Cryptic Diversity in the Monotypic Neotropical Micromoth Genus Angelabella (Lepidoptera: Gracillariidae) in the Peru-Chile Desert
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
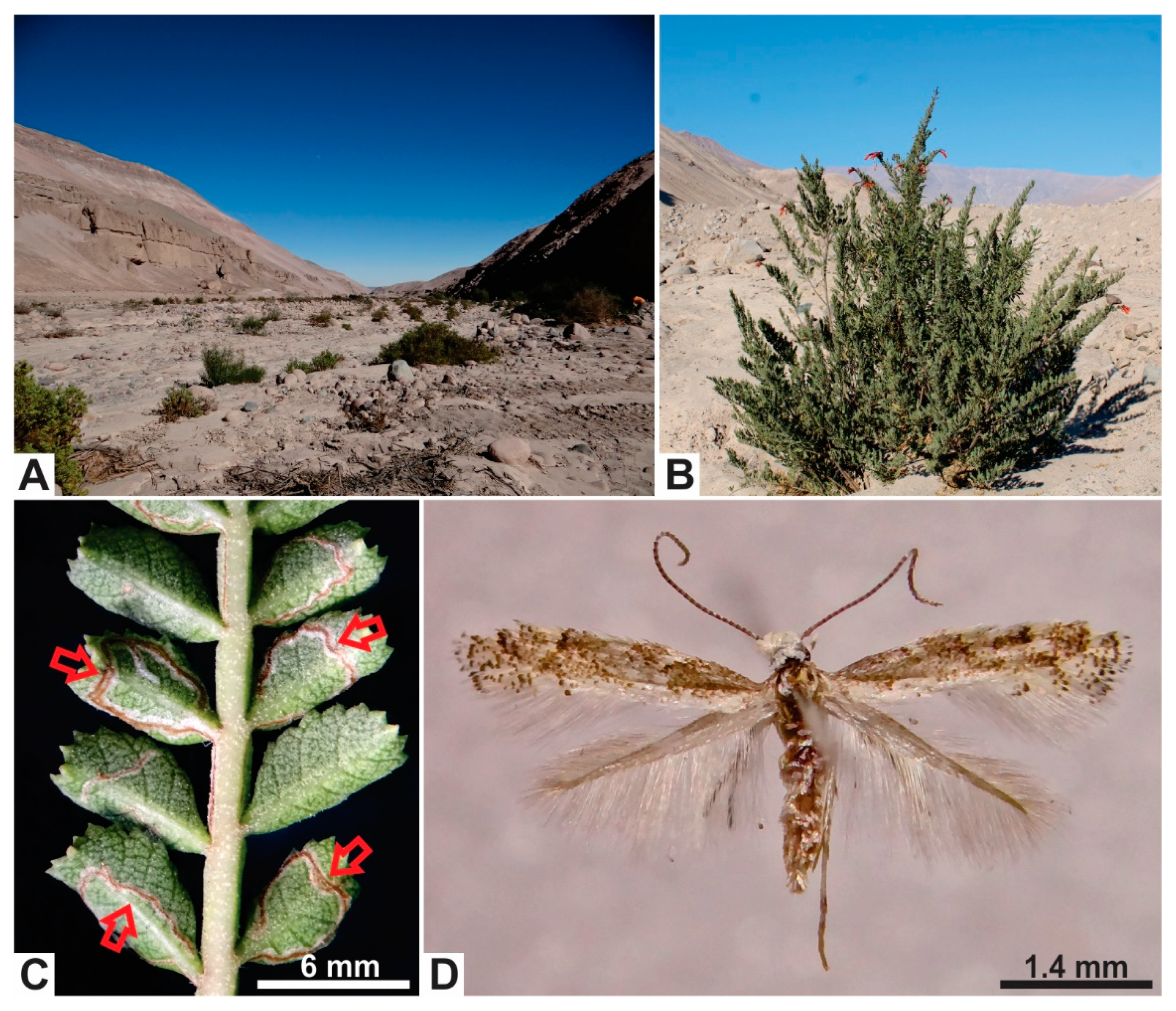
2.1. Sample Collection
2.2. DNA Extraction and Sequencing
2.3. Sequence Analysis
3. Results
3.1. Genetic Diversity and Spatial Distribution
3.2. Phylogenetic Analysis and Species Delimitation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Vargas, H.A.; Parra, L.E. Un nuevo género y una nueva especie de oecophyllembiinae (Lepidoptera: Gracillariidae) de Chile. Neotrop. Entomol. 2005, 34, 227–233. [Google Scholar] [CrossRef]
- Storey-Palma, J.; Benítez, H.A.; Mundaca, E.A.; Vargas, H.A. Egg laying site selection by a host plant specialist leaf miner moth at two intra-plant levels in the northern chilean atacama desert. Rev. Bras. Entomol. 2014, 58, 280–284. [Google Scholar] [CrossRef]
- Vargas, H.A. Angelabella tecomae (Lepidoptera: Gracillariidae): An exotic hostplant in northern Chile and first record from Peru. Rev. Colomb. Entomol. 2010, 36, 340–341. [Google Scholar]
- Wood, J.R.I. A revision of tecoma juss. (Bignoniaceae) in Bolivia. Bot. J. Linn. Soc. 2008, 156, 143–172. [Google Scholar] [CrossRef]
- Maita-Maita, J.; Huanca-Mamani, W.; Vargas, H.A. First remarks on genetic variation of the little known leaf miner Angelabella tecomae vargas & parra (Gracillariidae) in the atacama desert of northern chile. J. Lepid. Soc. 2015, 69, 192–196. [Google Scholar]
- Kirichenko, N.; Huemer, P.; Deutsch, H.; Triberti, P.; Rougerie, R.; Lopez-Vaamonde, C. Integrative taxonomy reveals a new species of Callisto (Lepidoptera, Gracillariidae) in the Alps. ZooKeys 2015, 473, 157–176. [Google Scholar] [CrossRef] [Green Version]
- Kirichenko, N.; Triberti, P.; Ohshima, I.; Haran, J.; Byun, B.-K.; Li, H.; Augustin, S.; Roques, A.; Lopez-Vaamonde, C. From east to west across the Palearctic: Phylogeography of the invasive lime leaf miner phyllonorycter issikii (Lepidoptera: Gracillariidae) and discovery of a putative new cryptic species in East Asia. PLoS ONE 2017, 12, e0171104. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L.; deWaard, J.R. Biological identifications through DNA barcodes. Proc. Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef] [Green Version]
- Huemer, P.; Wieser, C.; Stark, W.; Hebert, P.D.N.; Wiesmair, B. DNA barcode library of megadiverse Austrian noctuoidea (Lepidoptera)-a nearly perfect match of linnean taxonomy. Biodivers. Data J. 2019, 7, e37734. [Google Scholar] [CrossRef]
- Huemer, P.; Karsholt, O.; Aarvik, L.; Berggren, K.; Bidzilya, O.; Junnilainen, J.; Landry, J.-F.; Mutanen, M.; Nupponen, K.; Segerer, A.; et al. DNA barcode library for European gelechiidae (Lepidoptera) suggests greatly underestimated species diversity. ZooKeys 2020, 921, 141–157. [Google Scholar] [CrossRef]
- Kim, S.; Lee, Y.; Mutanen, M.; Seung, J.; Lee, S. High functionality of DNA barcodes and revealed cases of cryptic diversity in Korean curved-horn moths (Lepidoptera: Gelechioidea). Sci. Rep. 2020, 10, 6208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawahara, A.Y.; Jones, M.; Jia, Q.; Lapointe, S.L.; Stansly, P.A. A synthetic pheromone for phyllocnistis citrella (Lepidoptera: Gracillariidae) attracts multiple leafminer species. Fla. Entomol. 2013, 96, 1213–1216. [Google Scholar] [CrossRef] [Green Version]
- Laštůvka, Z.; Laštůvka, A.; Lopez-Vaamonde, C. A revision of the phyllonorycter ulicicolella species group with description of a new species (Lepidoptera: Gracillariidae). SHILAP Revta. Lepid. 2013, 41, 251–265. [Google Scholar]
- Lees, D.C.; Kawahara, A.Y.; Rougerie, R.; Ohshima, I.; Kawakita, A.; Bouteleux, O.; De Prins, J.; Lopez-Vaamonde, C. DNA barcoding reveals a largely unknown fauna of gracillariidae leaf-mining moths in the neotropics. Mol. Ecol. Resour. 2014, 14, 286–296. [Google Scholar] [CrossRef]
- Kirichenko, N.; Triberti, P.; Akulov, E.; Ponomarenko, M.; Gorokhova, S.; Sheiko, V.; Ohshima, I.; Lopez-Vaamonde, C. Exploring species diversity and host plant associations of leaf-mining micromoths (Lepidoptera: Gracillariidae) in the Russian far east using DNA barcoding. Zootaxa 2019, 4652, 1–55. [Google Scholar] [CrossRef]
- Hausmann, A.; Scalercio, S. Host-plant relationships of 29 mediterranean lepidoptera species in forested ecosystems unveiled by DNA Barcoding (Insecta: Lepidoptera). SHILAP. Revta. Lepid. 2016, 44, 463–471. [Google Scholar]
- Hausmann, A.; Diller, J.; Moriniere, J.; Höcherl, A.; Floren, A.; Haszprunar, G. DNA barcoding of fogged caterpillars in peru: A novel approach for unveiling host-plant relationships of tropical moths (Insecta, Lepidoptera). PLoS ONE 2020, 15, e0224188. [Google Scholar] [CrossRef] [Green Version]
- Brito, R.; Gonçalves, G.L.; Vargas, H.A.; Moreira, G.R.P. A new brazilian passiflora leafminer: Spinivalva gaucha, gen. n., sp. n. (Lepidoptera, Gracillariidae, Gracillariinae), the first gracillariid without a sap-feeding instar. ZooKeys 2013, 291, 1–26. [Google Scholar] [CrossRef]
- Vargas-Ortiz, M.; Bobadilla, D.; Huanca-Mamani, W.; Vargas, H.A. Genetic divergence of isolated populations of the native micromoth bucculatrix mirnae (Lepidoptera: Bucculatricidae) in the arid environments of northern chile. Mitochondrial DNA Part A 2018, 29, 1139–1147. [Google Scholar] [CrossRef]
- Luo, A.; Ling, C.; Ho, S.Y.W.; Zhu, C.D. Comparison of methods for molecular species delimitation across a range of speciation scenarios. Syst. Biol. 2018, 67, 830–846. [Google Scholar] [CrossRef] [Green Version]
- Puillandre, N.; Lambert, A.; Brouillet, S.; Achaz, G. ABGD, automatic barcode gap discovery for primary species delimitation. Mol. Ecol. 2012, 21, 1864–1877. [Google Scholar] [CrossRef] [PubMed]
- Pons, J.; Barraclough, T.G.; Gomez-Zurita, J.; Cardoso, A.; Duran, D.P.; Hazell, S.; Vogler, A.P. Sequence-based species delimitation for the DNA taxonomy of undescribed insects. Syst. Biol. 2006, 55, 595–609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujisawa, T.; Barraclough, T.G. Delimiting species using single-locus data and the generalized mixed yule coalescent approach: A revised method and evaluation on simulated data sets. Syst. Biol. 2013, 62, 707–724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Kapli, P.; Pavlidis, P.; Stamatakis, A. A general species delimitation method with applications to phylogenetic placements. Bioinformatics 2013, 29, 2869–2876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dellicour, S.; Flot, J.F. Delimiting species-poor data sets using single molecular markers: A study of barcode gaps, haplowebs and GMYC. Syst. Biol. 2015, 64, 900–908. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dellicour, S.; Flot, J.F. The hitchhiker’s guide to single-locus species delimitation. Mol. Ecol. Resour. 2018, 18, 1234–1246. [Google Scholar] [CrossRef]
- Carstens, B.C.; Pelletier, T.A.; Reid, N.M.; Satler, J.D. How to fail at species delimitation. Mol. Ecol. 2013, 22, 4369–4383. [Google Scholar] [CrossRef]
- Huanca-Mamani, W.; Rivera-Cabello, D.; Maita-Maita, J. A simple, fast, and inexpensive CTAB-PVP-Silica based method for genomic DNA isolation from single, small insect larvae and pupae. Genet. Mol. Res. 2015, 14, 7990–8000. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Penton, E.H.; Burns, J.M.; Janzen, D.H.; Hallwachs, W. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly astraptes fulgerator. Proc. Natl. Acad. Sci. USA 2004, 101, 14812–14817. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [PubMed]
- McDonald, J.H.; Kreitman, M. Adaptive protein evolution at the Adh locus in Drosophila. Nature 1991, 351, 652–654. [Google Scholar] [CrossRef]
- Xia, X.; Xie, Z.; Salemi, M.; Chen, L.; Wang, Y. An index of substitution saturation and its application. Mol. Phylogenet. Evol. 2003, 26, 1–7. [Google Scholar] [CrossRef]
- Xia, X. DAMBE7: New and improved tools for data analysis in molecular biology and evolution. Mol. Biol. Evol. 2018, 35, 1550–1552. [Google Scholar] [CrossRef] [Green Version]
- Guillot, G.; Mortier, F.; Estoup, A. GENELAND: A computer package for landscape genetics. Mol. Ecol. Notes 2005, 5, 712–715. [Google Scholar] [CrossRef]
- Guillot, G.; Renaud, S.; Ledevin, R.; Michaux, J.; Claude, J. A unifying model for the analysis of phenotypic, genetic, and geographic data. Syst. Biol. 2012, 61, 897–911. [Google Scholar] [CrossRef] [Green Version]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Pagel, M.; Meade, A. A phylogenetic mixture model for detecting pattern-heterogeneity in gene sequence or character-state data. Syst. Biol. 2004, 53, 571–581. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, A.Y.; Plotkin, D.; Ohshima, I.; Lopez-Vaamonde, C.; Houlihan, P.; Breinholt, J.W.; Kawakita, A.; Xiao, L.; Regier, J.C.; Davis, D.R.; et al. A molecular phylogeny and revised higher-level classification for the leaf-mining moth family Gracillariidae and its implications for larval host use evolution. Syst. Entomol. 2017, 42, 60–81. [Google Scholar] [CrossRef]
- Meade, A.; Pagel, M. BayesTrees, Version 1.3. 2011. Available online: http://www.evolution.reading.ac.uk/BayesTrees.html/ (accessed on 20 June 2020).
- Reid, N.M.; Carstens, B.C. Phylogenetic estimation error can decrease the accuracy of species delimitation: A Bayesian implementation of the general mixed Yule-coalescent model. BMC Evol. Biol. 2012, 12, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hickerson, M.J.; Meyer, C.P.; Moritz, C. DNA barcoding will often fail to discover new animal species over broad parameter space. Syst. Biol. 2006, 55, 729–739. [Google Scholar] [CrossRef] [PubMed]
- Wilson, J.J. Assessing the value of DNA barcodes for molecular phylogenetics: Effect of increased taxon sampling in lepidoptera. PLoS ONE 2011, 6, e24769. [Google Scholar] [CrossRef] [Green Version]
- Bouckaert, R.; Vaughan, T.G.; Barido-Sottani, J.; Duchêne, S.; Fourment, M.; Gavryushkina, A.; Heled, J.; Jones, G.; Kühnert, D.; De Maio, N.; et al. BEAST 2.5: An advanced software platform for bayesian evolutionary analysis. PLoS Comput. Biol. 2019, 15, e1006650. [Google Scholar] [CrossRef] [Green Version]
- Brower, A.V.Z. Rapid morphological radiation and convergence among races of the butterfly Heliconius erato inferred from patterns of mitochondrial DNA evolution. Proc. Natl. Acad. Sci. USA 1994, 91, 6491–6495. [Google Scholar] [CrossRef] [Green Version]
- Nazari, V.; Sperling, F.A.H. Mitochondrial DNA divergence and phylogeography in western palaearctic parnassiinae (Lepidoptera: Papilionidae): How many species are there? Insect Syst. Evol. 2007, 38, 121–138. [Google Scholar] [CrossRef]
- Talavera, G.; Dincǎ, V.; Vila, R. Factors affecting species delimitations with the GMYC model: Insights from a butterfly survey. Methods Ecol. Evol. 2013, 4, 1101–1110. [Google Scholar] [CrossRef] [Green Version]
- Ahrens, D.; Fujisawa, T.; Krammer, H.J.; Eberle, J.; Fabrizi, S.; Vogler, A.P. Rarity and incomplete sampling in DNA-based species delimitation. Syst. Biol. 2016, 65, 478–494. [Google Scholar] [CrossRef] [Green Version]
- Kozlov, A.M.; Darriba, D.; Flouri, T.; Morel, B.; Stamatakis, A. RAxML-NG: A fast scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 2019, 35, 4453–4455. [Google Scholar] [CrossRef] [Green Version]
- Brito, R.; De Prins, J.; De Prins, W.; Mielke, O.H.H.; Gonçalves, G.L.; Moreira, G.R.P. Extant diversity and estimated number of gracillariidae (Lepidoptera) species yet to be discovered in the neotropical region. Rev. Bras. Entomol. 2016, 60, 275–283. [Google Scholar] [CrossRef] [Green Version]
- De Prins, J.; Arévalo-Maldonado, H.A.; Davis, D.R.; Landry, B.; Vargas, H.A.; Davis, M.M.; Brito, R.; Fochezato, J.; Ohshima, I.; Moreira, G.R.P. An illustrated catalogue of the neotropical gracillariidae (Lepidoptera) with new data on primary types. Zootaxa 2019, 4575, 1–110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawakita, A.; Sato, A.A.W.; Salazar, J.R.L.; Kato, M. Leafflower–leafflower moth mutualism in the neotropics: Successful transoceanic dispersal from the old world to the new world by actively pollinating leafflower moths. PLoS ONE 2019, 14, e0210727. [Google Scholar] [CrossRef] [PubMed]
- Davis, D.R.; Farfán, J.; Cerdeña, J.; Huanca-Mamani, W.; Vargas, H.A.; Vargas-Ortiz, M.; Gonçalves, G.L.; Moreira, G.R.P. Adenogasteria leguminivora Davis & Vargas gen. et sp. nov. (Lepidoptera: Gracillariidae): A new seed-feeding micromoth associated with fabaceae in Peru and Chile. Austral Entomol. 2020, 59, 37–51. [Google Scholar] [CrossRef]
- Huemer, P.; Hebert, P.D.N.; Mutanen, M.; Wieser, C.; Wiesmair, B.; Hausmann, A.; Yakovlev, R.; Möst, M.; Gottsberger, B.; Strutzenberger, P.; et al. Large geographic distance versus small DNA barcode divergence: Insights from a comparison of European to South Siberian Lepidoptera. PLoS ONE 2018, 13, e0206668. [Google Scholar] [CrossRef] [PubMed]
- Valade, R.; Kenis, M.; Hernandez-Lopez, A.; Augustin, S.; Mari Mena, N.; Magnoux, E.; Rougerie, R.; Lakatos, F.; Roques, A.; Lopez-Vaamonde, C. Mitochondrial and microsatellite DNA markers reveal a balkan origin for the highly invasive horse-chestnut leaf miner Cameraria ohridella (Lepidoptera, Gracillariidae). Mol. Ecol. 2009, 18, 3458–3470. [Google Scholar] [CrossRef] [PubMed]
- Tóth, V.; Lakatos, F. Phylogeographic pattern of the plane leaf miner, Phyllonorycter platani (Staudinger, 1870) (Lepidoptera: Gracillariidae) in Europe. BMC Evol. Biol. 2018, 18, 135. [Google Scholar] [CrossRef]
- Vargas-Ortiz, M.; Gonçalves, G.L.; Huanca-Mamani, W.; Vargas, H.A.; Moreira, G.R.P. Description, natural history and genetic variation of Caloptilia guacanivora sp. nov. vargas-ortiz & vargas (Lepidoptera: Gracillariidae) in the atacama desert, Chile. Austral Entomol. 2019, 58, 171–191. [Google Scholar] [CrossRef] [Green Version]
- Escobar-Suárez, S.; Huanca-Mamani, W.; Vargas, H.A. Genetic divergence of a newly documented population of the cecidogenous micromoth Eugnosta azapaensis vargas & moreira (Lepidoptera: Tortricidae) in the atacama desert. Rev. Bras. Entomol. 2017, 61, 266–270. [Google Scholar] [CrossRef]
- Espinoza-Donoso, S.; Bobadilla, D.; Huanca-Mamani, W.; Vargas-Ortiz, M.; Vargas, H.A. A new species of ithome chambers (Lepidoptera, Cosmopterigidae, Chrysopeleiinae) from the atacama desert revealed by morphology and DNA barcodes. ZooKeys 2020, 912, 125–138. [Google Scholar] [CrossRef]
- Clarke, J.F.G. Two new cryptophlebia walsingham from Chile (Lepidoptera: Tortricidae). Acta Entomol. Chil. 1987, 14, 7–12. [Google Scholar]
- Vargas, H.A. A new species of Alucita L. (Lepidoptera: Alucitidae) from northern Chile. Neotrop. Entomol. 2011, 40, 85–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farfán, J.; Cerdeña, J.; Arivilca, M.; Condori-Mamani, M.; Huanca-Mamani, W.; Vargas, H.A. First record of Alucita danunciae (Lepidoptera: Alucitidae) in Peru. Studies Neotrop. Fauna Environ. 2020, 55, 103–108. [Google Scholar] [CrossRef]
- Pereira, C.M.; Silva, D.S.; Gonçalves, G.L.; Vargas, H.A.; Moreira, G.R.P. A new species of Leurocephala davis & mckay (Lepidoptera, Gracillariidae) from the azapa valley, northern chilean atacama desert, with notes on life history. Rev. Bras. Entomol. 2017, 61, 6–15. [Google Scholar] [CrossRef] [Green Version]
- Brito, R.; Mielke, O.H.H.; Gonçalves, G.L.; Moreira, G.R.P. Description of three new species of Phyllocnistis Zeller, 1848 (Lepidoptera: Gracillariidae), from the Atlantic forest, south Brazil, with notes on natural history and phylogeny. Austral Entomol. 2019, 58, 27–51. [Google Scholar] [CrossRef] [Green Version]



| Species | BOLD Accession | GenBank Accession | n | Locality |
|---|---|---|---|---|
| Phyllocnistis citrella | GBGL7447-11 | AB614514 | 1 | Japan |
| Eumetriochroa sp. | WOGRA322-15 | 1 | Taiwan | |
| Eumetriochroa hederae | GRPAL933-12 | KF367707 | 1 | Japan |
| Metriochroa latifoliella | GRPAL127-11 | KX046527 | 1 | France |
| Metriochroa latifoliella | GRACI588-09 | HM392532 | 1 | Croatia |
| Angelabella tecomae H1 | GBGL18153-15 | KM983591 | 4 | Peru (Tacna) Chile (Arica) |
| Angelabella tecomae H2 | GBGL18152-15 | KM983592 | 9 | Chile (Arica) |
| Angelabella tecomae H3 | GBGL18147-15 | KM983597 | 8 | Peru (Tacna) Chile (Arica) |
| Angelabella tecomae H4 | MT804725 | 1 | Peru (Tacna) | |
| Angelabella tecomae H5 | MT804726 | 1 | Peru (Tacna) | |
| Angelabella tecomae H6 | MT804727 | 2 | Peru (Tacna) | |
| Angelabella sp.2 H7 | MT804728 | 2 | Peru (Moquegua) | |
| Angelabella sp.2 H8 | MT804729 | 4 | Peru (Moquegua) | |
| Angelabella sp.2 H9 | MT804730 | 3 | Peru (Moquegua) | |
| Angelabella sp.2 H10 | MT804731 | 1 | Peru (Moquegua) | |
| Angelabella sp.3 H11 | MT804732 | 1 | Peru (Arequipa) | |
| Angelabella sp.3 H12 | MT804733 | 3 | Peru (Arequipa) | |
| Angelabella sp.3 H13 | MT804734 | 1 | Peru (Arequipa) | |
| Angelabella sp.1 H14 | MT804735 | 1 | Peru (Lima) | |
| Angelabella sp.1 H15 | MT804736 | 3 | Peru (Lima) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vargas-Ortiz, M.; Aliaga-Pichihua, G.; Lazo-Rivera, A.; Cerdeña, J.; Farfán, J.; Huanca-Mamani, W.; Vargas, H.A. Cryptic Diversity in the Monotypic Neotropical Micromoth Genus Angelabella (Lepidoptera: Gracillariidae) in the Peru-Chile Desert. Insects 2020, 11, 677. https://doi.org/10.3390/insects11100677
Vargas-Ortiz M, Aliaga-Pichihua G, Lazo-Rivera A, Cerdeña J, Farfán J, Huanca-Mamani W, Vargas HA. Cryptic Diversity in the Monotypic Neotropical Micromoth Genus Angelabella (Lepidoptera: Gracillariidae) in the Peru-Chile Desert. Insects. 2020; 11(10):677. https://doi.org/10.3390/insects11100677
Chicago/Turabian StyleVargas-Ortiz, Marcelo, Guido Aliaga-Pichihua, Ana Lazo-Rivera, José Cerdeña, Jackie Farfán, Wilson Huanca-Mamani, and Héctor A. Vargas. 2020. "Cryptic Diversity in the Monotypic Neotropical Micromoth Genus Angelabella (Lepidoptera: Gracillariidae) in the Peru-Chile Desert" Insects 11, no. 10: 677. https://doi.org/10.3390/insects11100677
APA StyleVargas-Ortiz, M., Aliaga-Pichihua, G., Lazo-Rivera, A., Cerdeña, J., Farfán, J., Huanca-Mamani, W., & Vargas, H. A. (2020). Cryptic Diversity in the Monotypic Neotropical Micromoth Genus Angelabella (Lepidoptera: Gracillariidae) in the Peru-Chile Desert. Insects, 11(10), 677. https://doi.org/10.3390/insects11100677





