, A.; Floren, A.; Haszprunar, G. DNA barcoding of fogged caterpillars in peru: A novel approach for unveiling host-plant relationships of tropical moths (Insecta, Lepidoptera). PLoS ONE 2020, 15, e0224188. [Google Scholar] [CrossRef]
- Brito, R.; Gonçalves, G.L.; Vargas, H.A.; Moreira, G.R.P. A new brazilian passiflora leafminer: Spinivalva gaucha, gen. n., sp. n. (Lepidoptera, Gracillariidae, Gracillariinae), the first gracillariid without a sap-feeding instar. ZooKeys 2013, 291, 1–26. [Google Scholar] [CrossRef]
- Vargas-Ortiz, M.; Bobadilla, D.; Huanca-Mamani, W.; Vargas, H.A. Genetic divergence of isolated populations of the native micromoth bucculatrix mirnae (Lepidoptera: Bucculatricidae) in the arid environments of northern chile. Mitochondrial DNA Part A 2018, 29, 1139–1147. [Google Scholar] [CrossRef]
- Luo, A.; Ling, C.; Ho, S.Y.W.; Zhu, C.D. Comparison of methods for molecular species delimitation across a range of speciation scenarios. Syst. Biol. 2018, 67, 830–846. [Google Scholar] [CrossRef]
- Puillandre, N.; Lambert, A.; Brouillet, S.; Achaz, G. ABGD, automatic barcode gap discovery for primary species delimitation. Mol. Ecol. 2012, 21, 1864–1877. [Google Scholar] [CrossRef] [PubMed]
- Pons, J.; Barraclough, T.G.; Gomez-Zurita, J.; Cardoso, A.; Duran, D.P.; Hazell, S.; Vogler, A.P. Sequence-based species delimitation for the DNA taxonomy of undescribed insects. Syst. Biol. 2006, 55, 595–609. [Google Scholar] [CrossRef] [PubMed]
- Fujisawa, T.; Barraclough, T.G. Delimiting species using single-locus data and the generalized mixed yule coalescent approach: A revised method and evaluation on simulated data sets. Syst. Biol. 2013, 62, 707–724. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Kapli, P.; Pavlidis, P.; Stamatakis, A. A general species delimitation method with applications to phylogenetic placements. Bioinformatics 2013, 29, 2869–2876. [Google Scholar] [CrossRef] [PubMed]
- Dellicour, S.; Flot, J.F. Delimiting species-poor data sets using single molecular markers: A study of barcode gaps, haplowebs and GMYC. Syst. Biol. 2015, 64, 900–908. [Google Scholar] [CrossRef] [PubMed]
- Dellicour, S.; Flot, J.F. The hitchhiker’s guide to single-locus species delimitation. Mol. Ecol. Resour. 2018, 18, 1234–1246. [Google Scholar] [CrossRef]
- Carstens, B.C.; Pelletier, T.A.; Reid, N.M.; Satler, J.D. How to fail at species delimitation. Mol. Ecol. 2013, 22, 4369–4383. [Google Scholar] [CrossRef]
- Huanca-Mamani, W.; Rivera-Cabello, D.; Maita-Maita, J. A simple, fast, and inexpensive CTAB-PVP-Silica based method for genomic DNA isolation from single, small insect larvae and pupae. Genet. Mol. Res. 2015, 14, 7990–8000. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Penton, E.H.; Burns, J.M.; Janzen, D.H.; Hallwachs, W. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly astraptes fulgerator. Proc. Natl. Acad. Sci. USA 2004, 101, 14812–14817. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [PubMed]
- Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [PubMed]
- McDonald, J.H.; Kreitman, M. Adaptive protein evolution at the Adh locus in Drosophila. Nature 1991, 351, 652–654. [Google Scholar] [CrossRef]
- Xia, X.; Xie, Z.; Salemi, M.; Chen, L.; Wang, Y. An index of substitution saturation and its application. Mol. Phylogenet. Evol. 2003, 26, 1–7. [Google Scholar] [CrossRef]
- Xia, X. DAMBE7: New and improved tools for data analysis in molecular biology and evolution. Mol. Biol. Evol. 2018, 35, 1550–1552. [Google Scholar] [CrossRef]
- Guillot, G.; Mortier, F.; Estoup, A. GENELAND: A computer package for landscape genetics. Mol. Ecol. Notes 2005, 5, 712–715. [Google Scholar] [CrossRef]
- Guillot, G.; Renaud, S.; Ledevin, R.; Michaux, J.; Claude, J. A unifying model for the analysis of phenotypic, genetic, and geographic data. Syst. Biol. 2012, 61, 897–911. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Pagel, M.; Meade, A. A phylogenetic mixture model for detecting pattern-heterogeneity in gene sequence or character-state data. Syst. Biol. 2004, 53, 571–581. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, A.Y.; Plotkin, D.; Ohshima, I.; Lopez-Vaamonde, C.; Houlihan, P.; Breinholt, J.W.; Kawakita, A.; Xiao, L.; Regier, J.C.; Davis, D.R.; et al. A molecular phylogeny and revised higher-level classification for the leaf-mining moth family Gracillariidae and its implications for larval host use evolution. Syst. Entomol. 2017, 42, 60–81. [Google Scholar] [CrossRef]
- Meade, A.; Pagel, M. BayesTrees, Version 1.3. 2011. Available online: http://www.evolution.reading.ac.uk/BayesTrees.html/ (accessed on 20 June 2020).
- Reid, N.M.; Carstens, B.C. Phylogenetic estimation error can decrease the accuracy of species delimitation: A Bayesian implementation of the general mixed Yule-coalescent model. BMC Evol. Biol. 2012, 12, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Hickerson, M.J.; Meyer, C.P.; Moritz, C. DNA barcoding will often fail to discover new animal species over broad parameter space. Syst. Biol. 2006, 55, 729–739. [Google Scholar] [CrossRef] [PubMed]
- Wilson, J.J. Assessing the value of DNA barcodes for molecular phylogenetics: Effect of increased taxon sampling in lepidoptera. PLoS ONE 2011, 6, e24769. [Google Scholar] [CrossRef]
- Bouckaert, R.; Vaughan, T.G.; Barido-Sottani, J.; Duchêne, S.; Fourment, M.; Gavryushkina, A.; Heled, J.; Jones, G.; Kühnert, D.; De Maio, N.; et al. BEAST 2.5: An advanced software platform for bayesian evolutionary analysis. PLoS Comput. Biol. 2019, 15, e1006650. [Google Scholar] [CrossRef]
- Brower, A.V.Z. Rapid morphological radiation and convergence among races of the butterfly Heliconius erato inferred from patterns of mitochondrial DNA evolution. Proc. Natl. Acad. Sci. USA 1994, 91, 6491–6495. [Google Scholar] [CrossRef]
- Nazari, V.; Sperling, F.A.H. Mitochondrial DNA divergence and phylogeography in western palaearctic parnassiinae (Lepidoptera: Papilionidae): How many species are there? Insect Syst. Evol. 2007, 38, 121–138. [Google Scholar] [CrossRef]
- Talavera, G.; Dincǎ, V.; Vila, R. Factors affecting species delimitations with the GMYC model: Insights from a butterfly survey. Methods Ecol. Evol. 2013, 4, 1101–1110. [Google Scholar] [CrossRef]
- Ahrens, D.; Fujisawa, T.; Krammer, H.J.; Eberle, J.; Fabrizi, S.; Vogler, A.P. Rarity and incomplete sampling in DNA-based species delimitation. Syst. Biol. 2016, 65, 478–494. [Google Scholar] [CrossRef]
- Kozlov, A.M.; Darriba, D.; Flouri, T.; Morel, B.; Stamatakis, A. RAxML-NG: A fast scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 2019, 35, 4453–4455. [Google Scholar] [CrossRef]
- Brito, R.; De Prins, J.; De Prins, W.; Mielke, O.H.H.; Gonçalves, G.L.; Moreira, G.R.P. Extant diversity and estimated number of gracillariidae (Lepidoptera) species yet to be discovered in the neotropical region. Rev. Bras. Entomol. 2016, 60, 275–283. [Google Scholar] [CrossRef][Green Version]
- De Prins, J.; Arévalo-Maldonado, H.A.; Davis, D.R.; Landry, B.; Vargas, H.A.; Davis, M.M.; Brito, R.; Fochezato, J.; Ohshima, I.; Moreira, G.R.P. An illustrated catalogue of the neotropical gracillariidae (Lepidoptera) with new data on primary types. Zootaxa 2019, 4575, 1–110. [Google Scholar] [CrossRef] [PubMed]
- Kawakita, A.; Sato, A.A.W.; Salazar, J.R.L.; Kato, M. Leafflower–leafflower moth mutualism in the neotropics: Successful transoceanic dispersal from the old world to the new world by actively pollinating leafflower moths. PLoS ONE 2019, 14, e0210727. [Google Scholar] [CrossRef] [PubMed]
- Davis, D.R.; Farfán, J.; Cerdeña, J.; Huanca-Mamani, W.; Vargas, H.A.; Vargas-Ortiz, M.; Gonçalves, G.L.; Moreira, G.R.P. Adenogasteria leguminivora Davis & Vargas gen. et sp. nov. (Lepidoptera: Gracillariidae): A new seed-feeding micromoth associated with fabaceae in Peru and Chile. Austral Entomol. 2020, 59, 37–51. [Google Scholar] [CrossRef]
- Huemer, P.; Hebert, P.D.N.; Mutanen, M.; Wieser, C.; Wiesmair, B.; Hausmann, A.; Yakovlev, R.; Möst, M.; Gottsberger, B.; Strutzenberger, P.; et al. Large geographic distance versus small DNA barcode divergence: Insights from a comparison of European to South Siberian Lepidoptera. PLoS ONE 2018, 13, e0206668. [Google Scholar] [CrossRef] [PubMed]
- Valade, R.; Kenis, M.; Hernandez-Lopez, A.; Augustin, S.; Mari Mena, N.; Magnoux, E.; Rougerie, R.; Lakatos, F.; Roques, A.; Lopez-Vaamonde, C. Mitochondrial and microsatellite DNA markers reveal a balkan origin for the highly invasive horse-chestnut leaf miner Cameraria ohridella (Lepidoptera, Gracillariidae). Mol. Ecol. 2009, 18, 3458–3470. [Google Scholar] [CrossRef] [PubMed]
- Tóth, V.; Lakatos, F. Phylogeographic pattern of the plane leaf miner, Phyllonorycter platani (Staudinger, 1870) (Lepidoptera: Gracillariidae) in Europe. BMC Evol. Biol. 2018, 18, 135. [Google Scholar] [CrossRef]
- Vargas-Ortiz, M.; Gonçalves, G.L.; Huanca-Mamani, W.; Vargas, H.A.; Moreira, G.R.P. Description, natural history and genetic variation of Caloptilia guacanivora sp. nov. vargas-ortiz & vargas (Lepidoptera: Gracillariidae) in the atacama desert, Chile. Austral Entomol. 2019, 58, 171–191. [Google Scholar] [CrossRef]
- Escobar-Suárez, S.; Huanca-Mamani, W.; Vargas, H.A. Genetic divergence of a newly documented population of the cecidogenous micromoth Eugnosta azapaensis vargas & moreira (Lepidoptera: Tortricidae) in the atacama desert. Rev. Bras. Entomol. 2017, 61, 266–270. [Google Scholar] [CrossRef]
- Espinoza-Donoso, S.; Bobadilla, D.; Huanca-Mamani, W.; Vargas-Ortiz, M.; Vargas, H.A. A new species of ithome chambers (Lepidoptera, Cosmopterigidae, Chrysopeleiinae) from the atacama desert revealed by morphology and DNA barcodes. ZooKeys 2020, 912, 125–138. [Google Scholar] [CrossRef]
- Clarke, J.F.G. Two new cryptophlebia walsingham from Chile (Lepidoptera: Tortricidae). Acta Entomol. Chil. 1987, 14, 7–12. [Google Scholar]
- Vargas, H.A. A new species of Alucita L. (Lepidoptera: Alucitidae) from northern Chile. Neotrop. Entomol. 2011, 40, 85–88. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Farfán, J.; Cerdeña, J.; Arivilca, M.; Condori-Mamani, M.; Huanca-Mamani, W.; Vargas, H.A. First record of Alucita danunciae (Lepidoptera: Alucitidae) in Peru. Studies Neotrop. Fauna Environ. 2020, 55, 103–108. [Google Scholar] [CrossRef]
- Pereira, C.M.; Silva, D.S.; Gonçalves, G.L.; Vargas, H.A.; Moreira, G.R.P. A new species of Leurocephala davis & mckay (Lepidoptera, Gracillariidae) from the azapa valley, northern chilean atacama desert, with notes on life history. Rev. Bras. Entomol. 2017, 61, 6–15. [Google Scholar] [CrossRef][Green Version]
- Brito, R.; Mielke, O.H.H.; Gonçalves, G.L.; Moreira, G.R.P. Description of three new species of Phyllocnistis Zeller, 1848 (Lepidoptera: Gracillariidae), from the Atlantic forest, south Brazil, with notes on natural history and phylogeny. Austral Entomol. 2019, 58, 27–51. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).