Identification, Characterization and Expression Analysis of TRP Channel Genes in the Vegetable Pest, Pieris rapae
Abstract
:1. Introduction
2. Materials and Methods
2.1. Insect
2.2. Identification of TRP Channels
2.3. Total RNA Isolation, Reverse Transcription and Splice Variants Detection
2.4. Bioinformatic Analysis
2.5. Real-Time Quantitative PCR and Statistical Analysis
3. Results
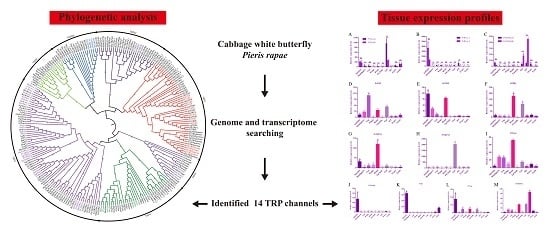
3.1. Identification and Phylogenetic Analysis of TRP Channels in P. Rapae and Other Lepidoptera Insects
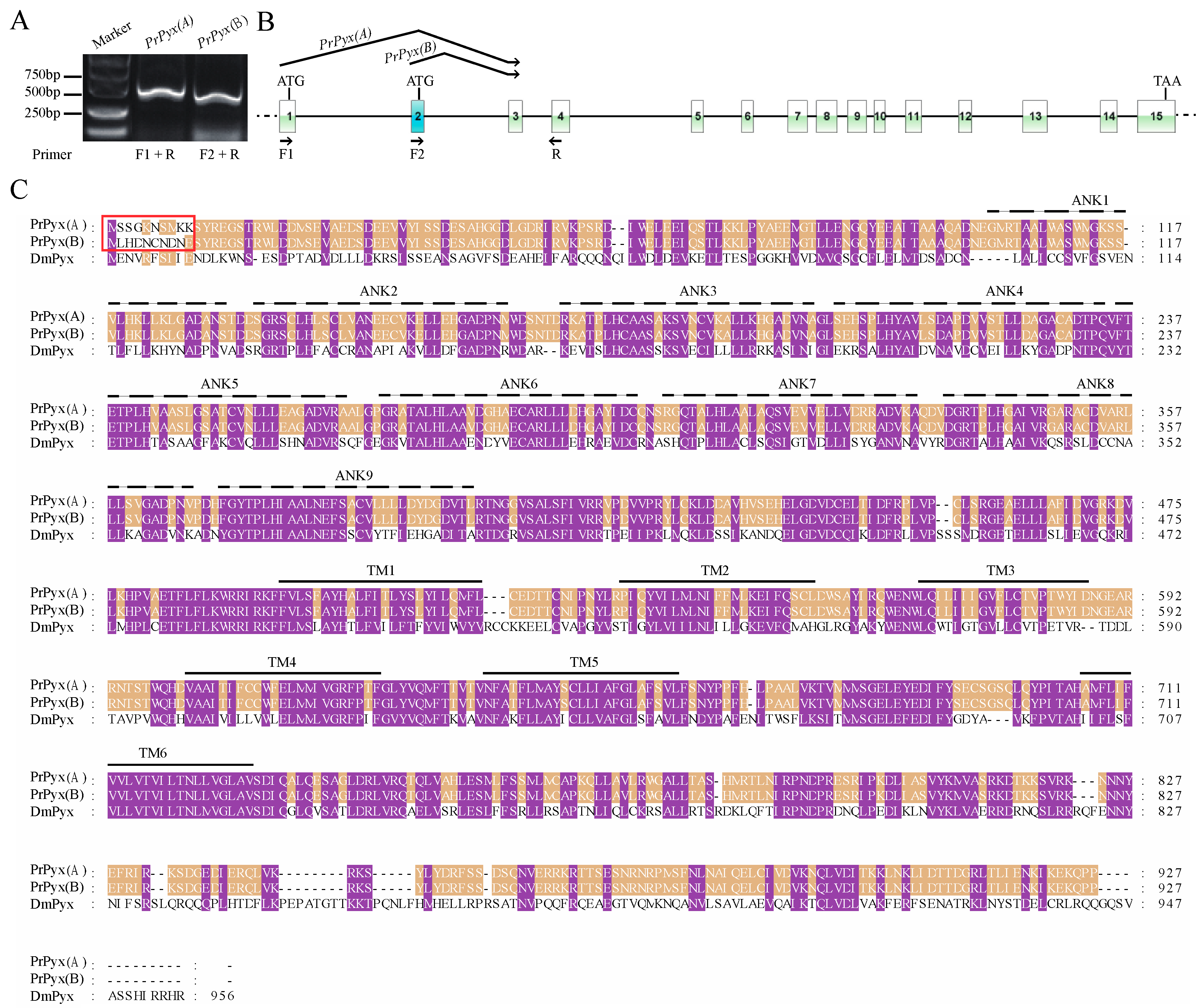
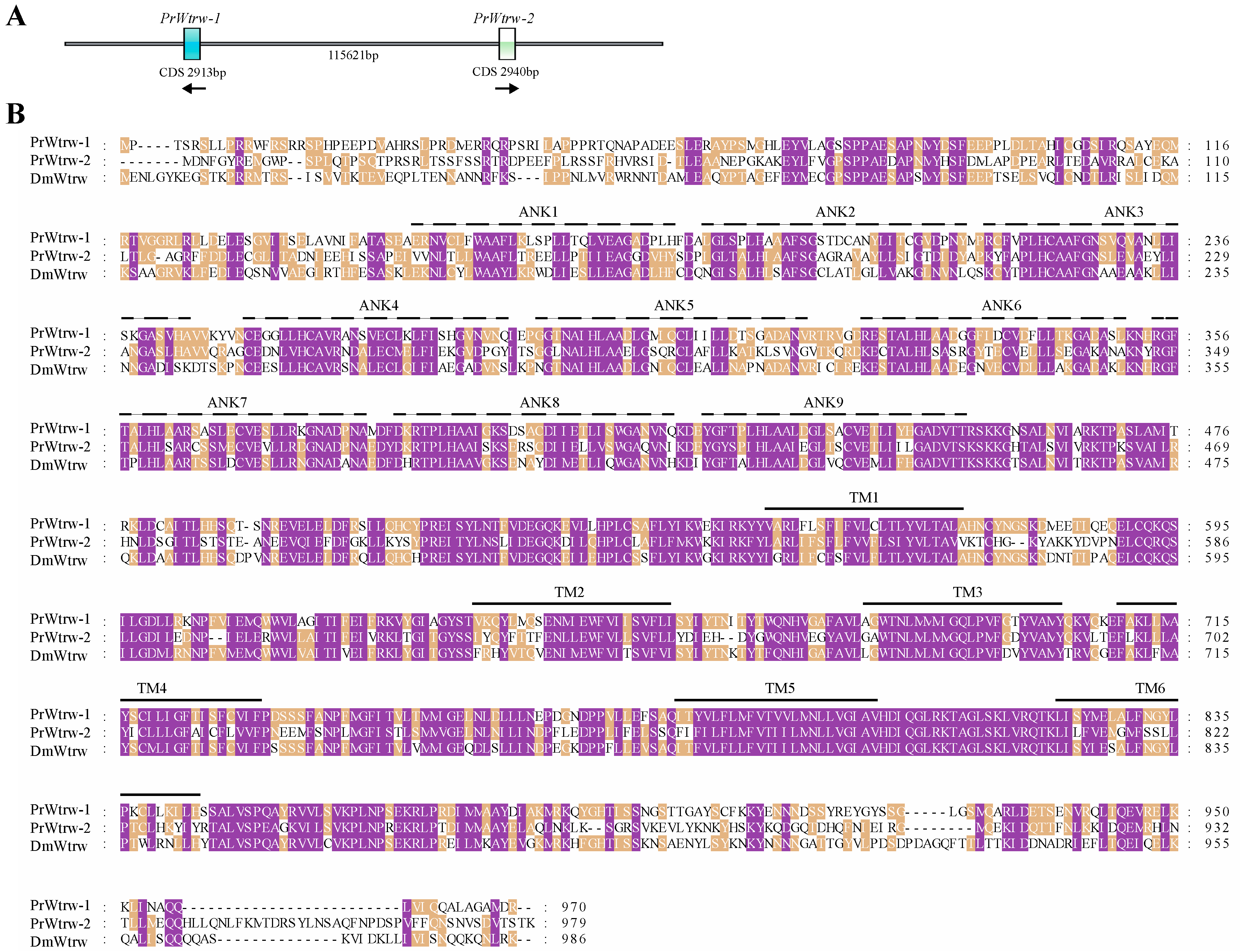
3.2. Sequence Analysis, Splice Variants and Gene Duplications of TRP Channelsin P. Rapae
3.3. Tissue Expression Profiles of TRP Channel in Adult P. Rapae
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Venkatachalam, K.; Montell, C. TRP channels. Annu. Rev. Biochem. 2007, 76, 387–417. [Google Scholar] [CrossRef] [Green Version]
- Fowler, M.A.; Montell, C. Drosophila TRP channels and animal behavior. Life Sci. 2013, 92, 394–403. [Google Scholar] [CrossRef] [Green Version]
- Cosens, D.J.; Manning, A. Abnormal electroretinogram from a Drosophila mutant. Nature 1969, 224, 285–287. [Google Scholar] [CrossRef]
- Montell, C.; Birnbaumer, L.; Flockerzi, V. The TRP channels, a remarkably functional family. Cell 2002, 108, 595–598. [Google Scholar] [CrossRef] [Green Version]
- Montell, C. Drosophila visual transduction. Trends Neurosci. 2012, 35, 356–363. [Google Scholar] [CrossRef] [Green Version]
- Caterina, M.J.; Schumacher, M.A.; Tominaga, M.; Rosen, T.A.; Levine, J.D.; Julius, D. The capsaicin receptor: A heat-activated ion channel in the pain pathway. Nature 1997, 389, 816–824. [Google Scholar] [CrossRef]
- Montell, C. The TRP superfamily of cation channels. Sci. Signal. 2005, 272, re3. [Google Scholar] [CrossRef] [Green Version]
- Sidi, S.; Friedrich, R.W.; Nicolson, T. NompC TRP channel required for vertebrate sensory hair cell mechanotransduction. Science 2003, 301, 96–99. [Google Scholar] [CrossRef]
- Montell, C. Drosophila TRP channels. Pflug. Arch. Eur. J. Physiol. 2005, 451, 19–28. [Google Scholar] [CrossRef]
- Bellemer, A. Thermotaxis, circadian rhythms, and TRP channels in Drosophila. Temperature 2015, 2, 227–243. [Google Scholar] [CrossRef] [Green Version]
- Kingsolver, J.G. Feeding, growth, and the thermal environment of cabbage white caterpillars, Pieris rapae L. Physiol. Biochem. Zool. 2000, 73, 621–628. [Google Scholar] [CrossRef]
- Xiang, M.; Zhang, X.; Deng, Y.; Li, Y.; Yu, J.; Zhu, J.; Huang, X.; Zhou, J.; Liao, H. Comparative transcriptome analysis provides insights of anti-insect molecular mechanism of Cassia obtusifolia trypsin inhibitor against Pieris rapae. Arch. Insect Biochem. Physiol. 2018, 97, e21417. [Google Scholar] [CrossRef] [Green Version]
- Zhong, G.H.; Liu, J.X.; Weng, Q.F.; Hu, M.Y.; Luo, J.J. Laboratory and field evaluations of rhodojaponin-III against the imported cabbage worm Pieris rapae (L.) (Lepidoptera: Pieridae). Pest Manag. Sci. 2006, 62, 976–981. [Google Scholar] [CrossRef]
- Kwon, Y.; Kim, S.H.; Ronderos, D.S.; Lee, Y.; Akitake, B.; Woodward, O.M.; Guggino, W.B.; Smith, D.P.; Montell, C. Drosophila TRPA1 channel is required to avoid the naturally occurring insect repellent citronellal. Curr. Biol. 2010, 20, 1672–1678. [Google Scholar] [CrossRef] [Green Version]
- Al-Anzi, B.; Tracey, W.D.; Benzer, S. Response of Drosophila to wasabi is mediated by painless, the fly homolog of mammalian TRPA1/ANKTM1. Curr. Biol. 2006, 16, 1034–1040. [Google Scholar] [CrossRef] [Green Version]
- Nesterov, A.; Spalthoff, C.; Kandasamy, R.; Katana, R.; Rankl, N.B.; Andres, M.; Jahde, P.; Dorsch, J.A.; Stam, L.F.; Braun, F.J.; et al. TRP channels in insect stretch receptors as insecticide targets. Neuron 2015, 86, 665–671. [Google Scholar] [CrossRef] [Green Version]
- Kim, S. Insect GPCRs and TRP channels: Putative targets for insect repellents. Interdiscip. Bio. Central. 2013, 5, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Lalitha, S. Primer premier 5. Biotech Softw. Internet Rep. 2000, 1, 270–272. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.F.; Haeseler, A.V.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoang, D.T.; Chernomor, O.; Haeseler, A.V.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Apte, A.; Singh, S. AlleleID. In PCR Primer Design, 2nd ed.; Yuryev, A., Ed.; Humana Press: Totowa, NJ, USA, 2007; Volume 402, pp. 329–345. [Google Scholar]
- Wu, S.F.; Wang, F.; Huang, J.; Fang, Q.; Shen, Z.C.; Ye, G.Y. Molecular and cellular analyses of a ryanodine receptor from hemocytes of Pieris rapae. Dev. Comp. Immunol. 2013, 41, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Peng, G.D.; Shi, X.; Kadowaki, T. Evolution of TRP channels inferred by their classification in diverse animal species. Mol. Phylogenet. Evol. 2015, 84, 145–157. [Google Scholar] [CrossRef]
- Su, H.A.; Bai, X.; Zeng, T.; Lu, Y.Y.; Qi, Y.X. Identification, characterization and expression analysis of transient receptor potential channel genes in the oriental fruit fly, Bactrocera dorsalis. BMC Genom. 2018, 19, 674. [Google Scholar] [CrossRef] [Green Version]
- Matsuura, H.; Sokabe, T.; Kohno, K.; Tominaga, M.; Kadowaki, T. Evolutionary conservation and changes in insect TRP channels. BMC Evol. Biol. 2009, 9, 228. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.L.; Batiza, A.F.; Loukin, S.H.; Palmer, C.P.; Kung, C.; Saimi, Y. The transient receptor potential channel on the yeast vacuole is mechanosensitive. Proc. Natl. Acad. Sci. USA 2003, 100, 7105–7110. [Google Scholar] [CrossRef] [Green Version]
- Gao, Z.Q.; Joseph, E.; Ruden, D.M.; Lu, X.Y. Drosophila Pkd2 is haploid-insufficient for mediating optimal smooth muscle contractility. J. Biol. Chem. 2004, 279, 14225–14231. [Google Scholar] [CrossRef] [Green Version]
- Gao, Z.Q.; Ruden, D.M.; Lu, X.Y. PKD2 cation channel is required for directional sperm movement and male fertility. Curr. Biol. 2003, 13, 2175–2178. [Google Scholar] [CrossRef]
- Kohno, K.; Sokabe, T.; Tominaga, M.; Kadowaki, T. Honey bee thermal/chemical sensor, AmHsTRPA, reveals neofunctionalization and loss of transient receptor potential channel genes. J. Neurosci. 2010, 30, 12219–12229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wicher, D.; Agricola, H.J.; Schonherr, R.; Heinemann, S.H.; Derst, C. TRPgamma channels are inhibited by cAMP and contribute to pacemaking in neurosecretory insect neurons. J. Biol. Chem. 2006, 281, 3227–3236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phua, S.C.; Lin, Y.C.; Inoue, T. An intelligent nano-antenna: Primary cilium harnesses TRP channels to decode polymodal stimuli. Cell Calcium 2015, 58, 415–422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davis, J.; Burr, A.R.; Davis, G.F.; Birnbaumer, L.; Molkentin, J.D. A TRPC6-dependent pathway for myofibroblast transdifferentiation and wound healing in vivo. Dev. Cell 2012, 23, 705–715. [Google Scholar] [CrossRef] [Green Version]
- Tang, X.; Platt, M.D.; Lagnese, C.M.; Leslie, J.R.; Hamada, F.N. Temperature integration at the AC thermosensory neurons in Drosophila. J. Neurosci. 2013, 33, 894–901. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.S.; Liu, L.; Ben-Shahar, Y.; Jacobs, J.S.; Eberl, D.F.; Welsh, M.J. TRPA channels distinguish gravity sensing from hearing in Johnston’s organ. Proc. Natl. Acad. Sci. USA 2009, 106, 13606–13611. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.X.; Niu, C.D.; Zhang, Y.; Jia, Y.L.; Zhang, Y.J.; Zhang, Y.; Zhang, Y.Q.; Gao, C.F.; Wu, S.F. The NompC channel regulates Nilaparvata lugens proprioception and gentle-touch response. Insect Biochem. Mol. Biol. 2019, 106, 55–63. [Google Scholar] [CrossRef]
- Liu, L.; Li, Y.H.; Wang, R.O.; Yin, C.; Dong, Q.; Hing, H.; Kim, C.; Welsh, M.J. Drosophila hygrosensation requires the TRP channels water witch and nanchung. Nature 2007, 450, 294–298. [Google Scholar] [CrossRef]
- Tracey, W.D.; Wilson, R.I.; Laurent, G.; Benzer, S. painless, a Drosophila gene essential for nociception. Cell 2003, 113, 261–273. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Chung, Y.D.; Park, D.Y.; Choi, S.K.; Shin, D.W.; Soh, H.; Lee, H.W.; Son, W.; Yim, J.; Park, C.S.; et al. A TRPV family ion channel required for hearing in Drosophila. Nature 2003, 424, 81–84. [Google Scholar] [CrossRef]
- Mao, F.; Guo, L.; Jin, M.; Qiao, X.M.; Ye, G.Y.; Huang, J. Molecular cloning and characterization of TRPVs in two rice pests: Nilaparvata lugens (Stål) and Nephotettix cincticeps (Uhler). Pest Manag. Sci. 2019, 75, 1361–1369. [Google Scholar] [CrossRef] [PubMed]
- Tuthill, J.C.; Wilson, R.I. Parallel transformation of tactile signals in central circuits of Drosophila. Cell 2016, 164, 1046–1059. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Faussone-Pellegrini, M.S.; Taddei, A.; Bizzoco, E.; Lazzeri, M.; Vannucchi, M.G.; Bechi, P. Distribution of the vanilloid (capsaicin) receptor type 1 in the human stomach. Histochem. Cell Biol. 2005, 124, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Georgiev, P.; Okkenhaug, H.; Drews, A.; Wright, D.; Lambert, S.; Flick, M.; Carta, V.; Martel, C.; Oberwinkler, J.; Raghu, P. TRPM channels mediate zinc homeostasis and cellular growth during Drosophila larval development. Cell Metab. 2010, 12, 386–397. [Google Scholar] [CrossRef] [Green Version]
- Hofmann, T.; Chubanov, V.; Chen, X.D.; Dietz, A.S.; Gudermann, T.; Montell, C. Drosophila TRPM channel is essential for the control of extracellular magnesium levels. PLoS ONE 2010, 5, e10519. [Google Scholar] [CrossRef] [Green Version]
- Cuajungco, M.P.; Samie, M.A. The varitint-waddler mouse phenotypes and the TRPML3 ion channel mutation: Cause and consequence. Pflug. Arch. Eur. J. Physiol. 2008, 457, 463–473. [Google Scholar] [CrossRef]
- Remis, N.N.; Wiwatpanit, T.; Castiglioni, A.J.; Flores, E.N.; Cantu, J.A.; Garcia-Anoveros, J. Mucolipin co-deficiency causes accelerated endolysosomal vacuolation of enterocytes and failure-to-thrive from birth to weaning. PLoS Genet. 2014, 10, e1004833. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Subfamily | Gene Name | Genomic Sequence ID | NCBI Accession No. (Transcripts) | Length (Amino Acids) | Protein Region Identified | Number Of Ankyrin Repeats | CG No. Of The Drosophila Melanogaster Orthologue | Sequence Identity |
|---|---|---|---|---|---|---|---|---|
| Group-1 TRPs | ||||||||
| TRPC | PrTRPγ | NW_019093512.1 | XM_022266966.1 | 1120 | TM1–6 | 2 | CG5996 | 65.81% |
| PrTRPL | NW_019093246.1 | XM_022259226.1 | 1158 | TM1–6 | 2 | CG18345 | 62.72% | |
| PrTRP | NW_019093246.1 | XM_022259230.1 | 788 | TM1–6 | 5 | CG7875 | 51.81% | |
| TRPA | PrPain | NW_019093434.1 | XM_022264961.1 | 950 | TM1–6 | 9 | CG15860 | 36.86% |
| PrPyx | NW_019093274.1 | XM_022260547.1 | 928 | TM1–6 | 9 | CG17142 | 52.06% | |
| PrTRPA1 | NW_019093159.1 | XM_022269136.1 | 1134 | TM1–6 | 14 | CG5751 | 66.23% | |
| PrWtrw-1 | NW_019093349.1 | XM_022263017.1 | 971 | TM1–6 | 9 | CG31284 | 63.26% | |
| PrWtrw-2 | NW_019093349.1 | XM_022263000.1 | 980 | TM1–6 | 8 | CG31284 | 49.24% | |
| PrTRPA5 | NW_019093774.1 | XM_022269806.1 | 2350 | TM1–6 | 16 | - | - | |
| TRPN | PrNompC | NW_019099827.1 | XM_022272967.1 | 1607 | TM1–6 | 29 | CG11020 | 77.82% |
| TRPM | PrTRPM | NW_019099824.1 | XM_022272780.1 | 1137 | TM1–6 | 0 | CG44240 | 61.22% |
| TRPV | PrIav | NW_019093844.1 | XM_022270177.1 | 1061 | TM1–6 | 5 | CG4536 | 72.71% |
| PrNan | NW_019093483.1 | XM_022266197.1 | 584 | TM1–4 | 5 | CG5842 | 64.03% | |
| Group–2TRPs | ||||||||
| TRPML | PrTRPML | NW_019093323.1 | XM_022262542.1 | 600 | TM1–6 | 0 | CG8743 | 61.04% |
| Species Name | Channel type | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TRPC | TRPA | TRPN | TRPV | TRPM | TRPP | TRPML | Total | ||||||||||
| TRP | TRPL | TRPγ | TRPA1 | TRPA5 | HsTRPA | Pain | Pyx | Wtrw | NompC | Iav | Nan | TRPM | Brv | Pkd2 | TRPML | ||
| Lepidoptera | |||||||||||||||||
| B. mori | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 14 |
| D. plexippus | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 14 |
| M. sexta | 1 | 1 | 1 | 1 | 2 | 0 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 15 |
| P. Xuthus | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 14 |
| P. rapae | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 14 |
| Isoptera | |||||||||||||||||
| Z. nevadensis | 1 | 1 | 1 | 1 | 1 | 0 | 7 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 22 |
| Blattodea | |||||||||||||||||
| B. germanica | 1 | 1 | 1 | 1 | 2 | 0 | 7 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 2 | 1 | 22 |
| Hemiptera | |||||||||||||||||
| N. lugens | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 14 |
| Hymenoptera | |||||||||||||||||
| A. mellifera | 1 | 1 | 1 | 0 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 13 |
| Diptera | |||||||||||||||||
| A. gambiae | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 13 |
| D. melanogaster | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 3 | 1 | 1 | 16 |
| Coleoptera | |||||||||||||||||
| T. castaneum | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0(1) | 1 | 1 | 14 (15) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mao, F.; Lu, W.-j.; Yang, Y.; Qiao, X.; Ye, G.-y.; Huang, J. Identification, Characterization and Expression Analysis of TRP Channel Genes in the Vegetable Pest, Pieris rapae. Insects 2020, 11, 192. https://doi.org/10.3390/insects11030192
Mao F, Lu W-j, Yang Y, Qiao X, Ye G-y, Huang J. Identification, Characterization and Expression Analysis of TRP Channel Genes in the Vegetable Pest, Pieris rapae. Insects. 2020; 11(3):192. https://doi.org/10.3390/insects11030192
Chicago/Turabian StyleMao, Fen, Wan-jun Lu, Yi Yang, Xiaomu Qiao, Gong-yin Ye, and Jia Huang. 2020. "Identification, Characterization and Expression Analysis of TRP Channel Genes in the Vegetable Pest, Pieris rapae" Insects 11, no. 3: 192. https://doi.org/10.3390/insects11030192
APA StyleMao, F., Lu, W.-j., Yang, Y., Qiao, X., Ye, G.-y., & Huang, J. (2020). Identification, Characterization and Expression Analysis of TRP Channel Genes in the Vegetable Pest, Pieris rapae. Insects, 11(3), 192. https://doi.org/10.3390/insects11030192