Characterization of the Complete Mitochondrial Genomes of Two Species with Preliminary Investigation on Phylogenetic Status of Zyginellini (Hemiptera: Cicadellidae: Typhlocybinae)
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Preparation and DNA Extraction
2.2. Sequence Analysis and Gene Annotation
2.3. Phylogenetic Analysis
3. Results and Discussion
3.1. Genome Organization and Base Composition
3.2. Protein-Coding Genes (PCGs)
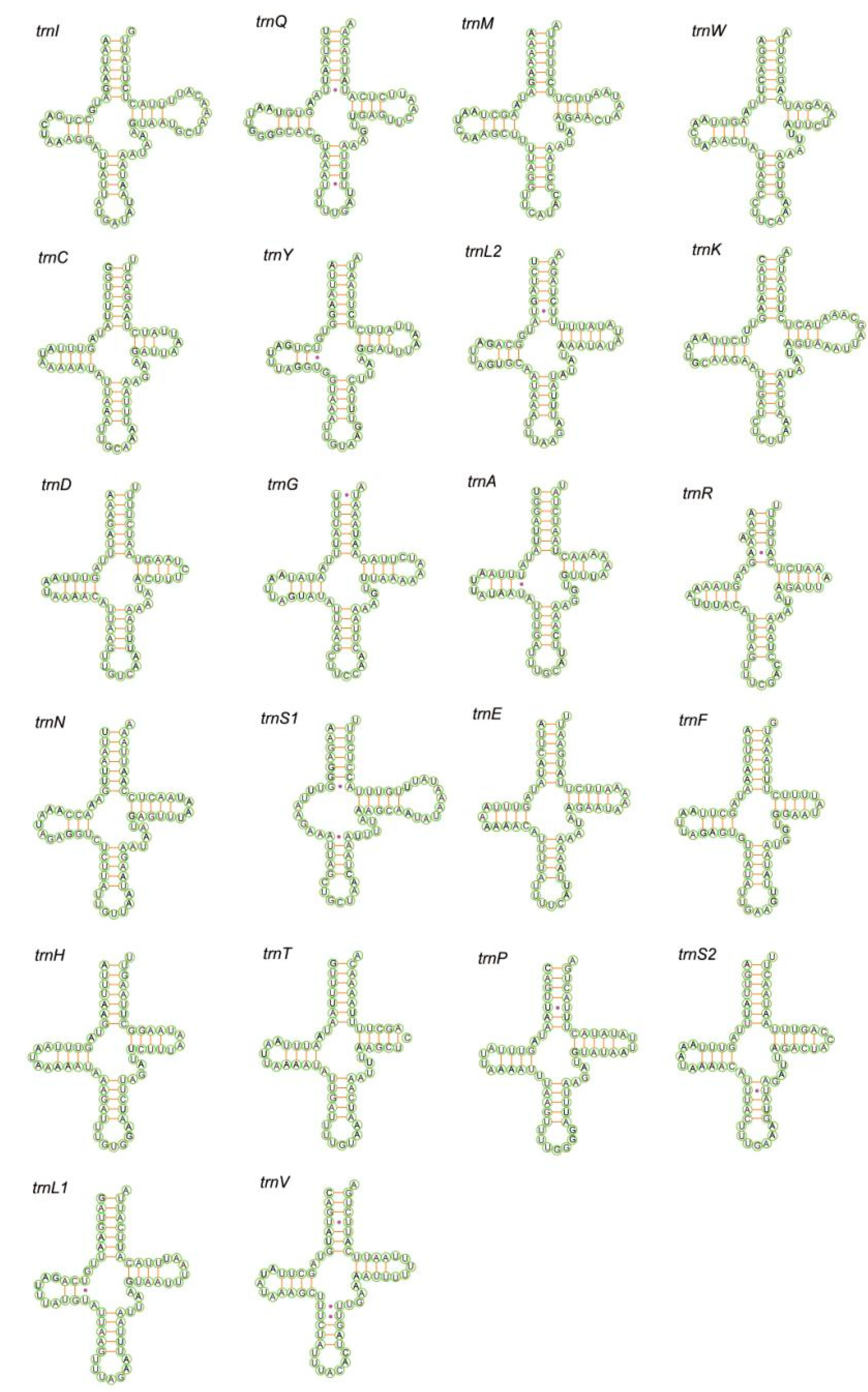
3.3. Transfer and Ribosomal RNA Genes
3.4. Gene Overlaps and Intergenic Spacers
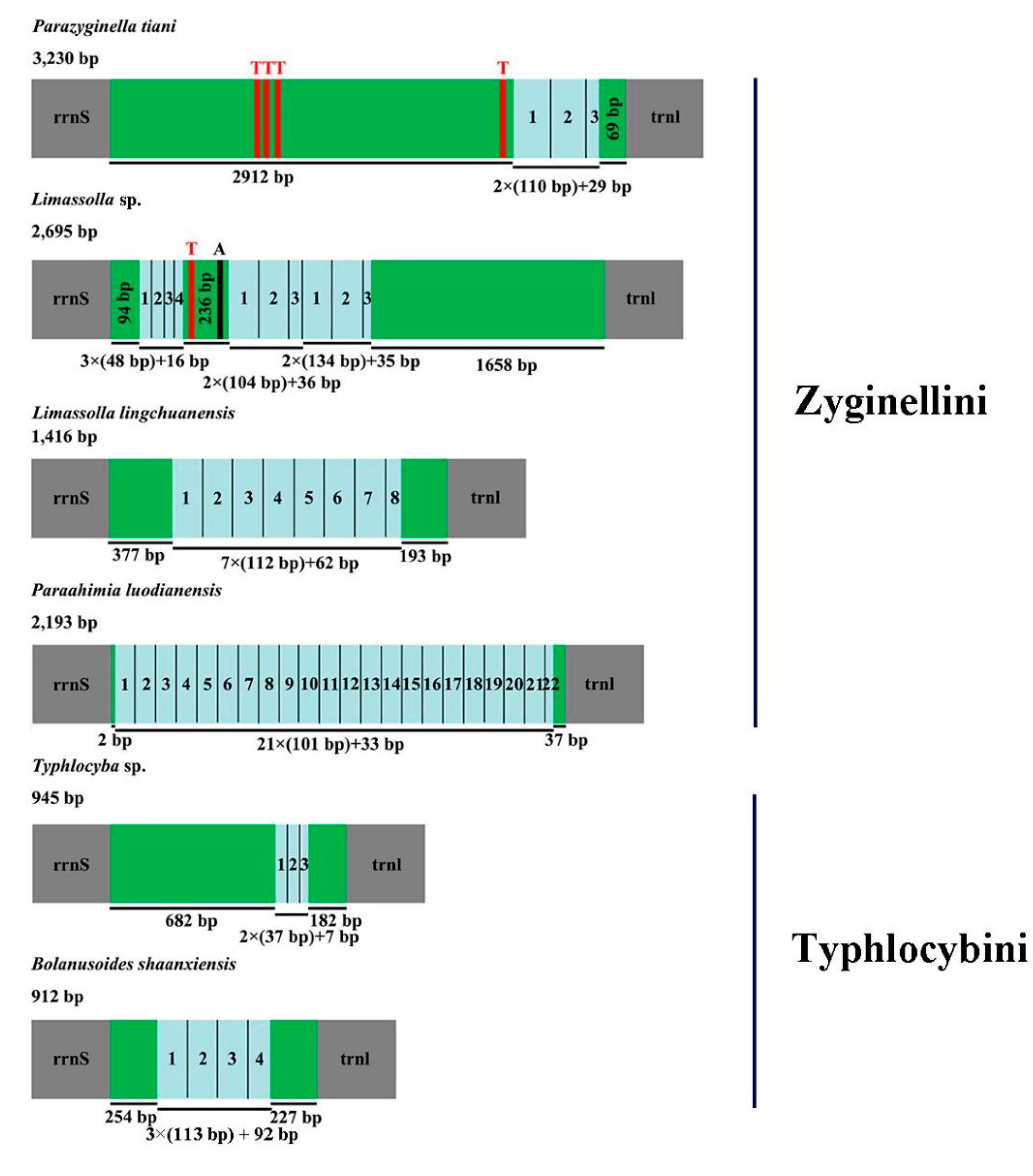
3.5. A + T-Rich Region
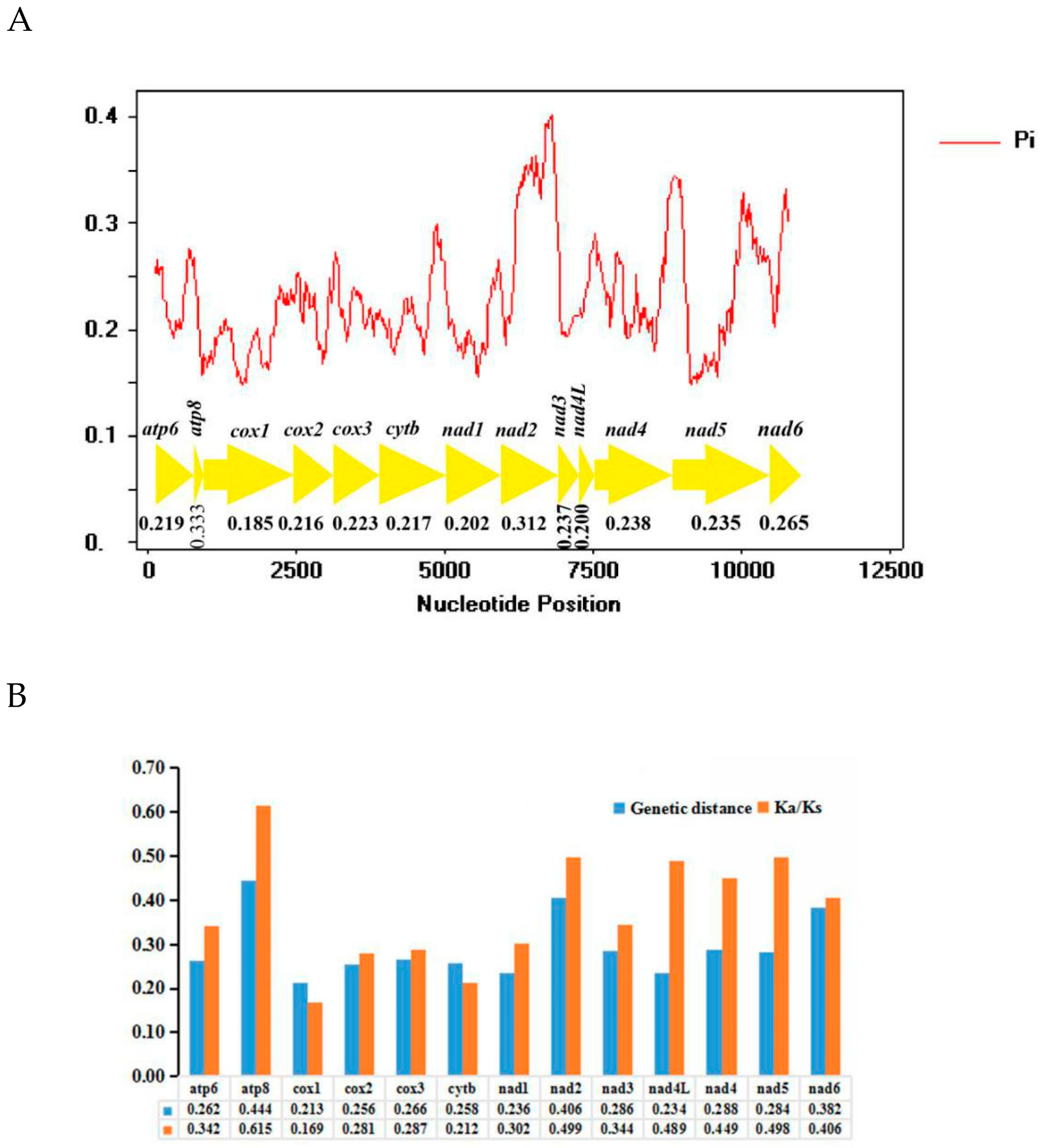
3.6. Nucleotide Diversity
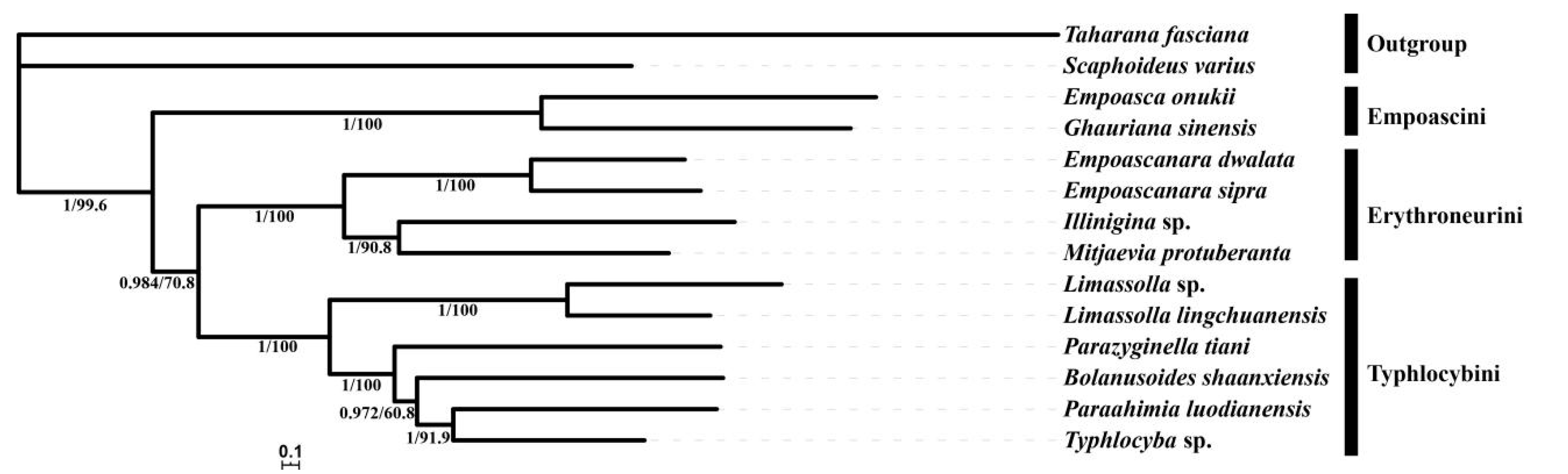
3.7. Phylogenetic Analyses
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- McKamey, S.H. Taxonomic catalogue of the Membracoidea (exclusive of leafhoppers); second supplement to Fascicle 1-Membracidae of the General Catalogue of the Hemiptera. Mem. Am. Entomol. Inst. 1998, 60, 1–377. [Google Scholar]
- McKamey, S.H. Leafhoppers of the World Database: Progress Report. In Proceedings of the 11th International Auchenorrhyncha Congress, Potsdam/Berlin, Germany, 5–9 August 2002; Museum für Naturkunde, Humboldt-Universität: Berlin, Germany, 2002; p. 85. [Google Scholar]
- Hamilton, K.G.A. New world species of Chlorita, Notus, and Forcipata (Rhynchota: Homoptera: Cicadellidae: Typhlocybinae) with a new tribe Forcipatini. Can. Entomol. 1998, 130, 491–507. [Google Scholar] [CrossRef]
- Balme, G.R. Phylogeny and Systematics of the Leafhopper Subfamily Typhlocybinae (Insecta: Hemiptera: Typhlocybinae). Ph.D. Thesis, North Carolina State University, Raleigh, NC, USA, 2007; pp. 1–149. [Google Scholar]
- Dietrich, C.H. South American leafhoppers of the tribe Typhlocybini (Hemiptera: Cicadellidae: Typhlocybinae). Zoologia 2013, 30, 519–568. [Google Scholar] [CrossRef]
- Cai, P.; Liang, L.J.; Wang, F. A new species of Limassolla, pest of Diospyros Kaki (Homoptera: Cicadelloidea: Typhlocybidae). J. Anhui Agric. Univ. 1992, 19, 324–326. [Google Scholar]
- Zhang, Y.L. A Taxonomic Study of Chinese Cicadellidae (Homiptera); Tianze Eldonejo: Yangling, China, 1990; pp. IV + 218. [Google Scholar]
- Xu, Y.; Wang, Y.R.; Dietrich, C.H.; Fletcher, M.J.; Qin, D.Z. Review of Chinese species of the leafhopper genus Amrasca Ghauri (Hemiptera, Cicadellidae, Typhlocybinae), with description of a new species, species checklist and notes on the identity of the Indian cotton leafhopper. Zootaxa 2017, 4353, 360–370. [Google Scholar] [CrossRef]
- Oman, P.W.; Knight, W.J.; Nielson, M.W. Leafhoppers (Cicadellidae): A Bibliography, Generic Check-List and Index to the World Literature 1956–1985; C.A.B. International Institute of Entomology: London, UK, 1990; pp. 1–368. [Google Scholar]
- Dworakowska, I. The leafhopper tribe Zyginellini (Homoptera, Auchenorrhyncha, Cicadellidae, Typhlocybinae). Rev. Zool. Afr. 1979, 93, 299–331. [Google Scholar]
- Dietrich, C.H. Phylogeny of the leafhopper subfamily Evacanthinae with a review of Neotropical species and notes on related groups (Hemiptera: Membracoidea: Cicadellidae). Syst. Entomol. 2004, 29, 455–487. [Google Scholar] [CrossRef]
- Dietrich, C.H.; Allen, J.M.; Lemmon, A.R.; Lemmon, E.M.; Takiya, D.M.; Evangelista, O.; Walden, K.K.; Grady, P.G.; Johnson, K.P.; Wiegmann, B. Anchored Hybrid Enrichment-Based Phylogenomics of Leafhoppers and Treehoppers (Hemiptera: Cicadomorpha: Membracoidea). Insect Syst. Divers. 2017, 1, 57–72. [Google Scholar] [CrossRef]
- Dietrich, C.H.; Rakitov, R.A.; Holmes, J.L.; Black, W.C. Phylogeny of the major lineages of Membracoidea (Insecta: Hemiptera: Cicadomorpha) based on 28S rDNA Sequences. Mol. Phylogenet. Evol. 2001, 18, 293–305. [Google Scholar] [CrossRef]
- Distant, W.L. Rhynchota—Homoptera. In The Fauna of British India including Ceylon and Burma; Secretary of State for India in Council: London, UK, 1908; Volume 4, p. 501. [Google Scholar]
- Boore, J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef]
- Cameron, S.L. Insect mitochondrial genomics: Implications for evolution and phylogeny. Annu Rev. Entomol. 2014, 59, 95–117. [Google Scholar] [CrossRef] [PubMed]
- Wolstenholme, D.R. Geneticnovelties inmitochondrial genomes of multicellular animals. Curr. Opin. Genet. Dev. 1992, 2, 918–925. [Google Scholar] [CrossRef]
- Kim, M.J.; Kang, A.R.; Jeong, H.C.; Kim, K.G.; Kim, I. Reconstructing intraordinal relationships in Lepidoptera using mitochondrial genome data with the description of two newly sequenced Lycaenids, Spindasis takanonis and Protantigius superans (Lepidoptera: Lycaenidae). Mol. Phylogenet. Evol. 2011, 61, 436–445. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Penton, E.H.; Burns, J.M.; Janzen, D.H.; Hallwachs, W. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proc. Natl. Acad. Sci. India Sect. A 2004, 101, 14812–14817. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.P.; Song, W.; Shi, S.L.; Liu, Y.Q.; Pan, M.H.; Dai, F.Y.; Lu, C.; Xiang, Z.H. Mitochondrial genome nucleotide substitution pattern between domesticated silkmoth, Bombyx mori, and its wild ancestors, Chinese Bombyx mandarina and Japanese Bombyx mandarina. Genet. Mol. Biol. 2010, 33, 186–189. [Google Scholar] [CrossRef] [PubMed]
- Simon, C.; Hadrys, H. A comparative analysis of complete mitochondrial genomes among Hexapoda. Mol. Phylogenet. Evol. 2013, 69, 393–403. [Google Scholar] [CrossRef]
- Simon, C.; Buckley, T.R.; Frati, F.; Stewart, J.B.; Beckenbach, A.T. Incorporating molecular evolution into phylogenetic analysis, and a new compilation of conserved polymerase chain reaction primers for animal mitochondrial DNA. Annu. Rev. Ecol. Evol. Syst. 2006, 37, 545–579. [Google Scholar] [CrossRef]
- Saccone, C.; Giorgi, C.D.; Gissi, C.; Pesole, G.; Reyes, A. Evolutionary genomics in Metazoa: The mitochondrial DNA as a model system. Gene 1999, 238, 195–209. [Google Scholar] [CrossRef]
- Yuan, X.W.; Xiong, K.N.; Li, C.; Song, Y.H. The complete mitochondrial genome of Limassolla lingchuanensis (Hemiptera: Cicadellidae: Typhlocybinae). Mitochondrial DNA Part B 2020, 5, 229–230. [Google Scholar] [CrossRef]
- Song, Y.H.; Yuan, X.W.; Li, C. The mitochondrial genome of Paraahimia luodianensis (Hemiptera: Cicadellidae: Typhlocybinae), a new genus and species from China. Mitochondrial DNA Part B 2020, 5, 1351–1352. [Google Scholar] [CrossRef]
- Tan, C.; Chen, X.X.; Li, C.; Song, Y.H. The complete mitochondrial genome of Empoascanara sipra (Hemiptera: Cicadellidae: Typhlocybinae) with phylogenetic consideration. Mitochondrial DNA Part B 2020, 5, 260–261. [Google Scholar] [CrossRef]
- Song, N.; Cai, W.; Li, H. Deep-level phylogeny of Cicadomorpha inferred from mitochondrial genomes sequenced by NGS. Sci. Rep. 2017, 7, 10429. [Google Scholar] [CrossRef] [PubMed]
- Shi, R.; Yu, X.F.; Yang, M.F. Complete mitochondrial genome of Ghauriana sinensis (Hemiptera: Cicadellidae: Typhlocybinae). Mitochondrial DNA Part B 2020, 5, 1367–1368. [Google Scholar] [CrossRef]
- Zhou, N.N.; Wang, M.X.; Cui, L.; Chen, X.X.; Han, B.Y. Complete mitochondrial genome of Empoasca vitis (Hemiptera: Cicadellidae). Mitochondrial DNA Part A DNA Mapp. Seq. Anal. 2016, 27, 1052–1053. [Google Scholar]
- Yuan, X.; Li, C.; Song, Y.H. Characterization of the complete mitochondrial genome of Mitjaevia protuberanta (Hemiptera: Cicadellidae: Typhlocybinae). Mitochondrial DNA Part B 2020, 5, 601–602. [Google Scholar] [CrossRef]
- Luo, X.; Chen, Y.; Chen, C.; Pu, D.Q.; Tang, X.B.; Zhang, H.; Lu, D.H.; Mao, J.H. Characterization of the complete mitochondrial genome of Empoasca sp. (Cicadellidae: Hemiptera). Mitochondrial DNA Part B 2019, 4, 1477–1478. [Google Scholar] [CrossRef]
- Liu, J.H.; Sun, C.Y.; Long, J.; Guo, J.J. Complete mitogenome of tea green leafhopper, Empoasca onukii (Hemiptera: Cicadellidae) from Anshun, Guizhou Province in China. Mitochondrial DNA Part B 2017, 2, 808–809. [Google Scholar] [CrossRef]
- Chen, X.X.; Yuan, Z.W.; Li, C.; Song, Y.H. Complete mitochondrial genome sequence of Empoascanara dwalata (Hemiptera: Cicadellidae: Typhlocybinae). Mitochondrial DNA Part B 2020, 5, 2260–2261. [Google Scholar] [CrossRef]
- Yang, X.; Yuan, Z.W.; Li, C.; Song, Y.H. Complete mitochondrial genome of Eupteryx (stacla) minusula (Hemiptera: Cicadellidae: Typhlocybinae) from China. Mitochondrial DNA Part B 2020, 5, 2375–2376. [Google Scholar] [CrossRef]
- Jiang, J.; Yuan, X.; Yuan, Z.; Song, Y.H. The complete mitochondrial genome of Parathailocyba orla (Hemiptera: Cicadellidae: Typhlocybinae). Mitochondrial DNA Part B 2020, 5, 1981–1982. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C. Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Grant, J.R.; Stothard, P. The CG View Server: A comparative genomics tool for circular genomes. Nucleic Acids Res. 2008, 36, 181–184. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gao, F.L.; Li, W.X.; Jakovlić, I.; Zou, H.; Zhang, J.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. bioRxiv 2018, 20, 489088. [Google Scholar] [CrossRef] [PubMed]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef] [PubMed]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870. [Google Scholar] [CrossRef]
- Du, Y.M.; Dai, W.; Dietrich, C.H. Mitochondrial genomic variation and phylogenetic relationships of three groups in the genus Scaphoideus (Hemiptera: Cicadellidae: Deltocephalinae). Sci. Rep. 2017, 7, 16908. [Google Scholar] [CrossRef]
- Wang, J.J.; Li, H.; Dai, R.H. Complete mitochondrial genome of Taharana fasciana (Insecta, Hemiptera: Cicadellidae) and comparison with other Cicadellidae insects. Genetica 2017, 145, 593–602. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements inperformance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef] [PubMed]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. Partition Finder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2016, 34, 772–773. [Google Scholar]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2014, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Clary, D.O.; Wolstenholme, D.R. The mitochondrial DNA molecule of Drosophila yakuba nucleotide sequence, gene organization, and genetic code. J. Mol. Evol. 1985, 22, 252–271. [Google Scholar] [CrossRef]
- Ojala, D.; Montoya, J.; Attardi, G. tRNA punctuation model of RNA processing in human mitochondria. Nature 1981, 290, 470. [Google Scholar] [CrossRef]
- Li, H.; Leavengood, J.M.; Chapman, E.G.; Burkhardt, D.; Song, F.; Jiang, P.; Liu, J.P.; Zhou, X.G.; Cai, W.Z. Mitochondrial phylogenomics of Hemiptera reveals adaptive innovations driving the diversification of true bugs. Proc. Biol. Sci. 2017, 284, 20171223. [Google Scholar] [CrossRef]
- Zhang, K.J.; Zhu, W.C.; Rong, X.; Ding, X.L.; Zhang, Y.K.; Liu, J.; Chen, D.S.; Du, Y.; Hong, X.Y. The complete mitochondrial genomes of two rice planthoppers, Nilaparvata lugens and Laodelphax striatellus: Conserved genome rearrangement in Delphacidae and discovery of new characteristics of atp8 and tRNA genes. BMC Genom. 2013, 14, 417. [Google Scholar] [CrossRef]
- Guo, Z.L.; Yuan, M.L. Research progress of mitochondrial genomes of Hemiptera insects. Sci. Sin. Vitae 2016, 46, 151–166. [Google Scholar]
- Doublet, V.; Ubrig, E.; Alioua, A.; Bouchon, D.; Marcadé, I.; Maréchal-Drouard, L. Large gene overlaps and tRNA processing in the compact mitochondrial genome of the crustacean Armadillidium vulgare. RNA Biol. 2015, 12, 1159–1168. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.X.; Szymura, J.M.; Hewitt, G.M. Evolution and structural conservation of the control region of insect mitochondrial DNA. J. Mol. Evol. 1995, 40, 382–391. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Zheng, J.; He, Y.; Luo, C. Tandem repeat modification during double-strand break repair induced by an engineered TAL effector nuclease in zebrafish genome. PLoS ONE 2013, 8, e84176. [Google Scholar] [CrossRef] [PubMed]
- Levinson, G.; Gutman, G.A. Slipped-strand mispairing: A major mechanism for DNA sequence evolution. Mol. Biol. Evol. 1987, 4, 203–221. [Google Scholar]
- Hurst, L.D. The Ka/Ks ratio: Diagnosing the form of sequence evolution. Trends Genet. 2002, 18, 486–487. [Google Scholar] [CrossRef]
- Mori, S.; Matsunami, M.J.G. Signature of positive selection in mitochondrial DNA in Cetartiodactyla. Genes Genet. Syst. 2018, 93, 65–73. [Google Scholar] [CrossRef]
- Jia, W.Z.; Yan, H.B.; Guo, A.J.; Zhu, X.Q.; Wang, Y.C.; Shi, W.G.; Chen, H.T.; Zhan, F.; Zhang, S.H.; Fu, B.Q. Complete mitochondrial genomes of Taenia multiceps, T. hydatigena and T. pisiformis: Additional molecular markers for a tapeworm genus of human and animal health significance. BMC Genom. 2010, 11, 447. [Google Scholar] [CrossRef]
- Ma, L.Y.; Liu, F.F.; Chiba, H.; Yuan, X.Q. The mitochondrial genomes of three skippers: Insights into the evolution of the family Hesperiidae (Lepidoptera). Genomics 2019, 112, 422–441. [Google Scholar] [CrossRef]
- Brabec, J.; Kostadinova, A.; Scholz, T.; Littlewood, D.T.J. Complete mitochondrial genomes and nuclear ribosomal RNA operons of two species of Diplostomum (Platyhelminthes: Trematoda): A molecular resource for taxonomy and molecular epidemiology of important fish pathogens. Parasit. Vectors 2015, 8, 336. [Google Scholar] [CrossRef]
- Demari-Silva, B.; Foster, P.G.; Oliveira-de, T.M.P.; Bergo, E.S.; Sanabani, S.S.; Pessôa, R.; Sallum, M.A.M. Mitochondrial genomes and comparative analyses of Culex camposi, Culex coronator, Culex usquatus and Culex usquatissimus (Diptera: Culicidae), members of the Coronator group. BMC Genom. 2015, 16, 831. [Google Scholar] [CrossRef]
- Young, D.A.J. A reclassification of western Hemisphere Typhlocybinae (Homoptera: Cicadellidae). Univ. Kans. Sci. Bull. 1952, 35, 3–217. [Google Scholar] [CrossRef]
- Dworakowska, I. On some Typhlocybinae from Vietnam (Homoptera: Cicadellidae). Folia Entomol. Hung. 1977, 30, 9–47. [Google Scholar]
- Hamilton, K.G.A. Classification, morphology and phylogeny of the family Cicadellidae (Rhynchota: Homoptera). In Proceedings of the 1st International Workshop on Biotaxonomy, Classification and Biology of Leafhoppers and Planthoppers (Auchenorrhyncha) of Economic Importance, London, UK, 4–7 October 1983; pp. 15–37. [Google Scholar]
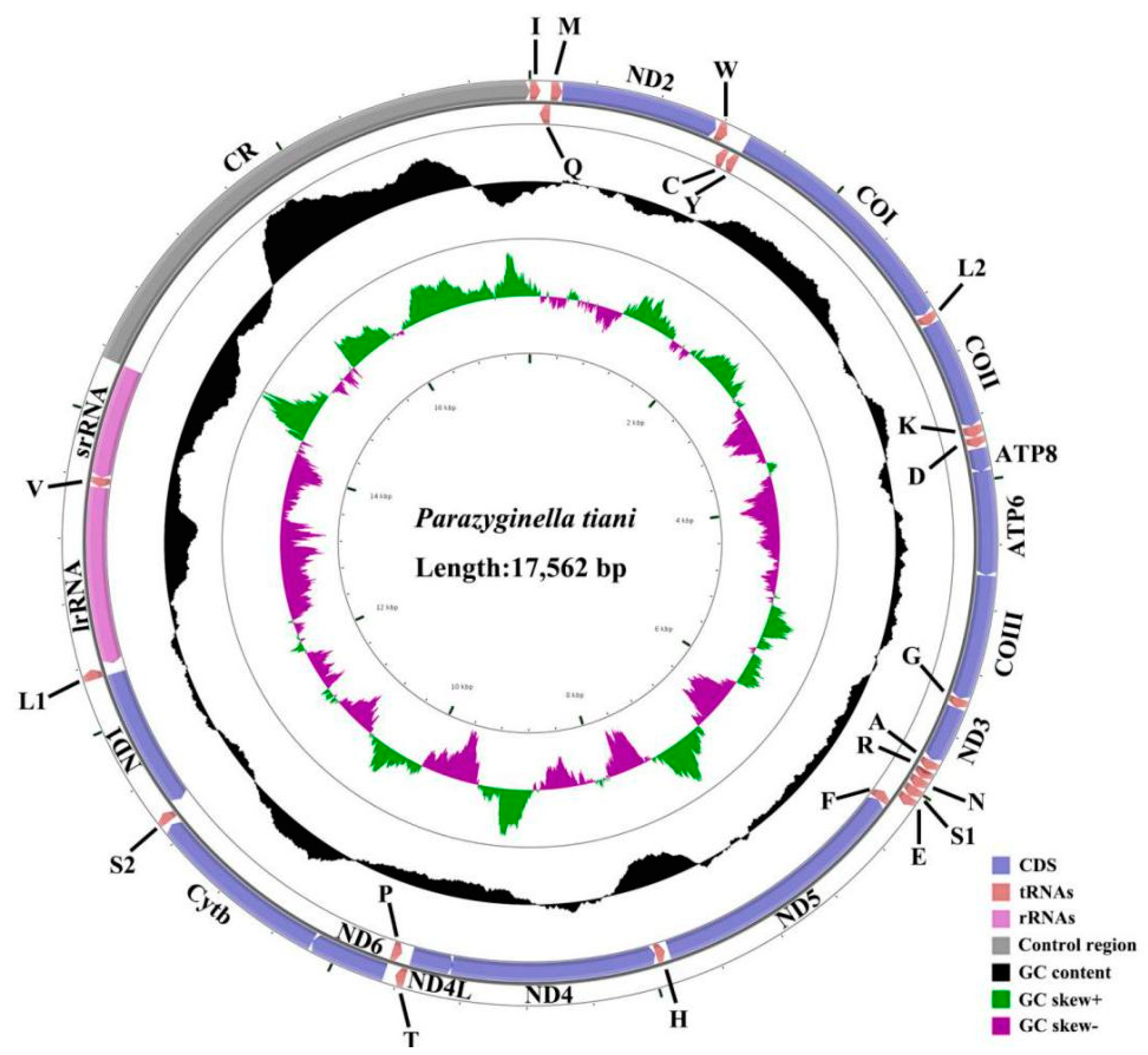
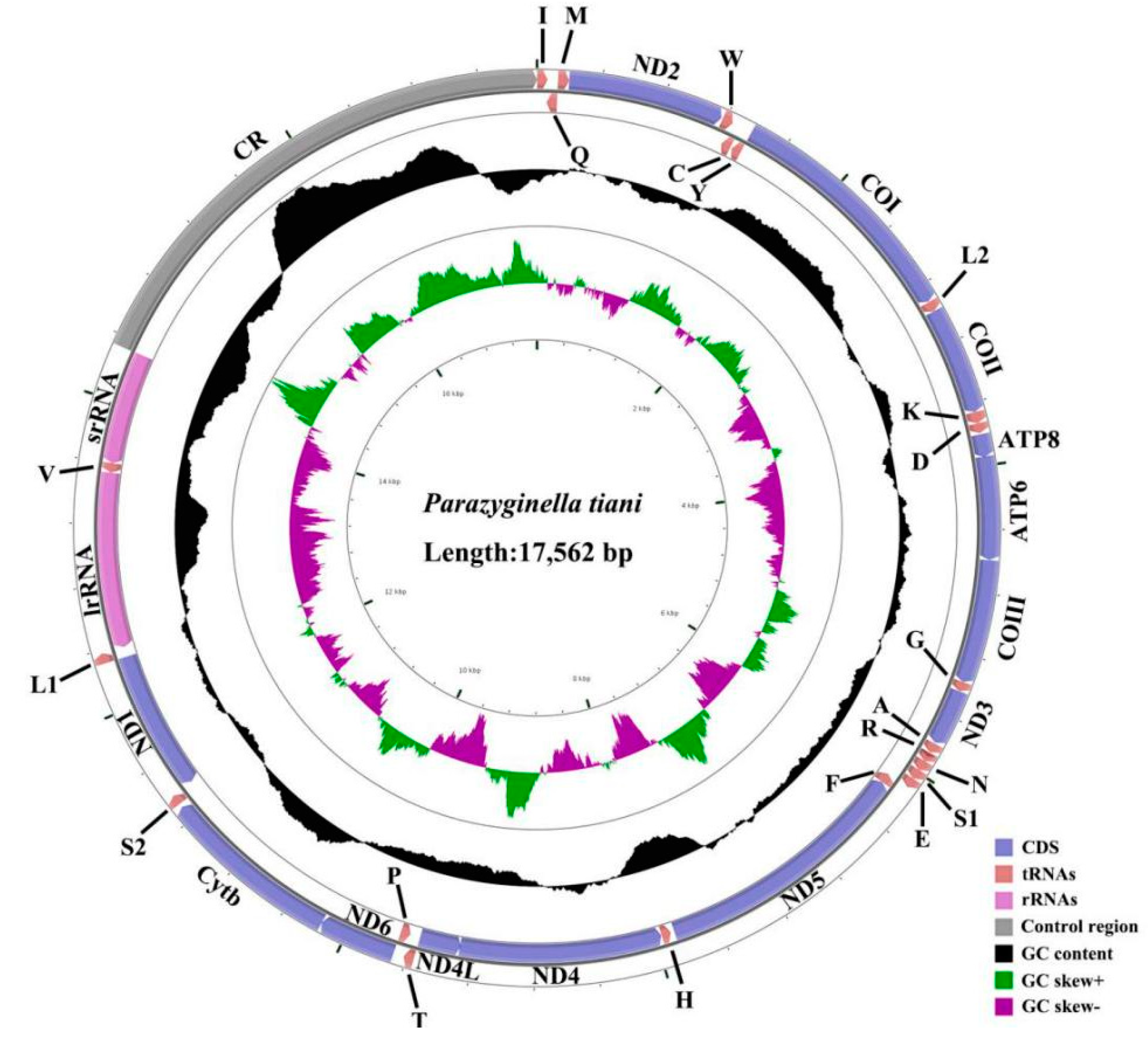
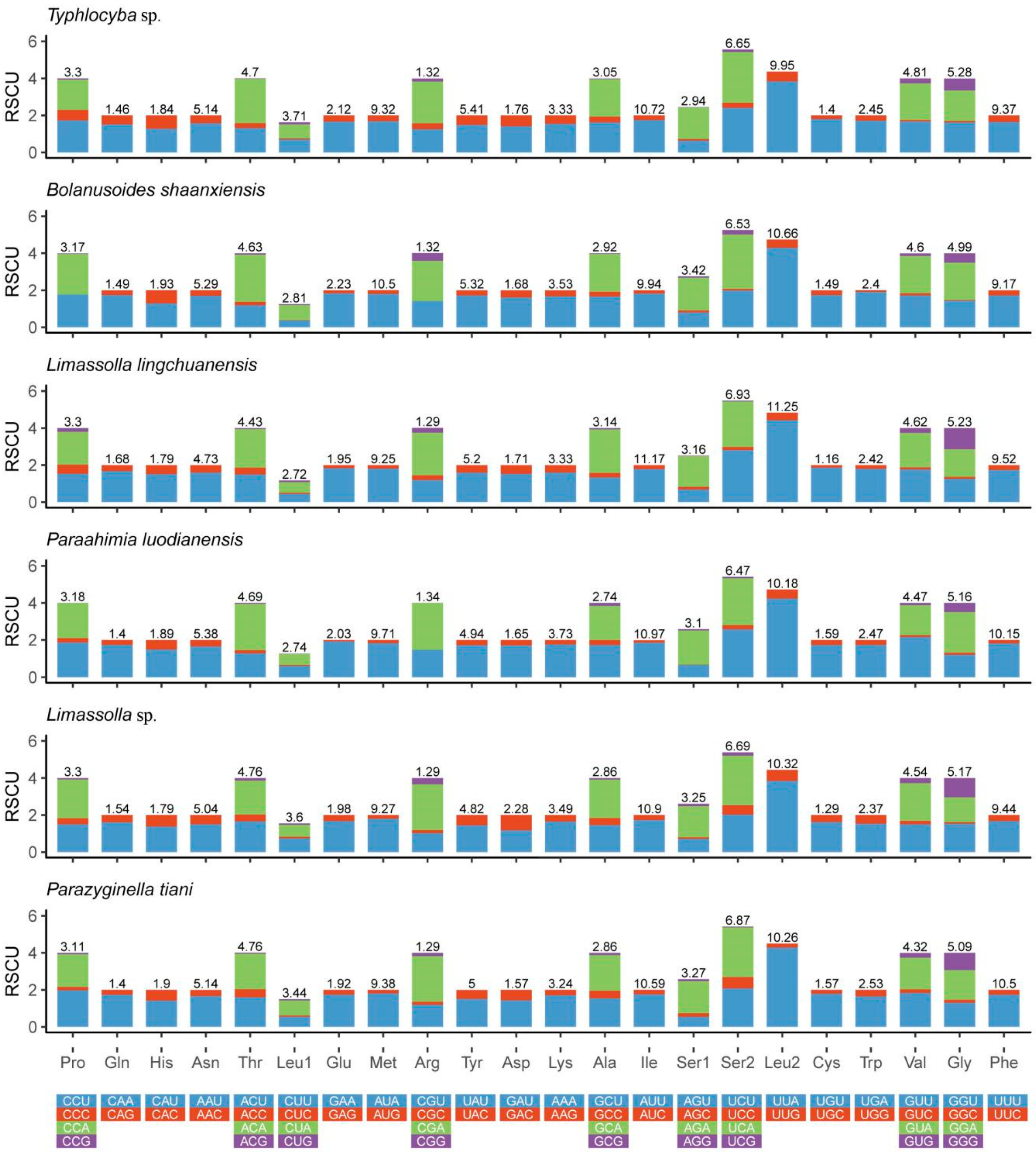
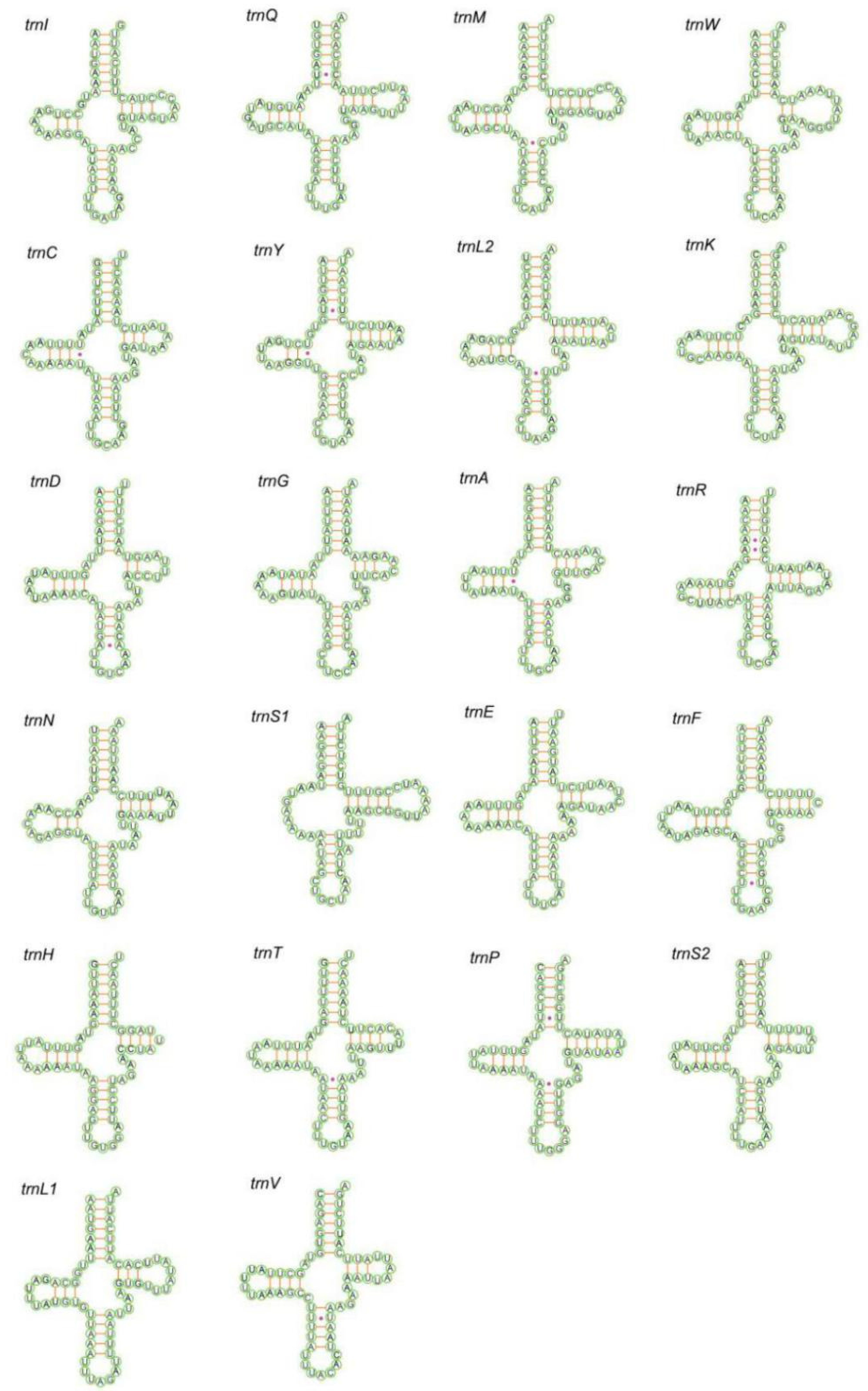
| Subfamily | Tribe | Species | Accession Number | Reference |
|---|---|---|---|---|
| Coelidiinae | Coelidiini | Taharana fasciana | NC_036015 | [44] |
| Deltocephalinae | Scaphoideini | Scaphoideus varius | KY817245 | [43] |
| Typhlocybinae | Empoascini | Ghauriana sinensis | MN699874 | [28] |
| Empoasca onukii | NC_037210 | [32] | ||
| Erythroneurini | Mitjaevia protuberanta | NC_047465 | [30] | |
| Illinigina sp. | KY039129 | [27] | ||
| Empoascanara dwalata | MT350235 | [33] | ||
| Empoascanara sipra | MN604278 | [26] | ||
| Typhlocybini | Typhlocyba sp. | KY039138 | [27] | |
| Bolanusoides shaanxiensis | MN661136 | Unpublished | ||
| Zyginellini | Limassolla lingchuanensis | MN605256 | [24] | |
| Paraahimia luodianensis | NC_047464 | [25] | ||
| Limassolla sp. | MT683892 | This study | ||
| Parazyginella tiani | MT683891 | This study |
| Regions | Size (bp) | T(U) | C | A | G | AT (%) | GC (%) | AT Skew | GC Skew |
|---|---|---|---|---|---|---|---|---|---|
| Full genome | 17,562/17,053 | 34.3/33.3 | 13.2/14.0 | 42.1/41.7 | 10.4/11.0 | 76.4/75.0 | 23.6/25.0 | 0.103/0.112 | −0.117/−0.119 |
| PCGs | 10,938/10,926 | 42.9/41.5 | 12.4/12.9 | 33.3/33.3 | 11.4/12.3 | 76.2/74.8 | 23.8/25.2 | −0.126/−0.110 | −0.041/−0.025 |
| 1st codon position | 3646/3642 | 36.9/35.1 | 13.4/11.5 | 36.3/36.6 | 15.7/16.8 | 73.2/71.7 | 26.8/28.3 | −0.008/0.022 | 0.172/0.187 |
| 2nd codon position | 3646/3642 | 48.4/48.0 | 14.4/17.6 | 20.4/21.1 | 13.7/13.3 | 68.8/69.1 | 31.3/30.9 | −0.407/−0.389 | −0.123/−0.137 |
| 3rd codon position | 3646/3642 | 43.4/41.4 | 15.4/9.7 | 43.3/42.1 | 4.8/6.7 | 86.7/83.5 | 13.3/16.4 | −0.002/0.008 | −0.278/−0.182 |
| tRNAs | 1422/1432 | 36.6/40.2 | 16.4/8.4 | 40.6/39.7 | 12.2/11.8 | 77.2/79.9 | 22.7/20.2 | 0.052/0.006 | 0.077/0.170 |
| rRNAs | 1892/1976 | 47.1/46.1 | 17.4/8.5 | 35.6/32.4 | 10.8/13.0 | 82.7/78.5 | 17.4/21.5 | −0.139/−0.173 | 0.244/0.209 |
| A + T rich-region | 3230/2695 | 38.6/35.0 | 18.4/14.7 | 34.0/36.3 | 13.4/14.0 | 72.6/71.3 | 27.4/28.7 | −0.063/0.019 | −0.020/−0.026 |
| Species | Whole Genome | AT Skew | GC Skew | PCGs | tRNAs | rRNAs | A + T-Rich Region | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Size (bp) | AT (%) | Size (bp) | AT (%) | Size (bp) | AT (%) | Size (bp) | AT (%) | Size (bp) | AT (%) | |||
| B. | 15,274 | 78.9 | 0.158 | −0.135 | 10,917 | 77.2 | 1469 | 79.7 | 1963 | 83.6 | 912 | 88.7 |
| T. | 15,223 | 77.1 | 0.138 | −0.138 | 10,950 | 75.2 | 1436 | 77.9 | 1866 | 82.3 | 945 | 85.9 |
| L1. | 15,716 | 78.8 | 0.096 | −0.098 | 10,932 | 76.3 | 1426 | 80.1 | 1911 | 83.1 | 1416 | 91.4 |
| P1. | 16,497 | 80.0 | 0.155 | −0.162 | 10,962 | 77.7 | 1427 | 79.0 | 1898 | 83.7 | 2193 | 88.4 |
| L2. | 17,053 | 75.0 | 0.112 | −0.119 | 10,926 | 74.8 | 1432 | 79.9 | 1976 | 78.5 | 2695 | 71.3 |
| P2. | 17,562 | 76.4 | 0.103 | −0.117 | 10,938 | 76.2 | 1422 | 77.2 | 1892 | 82.7 | 3230 | 72.6 |
| Gene | Start Codon/Stop Codon | |||||
|---|---|---|---|---|---|---|
| T. | B. | L1. | P1. | L2. | P2. | |
| nad2 | ATA/TAA | ATT/TAA | ATT/TAG | ATT/TAA | ATA/TAG | ATA/TAA |
| cox1 | ATG/TAA | ATG/TAA | ATG/T | ATG/T | ATG/T | ATG/TAA |
| cox2 | ATT/T | ATT/T | ATT/T | ATG/T | ATC/T | ATA/T |
| atp8 | TTG/TAA | TTG/TAA | TTG/TAA | TTG/TAA | ATC/TAA | ATA/TAA |
| atp6 | ATG/TAA | ATA/TAA | ATG/TAA | ATG/TAA | ATG/TAA | ATA/TAA |
| cox3 | ATG/TAA | ATG/T | ATG/TAA | ATG/T | ATG/T | ATG/T |
| nad3 | ATT/TAA | ATT/TAA | ATA/TAA | ATA/TAA | ATA/TAA | ATA/TAA |
| nad5 | ATG/T | TTG/T | ATA/TAA | ATG/T | ATT/TAA | ATT/T |
| nad4 | ATG/TAA | ATA/T | ATG/T | ATG/TAA | ATG/T | ATG/TAG |
| nad4L | ATG/TAA | ATG/TAA | ATG/TAA | ATG/TAA | ATG/TAA | ATG/TAA |
| nad6 | ATT/TAA | ATT/TAA | ATA/TAA | ATT/TAA | ATA/T | ATT/TAA |
| cytb | ATG/TAG | ATT/TAA | ATG/TAA | ATG/TAA | ATG/TAA | ATG/TAA |
| nad1 | ATT/TAA | ATT/TAA | ATT/TAA | ATT/TAA | ATT/TAA | ATA/T |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, X.; H. Dietrich, C.; Huang, M. Characterization of the Complete Mitochondrial Genomes of Two Species with Preliminary Investigation on Phylogenetic Status of Zyginellini (Hemiptera: Cicadellidae: Typhlocybinae). Insects 2020, 11, 684. https://doi.org/10.3390/insects11100684
Zhou X, H. Dietrich C, Huang M. Characterization of the Complete Mitochondrial Genomes of Two Species with Preliminary Investigation on Phylogenetic Status of Zyginellini (Hemiptera: Cicadellidae: Typhlocybinae). Insects. 2020; 11(10):684. https://doi.org/10.3390/insects11100684
Chicago/Turabian StyleZhou, Xian, Christopher H. Dietrich, and Min Huang. 2020. "Characterization of the Complete Mitochondrial Genomes of Two Species with Preliminary Investigation on Phylogenetic Status of Zyginellini (Hemiptera: Cicadellidae: Typhlocybinae)" Insects 11, no. 10: 684. https://doi.org/10.3390/insects11100684
APA StyleZhou, X., H. Dietrich, C., & Huang, M. (2020). Characterization of the Complete Mitochondrial Genomes of Two Species with Preliminary Investigation on Phylogenetic Status of Zyginellini (Hemiptera: Cicadellidae: Typhlocybinae). Insects, 11(10), 684. https://doi.org/10.3390/insects11100684




