Abstract
We analyzed the transcriptomes of Romalea microptera grasshoppers after 8 years of artificial selection for either long or short thoraces. Evolution proceeded rapidly during the experiment, with a 13.3% increase and a 32.2% decrease in mean pronotum lengths (sexes combined) in the up- and down-selected colonies, respectively, after only 11 generations. At least 16 additional traits also diverged between the two colonies during the selection experiment. Transcriptomic analysis identified 693 differentially expressed genes, with 386 upregulated and 307 downregulated (55.7% vs. 44.3%), including cellular process, metabolic process, binding, general function prediction only, and signal transduction mechanisms. Many of the differentially expressed genes (DEGs) are known to influence animal body size.
1. Introduction
Artificial selection is a valuable tool for testing and understanding natural selection, evolution, and the genetics underlying these processes [1,2,3]. Indeed, knowledge of artificial selection was foundational for the cognitive development of Darwin’s evolutionary theory [4], as shown by the fact that he devoted his first chapter in On the Origin of Species by Natural Selection to domestication and selective breeding [5]. Today, “artificial selection” generally refers to active selection (culling) by humans, whereas “experimental evolution” refers to experiments where humans do not actively select individuals for further breeding, but instead where populations are placed into specific environmental/experimental conditions selected by humans, and thereafter allowed to evolve on their own over successive generations [6,7]. Domestication is a type of long-term artificial selection. All three represent selection and evolution directed by humans.
The great value of artificial selection is that it represents applied evolution in real time. Given some background genetic variation in the experimental population, virtually any trait can be modified by artificial selection [8,9,10]. Like natural selection, artificial selection can alter allele frequencies in populations, resulting in altered, inherited phenotypes [11,12,13,14]. Artificial selection, domestication, and experimental evolution can produce substantial phenotypic change [15,16,17] and can be stunningly fast, with clear divergence sometimes in as little as three to five generations [18,19,20,21,22,23,24,25]. In fact, evolution by artificial selection can be instantaneous when an experimenter removes from a given population all individuals possessing a phenotype determined by a dominant allele. As such, artificial selection allows the rapid testing of evolutionary hypotheses and the elucidation of the processes and molecular mechanisms underlying evolution and trait expression.
Today, modern molecular biology allows us to quickly know the genetic changes resulting from artificial selection—genetic changes that alter phenotypes. When combined with molecular and other analysis, comprehensive artificial selection experiments can help reveal the entire multilevel, multisystem sequence of events that occurs during and as a consequence of evolution, from selection to changes in genomes → transcripts → proteins → metabolism → cellular physiology → system physiology → development, and on to morphology, life-history, and behavior [6,7]. Additional research can explore the fitness and ecological consequences of phenotypic changes produced by artificial selection. Such broad and deep understanding has been achieved in a number of model study organisms, most notably in Drosophila [26,27,28,29,30].
One surprising finding of artificial selection research is that focused selection on a single trait usually also alters numerous other traits, some of which were previously unknown and unexpected to be related to the focus-trait [2,6,10,24,25,31,32]. Examples of these are observed in silver foxes selected only for tameness (a behavioral trait), regarding the evolution of light fur color, spotted coats, droopy ears, short snouts, narrow skulls, shorter, curled tails, under- and over-bites, shorter legs, tail wagging, loss of musky smell, and larger litter size (mostly morphological traits) [33,34]. Such “side-effects” of artificial selection were recognized by Darwin [5] and can be caused by pleiotropy, linkage disequilibrium, genetic drift, developmental, phenotypic, and functional correlations, and (in long-running or high-population experiments) mutation [3,14].
In this paper, we compare transcriptomes of grasshoppers after laboratory selection for long vs. short pronotum length, in order to understand molecular mechanisms underlying evolution and genes and pathways that determine body size. Transcriptomic analysis following artificial selection allows us to link phenotypic change to gene expression, and, thus can help identify genes and pathways that contribute to selection-caused phenotypic changes. We hypothesize that many of these transcriptional differences represent genes active in constructing and directing divergent phenotypes resulting from up- vs. down-artificial selection on body size.
An unanticipated discovery from early transcriptomic analysis was the sometimes very large number of transcriptional changes that resulted from artificial selection, including many influencing unexpected proteins/systems/traits. Although some of these undoubtedly represent nonadaptive hitchhiking, genetic drift, and mutation, many represent evolved pleiotropy. As such, transcriptomic analysis has helped scientists to understand the polygenic nature of complex quantitative traits, discover new pleiotropic relationships, assign functions to previously unstudied or unannotated genes, understand the complex and extensive nature of pleiotropy, and to observe genome evolution and epigenetics in real time, thus further revealing how biological systems work [14,34,35].
The subject of our study, body size, is an excellent focus trait for artificial selection and evolutionary studies, because, for nearly all species, body size is highly variable, easily measured and selected, polygenic and correlated with numerous other traits, has high heritability, and is fundamental to nearly all other traits. Size influences physiology, ecology, life history, behavior, performance, fitness, and evolution [36,37]. This current paper is part of a long-term study of body size in Orthoptera in our laboratories [36,37,38,39,40,41,42,43,44,45,46,47,48].
2. Materials and Methods
2.1. Ethics Statement
Romalea microptera (Beauvois) were fed in the biological science laboratory in Illinois State University. R. microptera is an agricultural pest and is not on the ‘List of Protected Animals in America‘. No permits were required for the field studies.
2.2. The Insect
The eastern lubber grasshopper, R. microptera (Beauvois) (Orthoptera; Romaleidae) ranges across the southeastern USA [49,50]. It is a model study organism due to its large size (up to 7 cm long and 21 g), long life, calm demeanor, flightlessness, interesting biology, and ease of laboratory culture [51]. This species, and Orthoptera in general, exhibit considerable inter-and intrapopulational variation in body size in nature, and this variation has both genetic and environmental origins [37,41,43,46].
2.3. The Artificial Selection Experiment
We selected for long- vs. short-pronotum length for 8 years (11 generations) in two laboratory colonies of R. microptera that originated from 80 adults collected from the field at Shark Valley, Everglades National Park, Florida, USA in 2005 [43,44]. These wild insects were transported to Illinois State University and maintained and allowed to mate at will for one generation in a large communal cage [51], producing ~80 egg pods. The resulting hatchlings gave rise to ~230 adults. From this single population (referred to as the Parent Lab Population), we selected the 30 adults with the longest pronotum length and the 30 adults with the shortest pronotum length (15 of each sex/treatment) to initiate our artificial selection experiment. These animals formed Generation 1 of the Large Colony and the Small Colony, respectively. For the next 8 years we maintained these two colonies in separate cages, while always selecting the largest ~15% and smallest ~15% of individuals of each generation for further breeding. Throughout the experiment, we used Mitutoyo Inc. Model CD-6 digital calipers to measure the lengths of their thoraces along the dorsal mid-line. Each individual was measured by two different researchers. If the two measurements were not identical, the individual was remeasured until a consensus was found.
2.4. RNA Extraction and Sequencing
Molecular analysis was undertaken by the State Key Laboratory for Biology of Plant Diseases and Insect Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences. Total RNA was isolated from the whole body of five female adults of the Large Colony and from the same number and sex of Small Colony individuals with Trizol (Invitrogen) as per the manufacturer’s protocol. Environmental effects on transcription were minimized by rearing the two colonies side-by-side under identical conditions. Quality and quantity of RNA samples were determined with a Thermo Scientific NanoDropTM ND-1000 spectrophotometer (Implen, CA, USA). Total RNA was treated with RNase-free Dnase I (New England BioLabs, USA) to remove contaminants of residual DNA. The first complementary DNA (cDNA) strand was synthesized using random hexamer primers and M-MuLV Reverse Transcriptase (Rnase H-). The double-stranded cDNA fragments were processed by end repair using T4 DNA polymerase, Klenow DNA polymerase, and T4 polynucleotide kinase (NEB, USA), followed by a single adenine base addition using Klenow 39 to 59 exo-polymerase, and was concluded by ligation with Illumina’s adaptor. The products were purified using a QIAquick PCR extraction kit (Qiagen) and enriched by PCR amplification. The library products were sequenced on an Illumina Hiseq 4000 platform (Illumina, San Diego, CA, USA) and 150 bp paired-end reads were subsequently generated by Novogene Corporation (Novogene, Tianjin, China) as per the protocol [52].
2.5. Bioinformatics Analyses
In order to obtain clean and high-quality data, clean data (clean reads) were obtained by removing reads containing adapter, reads containing ploy-N and the low-quality reads from the raw data. At the same time, Q20, Q30, and the GC content and sequence duplication level of clean data were calculated. Later on, we used Trinity to assemble the clean data and took the transcriptome from Trinity assembled as the reference sequence (Ref), mapping the clean reads of each sample on the Ref. The longest transcript of each gene was taken as unigene for further analysis [53,54]. All unigenes assembled were compared respectively with the nonredundant protein database (NR) of the National Center for Biotechnology Information (NCBI), nonredundant nucleotide sequence (NT) of NCBI database, UniProt/Swiss-Prot, Gene Ontology (GO), and the Keeper of the Grove (KOG) database. Similarly, the Gene-list enrichment was completed using KOBAS 3.0 [53,54].
DEG analysis of two samples was performed using the DEGseq R package [53,54]. p value was adjusted using p value [54,55]. p value ≤ 0.05 and |log2(fold change)| ≥ 1 was set as the threshold for significantly differential expression. To enrich the identified genes, especially the differential genes involved in the canonical pathway, the Kyoto Encyclopedia of Genes and Genomes database and GO enrichment analysis were carried out as described previously through cluster Profiler R Package [53,54,55].
2.6. Accession Number(s)
R. microptera transcriptome datasets are available at NCBI, under project No. PRJNA392010 with accession number SRP110722, and at SRA with accession number SRS2324007. All other data are contained in the manuscript and in the Supplementary Materials.
3. Results
3.1. Adult Pronotum Length
During our study, adult pronotum length responded to both up- and down-selection (Table 1). After 11 generations, mean male and female pronotum length had increased in the Large Colony by 14.8% and 11.8%, respectively, and decreased in the Small Colony by 31.3% and 33.0%, respectively (Table 1). Interestingly, although the only selection criterion was pronotum length, at least 16 additional traits had changed by the end of the selection experiment, including development rate, number of molts, number and size of ovarioles and eggs, behavior, body color, and numerous other morphological traits. By year 8, adult body mass and volume in the Large Colony was almost double that of the Small Colony. A comprehensive description of the methods and results of the selection experiment, emphasizing these phenotypic changes, will be published in a separate paper.

Table 1.
Change in mean pronotum length of Romalea microptera grasshoppers during 8 years (11 generations) of artificial selection in the laboratory for either long- or short-pronotum length. See Section 2 for description of selection experiment and populations.
3.2. Unigenes and Magnitude of Expression Differences
Following de novo assembly [55], a total of 120,267 unigenes were generated from both samples with a total length of 82,796,396 bp and the N50 of 1149 bp (Figure S1). Among these, 693 genes were differentially expressed in samples from the Large Colony vs. the Small Colony (p < 0.05), with 386 upregulated and 307 downregulated (55.7% vs. 44.3%) (Tables S1 and S2). Overall, the magnitude of expression differences tended to be high, with 335 of 693 DEG (48.3%) exhibiting at least a 5-fold difference in expression level. However, up- and downregulated genes tended to differ in their levels of expression (Table 2). More upregulated genes exhibited >2-fold expression vs. downregulated genes (362 vs. 267, respectively). However, downregulated genes predominated (40 vs. 24) when DEG was less than 2-fold (Table 2).

Table 2.
Fold-change distribution of DEGs performing Illumina sequencing and de novo assembly for two colonies of Romalea microptera.
3.3. Functional Annotation of Unigenes
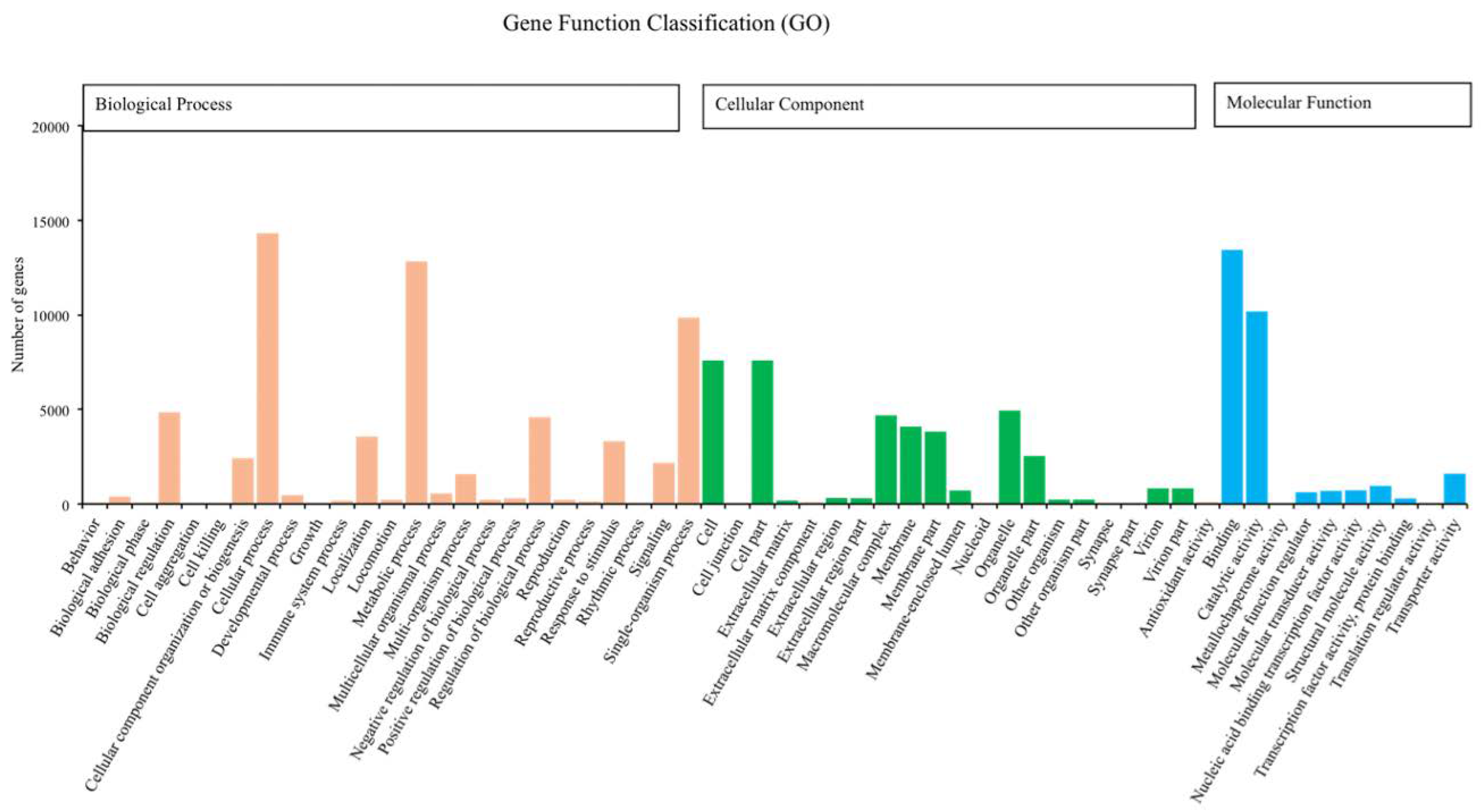
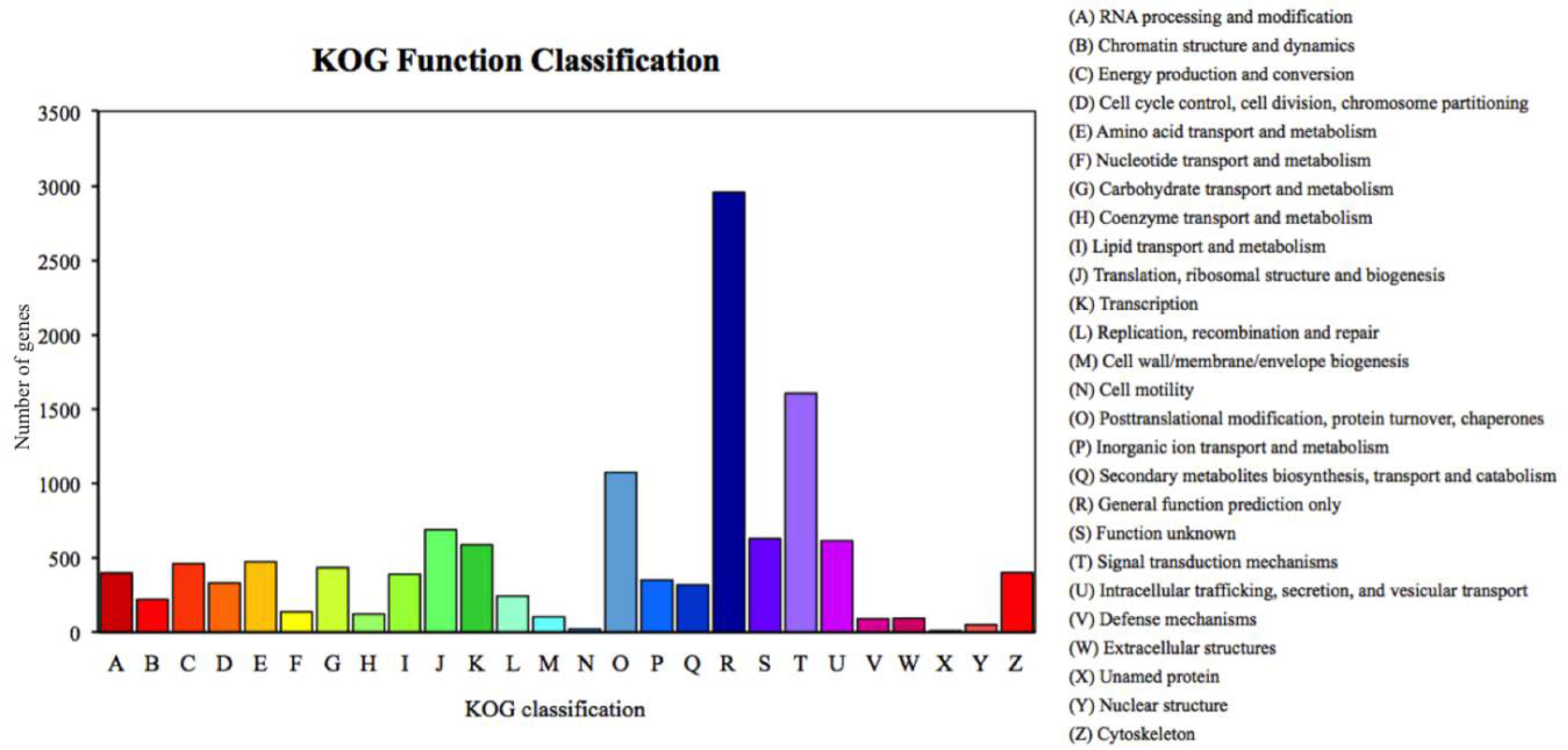
We annotated 39,395 of 120,267 unigenes via seven databases: NR (29,217) > GO (24,742) > PFAM (24,622) > SwissProt (17,521) > KOG (11,500) > KO (9,207) > NT (8,007). Among the most dominant functional classifications as explained by public database GO, were biological process designated as ‘cell process‘, ‘metabolic process‘ and ‘single organism process‘, followed by molecular function with terms ‘binding‘ and ‘catalytic activity‘, and cellular component with terms ‘cell‘ and ‘cell part‘ (Figure 1). In the category of biological process, a total of 12,826 genes were absolutely related with the metabolic process. On the other hand, the KOG functional classifications respectively explained ‘general function prediction only (R)’, ‘signal transduction mechanisms (T)’, ‘post-translational modification protein turnover chaperones (O)’ and ‘translation ribosomal structure and biogenesis (J)’ as the most dominated functions (Figure 2).
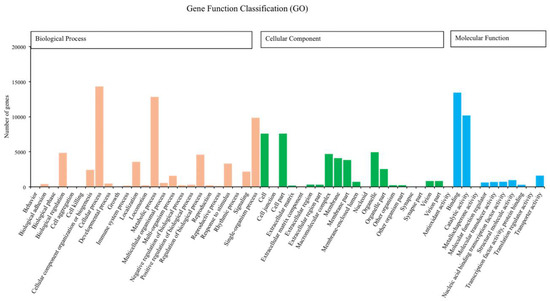
Figure 1.
Number of unigenes of Romalea microptera explained and classified by Gene Ontology (GO) data library in different functional groups within the categories of biological process, cellular component, and molecular function.
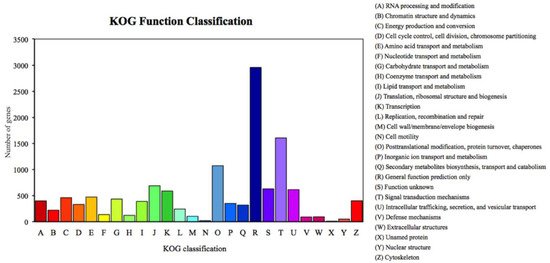
Figure 2.
Percent of Romalea microptera’s unigenes in each functional category for expressivity and dominancy, explained and classified by the Keeper of the Grove (KOG) database.
3.4. Functional Characterization
To understand the molecular changes resulting from size-selection in R. microptera, we used clusterProfiler R Package to analyze the DEGs enriched from GO and KEGG databases. The GO database revealed that seven terms were substantially enriched (p < 0.05), including DNA metabolic process (49; 25.7%), DNA replication (31; 16.2%), RNA-dependent DNA replication (27; 14.1%), DNA integration (16; 8.4%) under the category of biological processes and nucleotidyltransferase activity (32; 16.8%), DNA polymerase activity (30; 15.7%) and RNA-directed DNA polymerase activity (27; 14.1%) within the category of molecular function (Table 3). To test for genes that might influence size-regulation in R. microptera, we used KOBAS 3.0 to find statistically enriched DEGs of downregulated and upregulated features in KEGG pathways [53]. The 307 downregulated DEGs were mapped to 30 pathways in the KEGG database. Among them, only 10 pathways were substantially enriched (p < 0.05) involving single genes except for Huntington’s disease, which had two genes (Table 4). Besides, Huntington’s disease, phenylalanine metabolism, and other glycan degradation were among the top contributing pathway terms. Similarly, 386 upregulated DEGs were mapped to a total of 21 pathways, which were substantially richer (Table 5). Furthermore, the number of genes participating in most of these upregulated pathways was higher than those of DEGs negatively expressed. Fat digestion and absorption (2), glycerolipid metabolism (2), metabolic pathways (5), and ECM–receptor interaction (2) were among the highly represented pathway terms.

Table 3.
Substantially enriched GO terms of DEGs.

Table 4.
Number of substantially enriched pathways along with number of genes of downregulated DEGs computed by Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis.

Table 5.
Number of substantially enriched pathways along with number of genes of upregulated DEGs computed by KEGG pathway analysis.
4. Discussion
In our study, we selected for either long or short pronotum length in a grasshopper. After 8 years (11 generations) of artificial selection in the laboratory, pronotum length had increased ~13.3% in the population selected for long thoraces and decreased ~32.2% in the population selected for short thoraces (sexes combined) (Table 1). In addition, although we had selected for only a single trait, at least 16 other traits had diverged between the two populations, including overall body size and mass: adults from the Large Colony had become nearly twice the mass of those from the Small Colony (unpublished). These results suggest that artificial selection on a single trait influenced numerous genes, causing profound changes in multiple physiological processes across multiple systems, and induced multiple phenotypic changes. Multitrait changes are common in selection experiments and are attributable to pleiotropy, linkage disequilibrium, genetic drift, and (in long-term or high-population experiments) mutation [7,14,34].
Evolution proceeded rapidly during our experiment, altering numerous traits in only 11 generations. Indeed, both natural and artificial selection can be stunningly fast [23,25,56,57]. Our experiment employed relatively low population sizes. Generation 1 started with only 15 individuals of each sex/treatment, and subsequent generations started with between 20 and ~65 breeding individuals/treatment, after removing ~85% of individuals with inappropriate pronotum lengths. Hence, we employed a moderate level of selection, with ~15% of individuals selected for continued breeding during each generation. We do not know how our population size and level of selection influenced our results. Researchers have debated the effects of different population sizes and selection intensities on artificial selection, with some suggesting that small population size quickly reduces genetic variation and that intense selection acts as a bottleneck, eliminating important background genetic variation and variation for the focus trait, as well as lowering overall colony fitness [1,7,14].
In this experiment, we sought to characterize the transcriptomic responses to artificial selection as a first step in identifying putative genes and biochemical pathways involved in evolution and regulation of body size and other altered traits. We assume that the divergent phenotypes resulting from our selection experiment derived from altered genotypes. To begin to understand these possible genetic changes, and to identify possible biological pathways active in size regulation, we analyzed, annotated, and compared the transcriptomes of the two colonies. We found comparatively higher ratios for upregulated vs. downregulated DEGs in both groups. Similarly, log2 (fold change) at level 2–3 and >5 represented maximum contribution for DEGs. Among DEGs were those active in controlling metabolic process, cell process, and binding, including the phosphatidylinositol-3-kinase/protein kinase B (PI3K-Akt) pathway previously reported in other insects for cellular responses such as cell proliferation, apoptosis, DNA repair, and protein synthesis [58]—all related in some way to growth and development. KOG function analysis described maximum contribution of signal transduction mechanisms, post-translational modification, protein turnover and chaperones coupled with general function prediction for size variation in insects. Similarly, within the biological pathways, the active terms identified by the GO database associated with body size of R. microptera were DNA metabolic process and DNA replication, besides the other pathways, such as biological processes and cellular components. Laminin and collagen related with the formation of basement membrane are important genes of the PI3K pathway. In our results, collagen and laminin exhibited an upregulated expression pattern in the Large Colony (Table S1, S2). These collagens are reported to be structural proteins involved in the synthesis of a variety of extracellular matrices in animals [59,60]. For example, in nematodes, such as Caenorhabditis elegans, the role of collagen is associated with the formation of basement membrane and cuticle, and cuticle collagens can act as regulators of body size [61,62]. Laminins, involved in PI3K-Akt pathway, are a major component of the basement membrane and contribute in most biological and ecological functions [63,64]. As such, laminin and collagen might be involved in the body size regulation of lubber grasshoppers.
Leptin is another fat-cell-specific hormone influencing development, growth, metabolism and reproduction by binding to the leptin receptor (lepr) whereas leptin receptor gene-related protein negatively regulates leptin-receptor cell surface expression and thus decreases response to leptin [65,66]. In mammals, leptin receptor gene-related protein influences growth hormone signaling that regulates body size and metabolism [67]. In this study, we found that the leptin receptor gene-related protein in R. microptera has no such significant contribution in body size at the transcriptomic level. We found that ECM–receptor interaction had a positive role in upregulated DEGs of both samples. For example, Spondin-1 is a cell adhesion protein usually attached to the sensory neuron cells and outgrowth of neuritis that encode a secreted basement membrane molecule similar in function to F-spondin of vertebrate [68,69]. In our study, the population selected for long-thorax (Large Colony) exhibited an upregulated expression pattern for spondin-1, which could be involved in insect body size regulation. Similarly, we identified three upregulated carboxylesterases in the Large Colony. Carboxylesterase has several functions, including regulating development, metabolic detoxification of insecticides and exogenous substances, hormone degradation, and neurogenesis [70].
As discussed, above, numerous phenotypic traits and transcripts appeared to change during our artificial selection experiment. At this time, we cannot assign a direct cause-and-effect relationship between any specific altered transcription product and any specific altered trait. However, this paper is a necessary first step toward that elucidation.
5. Conclusions
Our experiment confirms that artificial selection on a single morphological trait (pronotum length) in small populations can quickly alter numerous transcripts and phenotypic traits, including nonmorphological traits, such as behavior and life-history traits. As such, our study demonstrates rapid genetic and phenotypic evolution, apparently accompanied by strong genetic correlations during artificial selection. Only 11 generations of selection produced evolution in at least 17 phenotypic traits and 693 DEGs, including many known to influence body size in animals, like cuticle protein 34, cuticle protein 8, structural constituent of cuticle, insect cuticle protein, and pro-resilin. We hypothesize that pleiotropy, linkage disequilibrium, and genetic drift during artificial selection in small populations induced these observed genomic and phenotypic changes.
Supplementary Materials
The following are available online at https://www.mdpi.com/2075-4450/11/3/176/s1, Figure S1: Unigene length distribution of transcriptome assembly in Romalea microptera, Table S1: Down Regulated Genes; Table S2: Up Regulated Genes
Author Contributions
Conceptualization, D.W.W. and Z.-H.Z.; Data curation, S.L.; Formal analysis, D.-N.C., S.L. and X.-B.T.; Investigation, D.-N.C., S.L. and X.-B.T.; Methodology, H.U. and D.W.W.; Project administration, Z.-H.Z.; Resources, J.C. and S.L.; Validation, D.-N.C.; Writing—original draft, D.-N.C., H.U. and X.-B.T.; Writing—review & editing, H.U., D.W.W. and Z.-H.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the China Agriculture Research System (CARS-34-07), the National Natural Science Foundation of China (31672485), and by the United States National Science Foundation Grant (DBI 044212).
Acknowledgments
The manuscript has undergone English language editing by MDPI.
Conflicts of Interest
The authors declare no competing financial interests.
Abbreviations
| DEGs | Differentially Expressed Genes |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| SC | Small colony |
| LG | Large colony |
| NR | NCBI nonredundant protein sequences |
| Pfam | Protein family |
| KOG | Keeper of the Grove database |
| KO | KEGG Ortholog database |
| Nt | NCBI nonredundant nucleotide sequences |
| Swiss-Prot | Manually annotated and reviewed protein sequence database |
| CAMs | Cell adhesion molecules |
| cDNA | Complementary DNA |
| DNA | Deoxyribonucleic acid |
| RNA | Ribonucleic acid |
| NCBI | National Center for Biotechnology Information |
References
- Fuller, R.C.; Baer, C.F.; Travis, J. How and when selection experiments might actually be useful. Integr. Comp. Biol. 2005, 45, 391–404. [Google Scholar] [CrossRef]
- Swallow, J.G.; Garland, T.J. Selection experiments as a stool in evolutionary and comparative physiology: Insights into complex traits—an introduction to the symposium. Integr. Comp. Biol. 2005, 45, 387–390. [Google Scholar] [CrossRef] [PubMed]
- Frankino, W.A.; Emlen, D.J.; Shingleton, A.W. Experimental approaches to studying the evolution of animal form. In Experimental Evolution: Concepts, Methods, and Applications of Selection Experiments; Garland, T., Rose, M.R., Eds.; U. California: Berkeley, CA, USA, 2009; pp. 419–478. [Google Scholar]
- Evans, L.T. Darwin’s use of the analogy between artificial and natural selection. J. Hist. Biol. 1984, 17, 113–140. [Google Scholar] [CrossRef] [PubMed]
- Darwin, C. On the origin of species by means of natural selection or the preservation of favoured races in the struggle for life; Milford, H., Ed.; Oxford University Press: Oxford, UK, 1859; pp. 1–194. [Google Scholar] [CrossRef]
- Conner, J.K. Artificial selection: A powerful tool of ecologists. Ecology 2003, 84, 1650–1660. [Google Scholar] [CrossRef]
- Garland, T.; Rose, M.R. Experimental Evolution: Concepts, Methods, and Applications of Selection Experiments; U. California: Berkeley, CA, USA, 2009; Volume 60, p. 762. [Google Scholar] [CrossRef]
- Falconer, D.S. Early selection experiments. Annu. Rev. Genetics. 1992, 26, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Hill, W.G.; Caballero, A. Artificial selection experiments. Annu. Rev. Ecol. Syst. 1992, 23, 287–310. [Google Scholar] [CrossRef]
- Bennett, A.F. Experimental evolution and the Krogh Principle: Generating biological novelty for functional and genetic analyses. Phys. Biol. Zool. 2003, 76, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Hays, B.J.; Chamberlain, A.J.; Macaechern, S.; Savin, K.; McPartlan, H.; MacLeod, I.; Sethurman, L.; Goddard, M.E. A genome map of divergent artificial selection between Bos taurus dairy cattle and Bos taurus beef cattle. Anim. Gen. 2009, 40, 176–184. [Google Scholar] [CrossRef] [PubMed]
- Long, A.; Liti, G.; Luptak, A.; Tenaillon, O. Elucidating the molecular architecture of adaptation via evolve and resequence experiments. Nat. Rev. Gen. 2015, 16, 567–582. [Google Scholar] [CrossRef]
- Ma, L.; Sonstegard, T.S.; Cole, J.B.; VanTassell, C.P.; Wiggans, G.R.; Crooker, B.A.; Tan, C.; Prakapenka, D.; Liu, G.E.; Da, Y. Genome changes due to artificial selection in U.S. Holstein cattle. BMC Genomics 2019, 20, 128. [Google Scholar] [CrossRef]
- Walsh, B.; Lynch, M. Evolution and Selection of Quantitative Traits; Oxford U. Press: Oxford, UK, 2018. [Google Scholar]
- Coile, D.C. Encyclopedia of Dog Breeds; Barron’s Educational Series; Simmon & Schuster: New York, NY, USA, 1998. [Google Scholar]
- Meyer, R.S.; DuVal, A.E.; Jensen, H.R. Patterns and processes in crop domestication: An historical review and quantitative analysis of 203 global food crops. New. Phytologist. 2012, 196, 29–48. [Google Scholar] [CrossRef] [PubMed]
- Zeder, M.A. The domestication of animals. J. Anthropol. Res. 2012, 68, 161–190. [Google Scholar] [CrossRef]
- Holthorp, H.E. Tricotyledony. Nature 1944, 153, 13–14. [Google Scholar] [CrossRef][Green Version]
- Partridge, L.; Fowler, K. Responses and correlated responses to artificial selection on thorax length in Drosophila melanogaster. Evolution 1993, 47, 213–226. [Google Scholar] [CrossRef] [PubMed]
- Fellowes, M.D.E.; Kraaijeveld, A.R.; Godfray, H.C.J. Cross-resistance following artificial selection for increased defense against parasitoids in Drosophila melanogaster. Evolution 1999, 53, 966–972. [Google Scholar] [CrossRef]
- Sauvage, C.; Derôme, N.; Normandeau, E.; Cyr, J.S.; Audet, C.; Bernatchez, L. Fast transcriptional responses to domestication in the brook charr Salvelinus fontinalis. Genetics 2010, 185, 105–112. [Google Scholar] [CrossRef]
- Barrett, R.D.H.; Paccard, A.; Healy, T.M.; Bergek, S.; Schulte, P.M.; Schluter, D.; Rogers, S.M. Rapid evolution of cold tolerance in stickleback. Proc. R. Soc. B. Biol. Sci. 2011, 278, 233–238. [Google Scholar] [CrossRef]
- Saavedra-Rodriguez, K.; Suarez, A.F.; Salas, I.F.; Strode, C.; Ranson, H.; Hemingway, J.; Black, W.C. Transcription of detoxification genes following permethrin selection in the mosquito Aedes aegypti. Insect. Mol. Biol. 2012, 21, 61–77. [Google Scholar] [CrossRef]
- Zu, P.; Blanckenhorn, W.U.; Schiestl, F.P. Heritability of floral volatiles and pleiotropic responses to artificial selection in Brassica rapa. New. Phytol. 2016, 209, 1208–1219. [Google Scholar] [CrossRef]
- Zu, P.; Schiestl, F.P. The effects of becoming taller: Direct and pleiotropic effects of artificial selection on plant height in Brassica rapa. Plant. J. 2017, 89, 1009–1019. [Google Scholar] [CrossRef]
- Powell, J.R. Progress and Prospects in Evolutionary Biology: The Drosophila Model; Oxford University Press: Oxford, UK, 1997. [Google Scholar]
- Arias, A.M. Drosophila melanogaster and the development of biology in the 20th century. In Drosophila methods and protocols; Dahmann, C., Ed.; Humana: Louisville, NJ, USA, 2008; pp. 1–25. [Google Scholar] [CrossRef]
- Wheeler, M.A. Studies in Genetics, V2: Research Reports on Drosophila Genetics, Taxonomy and Evolution. Literary Licensing; LLC: Whitefish, MT, USA, 2013; p. 566. [Google Scholar]
- Singh, P. Evolutionary Population Genetics of Drosophila ananassae; Springer: New Delhi, India, 2015; pp. 1–117. [Google Scholar] [CrossRef]
- Dahmann, C. Drosophila: Methods and Protocols; Humana: Louisville, NJ, USA, 2016. [Google Scholar]
- Cai, J.; Zu, P.; Schiestl, F.P. The molecular bases of floral scent evolution under artificial selection: Insights from a transcriptome analysis in Brassica rapa. Sci. Rep. 2016, 6, 36966. [Google Scholar] [CrossRef] [PubMed]
- Goldman, J.G. Mans’s new best friend? A forgotten Russian experiment in fox domestication. Scientific American Guest Blog. 2010. Available online: https://blogs.scientificamerican.com/guest-blog/mans-new-best-friend-a-forgotten-russian-experiment-in-fox-domestication/ (accessed on 16 September 2010).
- Dugatkin, L.A.; Trut, L.; Trut, L.N. How to Tame a Fox (and build a dog): Visionary Scientists and a Siberian Tale of Jump-Started Evolution; University of Chicago Press: Chicago, IL, USA, 2017. [Google Scholar]
- Conner, J.K.; Hartl, D.L. A Primer of Ecological Genetics; Sinauer Associates: Sunderland, MA, USA, 2004. [Google Scholar]
- Lu, X.; Fu, X.; Wang, D.; Wang, J.; Chen, X.; Hao, M.; Wang, J.; Gervers, K.A.; Guo, L.; Wang, S.; et al. Resequencing of cv CRI-12 family reveals haplotype block inheritance and recombination of agronomically important genes in artificial selection. Plant. Biotec. J. 2019, 17, 945–955. [Google Scholar] [CrossRef] [PubMed]
- Whitman, D.W. The significance of body size in the Orthoptera: A review. J. Orthoptera. Res. 2008, 17, 117–134. [Google Scholar] [CrossRef]
- Whitman, D.W.; Vincent, S. Body size in Orthoptera. J. Orthoptera. Res. 2008, 17, 113–375. [Google Scholar] [CrossRef]
- Whitman, D.W.; Vincent, S. Large size as an anti-predator defense in a grasshopper. J. Orthoptera. Res. 2008, 17, 253–371. [Google Scholar] [CrossRef]
- Whitman, D.W. Developmental thermal requirements for the grasshopper Taeniopoda eques (Orthoptera: Acrididae). Ann. Entomol. Soc. Amer. 1986, 79, 711–714. [Google Scholar] [CrossRef]
- Akman, O.; Gamage, J.; Juliano, S.; Thurman, A.; Whitman, D.W. A simple test for detection of length-biased sampling. JP J. Biostat. 2007, 1, 189–195. [Google Scholar]
- Akman, O.; Whitman, D.W. Analysis of body size and fecundity in a grasshopper. J. Orthoptera. Res. 2008, 17, 249–257. [Google Scholar] [CrossRef][Green Version]
- Fronstin, R.B.; Hatle, J.D. Interpopulation variation in the trade-off between body-mass gain and age at oviposition in the Eastern Lubber Grasshopper, Romalea microptera. J. Orthoptera. Res. 2008, 17, 273–277. [Google Scholar] [CrossRef]
- Huizenga, K.M.; Shaidle, M.D.; Brinton, J.S.; Gore, L.N.A.; Ebo, M.A.; Solliday, A.J.; Buguey, P.J.; Whitman, D.W.; Juliano, S.A. Geographic differences in the body sizes of adult Romalea microptera. J. Orthoptera. Res. 2008, 17, 135–139. [Google Scholar] [CrossRef]
- Vincent, S.E.; Lailvaux, S.P. Does phenotypic integration constrain sexual size dimorphism in Eastern Lubber grasshoppers (Romalea microptera)? J. Orthoptera. Res. 2008, 17, 219–225. [Google Scholar] [CrossRef][Green Version]
- Weissman, D.B.; Judge, K.A.; Williams, S.C.; Whitman, D.W. Small-male mating advantage in a species of Jerusalem cricket (Orthoptera: Stenopelmatinae: Stenopelmatus). J Orthoptera. Res. 2008, 17, 321–332. [Google Scholar] [CrossRef]
- Whitman, D.W.; Agrawal, A.A. What is phenotypic plasticity ad why is it important? In Phenotypic Plasticity of Insects; Whitman, D.W., Ananthakrishnan, T.N., Eds.; Science Publishers: Concord, NH, USA, 2009; pp. 1–63. [Google Scholar]
- Cui, B.Y.; Huang, X.B.; Li, S.; Hao, K.; Chang, B.H.; Tu, X.B.; Pang, B.P.; Zhang, Z.H. Quercetin affects the growth and development of the grasshopper Oedaleus asiaticus (Orthoptera: Acrididae). J. Econ. Ent. 2019, 112, 1175–1182. [Google Scholar] [CrossRef] [PubMed]
- Jarwar, A.R.; Hao, K.; Bitume, E.V.; Ullah, H.; Cui, D.N.; Nong, X.Q.; Wang, G.J.; Tu, X.B.; Zhang, Z.H. Comparative transcriptomic analysis reveals molecular profiles of central nervous system in maternal diapause induction of Locusta migratoria. G3-Genes Genom. Genet. 2019, g3-400475. [Google Scholar] [CrossRef] [PubMed]
- Rehn, J.A.G.; Grant, H.G. A review of the Romaleinae (Orthoptera: Acrididae) found in America north of Mexico. Proc. Acad. Nat. Sci. Phil. 1961, 111, 109–271. [Google Scholar]
- Capinera, J.L.; Scherer, C. Featured creatures: Eastern lubber grasshopper. University of Florida/IFAS Publication EENY-6, Florida. 2016. Available online: http://entnemdept.ufl.edu/creatures/orn/lubber.htm (accessed on 24 May 2016).
- Matusezek, J.V.; Whitman, D.W. Captive rearing of Eastern Lubber grasshoppers. In Conference Proceedings: Invertebrates in Captivity 2001; Sonoran Arthropod Studies Institute: Rio Rico, AZ, USA, 2002; pp. 56–63. [Google Scholar]
- Hao, K.; Wang, J.; Tu, X.B.; Whitman, D.W.; Zhang, Z.H. Transcriptomic and proteomic analysis of Locusta migratoria eggs at different embryonic stages: Comparison for diapause and non-diapause regimes. J. Integ. Agric. 2017, 16, 1777–1788. [Google Scholar] [CrossRef]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic. Acids. Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef]
- Tu, X.B.; Wang, J.; Hao, K.; Whitman, D.W.; Fan, Y.; Cao, G.C.; Zhang, Z.H. Transcriptomic and proteomic analysis of pre-diapause and non-diapause eggs of migratory locust, Locusta migratoria L. (Orthoptera: Acridoidea). Sci. Rep. 2015, 5, 11402. [Google Scholar] [CrossRef]
- Chen, S.; Yang, P.; Jiang, F.; Wei, Y.; Ma, Z.; Kang, L. de novo analysis of transcriptome dynamics in the migratory locust during the development of phase traits. PLoS ONE 2010, 5, e15633. [Google Scholar] [CrossRef]
- Little, J.B. Rapid evolution changes species in real time. Discover Magazine Issue 2 March, 2 March 2015. [Google Scholar]
- Hendry, A.P. Eco-Evolutionary Dynamics; Princeton University Press: Princeton, NJ, USA, 2016. [Google Scholar]
- Andrade, G.M.; da Silveira, J.C.; Perrini, C.; Del Collado, M.; Gebremedhn, S.; Tesfaye, D.; Meirelles, F.V.; Perecin, F. The role of the PI3K-Akt signaling pathway in the developmental competence of bovine oocytes. PLoS ONE 2017, 12, e0185045. [Google Scholar] [CrossRef] [PubMed]
- Prockop, D.J. What holds us together? Why do some of us fall apart? What can we do about it? Soci. Matrix. Biol. 1998, 16, 519–528. [Google Scholar] [CrossRef]
- Prockop, D.J.; Kivirikko, K.I. Collagens: Molecular biology, diseases, and potentials for therapy. Ann. Rev. Biochem. 1995, 64, 403–434. [Google Scholar] [CrossRef] [PubMed]
- Johnstone, I.L. Cuticle collagen genes: Expression in Caenorhabditis elegans. Trends Genet. 2000, 16, 21–27. [Google Scholar] [CrossRef]
- Madaan, U.; Yzeira, E.; Meade, M.; Clark, J.F.; Rushlow, C.A.; Savage-Dunn, C. BMP Signaling Determines Body Size via Transcriptional Regulation of Collagen Genes in Caenorhabditis elegans. Genetics 2018, 210, 1355–1367. [Google Scholar] [CrossRef]
- Lepage, M.; Seltana, A.; Thibault, M.P.; Tremblay, É.; Beaulieu, J.F. Knockdown of laminin α5 stimulates intestinal cell differentiation. Biochem. Biophys. Res. Comm. 2018, 495, 1510–1515. [Google Scholar] [CrossRef]
- Chang, B.H.; Cui, B.Y.; Ullah, H.; Li, S.; Kun, H.; Tu, X.B.; Wang, G.J.; Nong, X.Q.; McNeill, M.R.; Huang, X.B.; et al. Role of PTP/PTK trans activated insulin-like signalling pathway in regulation of grasshopper (Oedaleus asiaticus) development. Environ. Sci. Pollut. Res. 2019, 26, 8312–8324. [Google Scholar] [CrossRef]
- Tartaglia, L.A.; Dembski, M.; Weng, X.; Deng, N.; Culpepper, J.; Devos, R.; Richards, G.J.; Campfield, L.A.; Clark, F.T.; Deeds, J.; et al. Identification and expression cloning of a leptin receptor, OB-R. Cell 1995, 83, 1263–1271. [Google Scholar] [CrossRef]
- Couturier, C.; Sarkis, C.; Séron, K.; Belouzard, S.; Chen, P.; Lenain, A.; Corset, L.; Dam, J.; Vauthier, V.; Dubart, A.; et al. Silencing of OB-RGRP in mouse hypothalamic arcuate nucleus increases leptin receptor signaling and prevents diet-induced obesity. Proc. Natl. Acad. Sci. 2007, 104, 19476–19481. [Google Scholar] [CrossRef]
- Touvier, T.; Conte-Auriol, F.; Briand, O.; Cudejko, C.; Paumelle, R.; Caron, S.; Baugé, E.; Rouillé, Y.; Salles, J.P.; Staels, B.; et al. LEPROT and LEPROTL1 cooperatively decrease hepatic growth hormone action in mice. J. Clin. Invest. 2009, 119, 3830–3838. [Google Scholar] [CrossRef]
- Woo, W.M.; Berry, E.C.; Hudson, M.L.; Swale, R.E.; Goncharov, A.; Chisholm, A.D. The C. elegans F-spondin family protein SPON-1 maintains cell adhesion in neural and non-neural tissues. Development 2008, 135, 2747–2756. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.; Dong, T.; Ma, X.; Zhang, T.; Chen, Z.; Yang, Z.; Zhang, Y. Spondin 1 promotes metastatic progression through Fak and Src dependent pathway in human osteosarcoma. Biochem. Biophys. Res. Comm. 2015, 464, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.D.; Huang, M.X.; Zhang, Y.L.; Wang, X.C.; Du, J.; Li, B.; Chen, Y.H.; Xu, Y.X.; Wei, Z.G. Expression analysis and RNA interference of BmCarE-10 gene from Bombyx mori. Mol. Biol. Rep. 2014, 41, 1607–1616. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).