Trypanosomatids Detected in the Invasive Avian Parasite Philornis downsi (Diptera: Muscidae) in the Galapagos Islands
Abstract
:1. Introduction
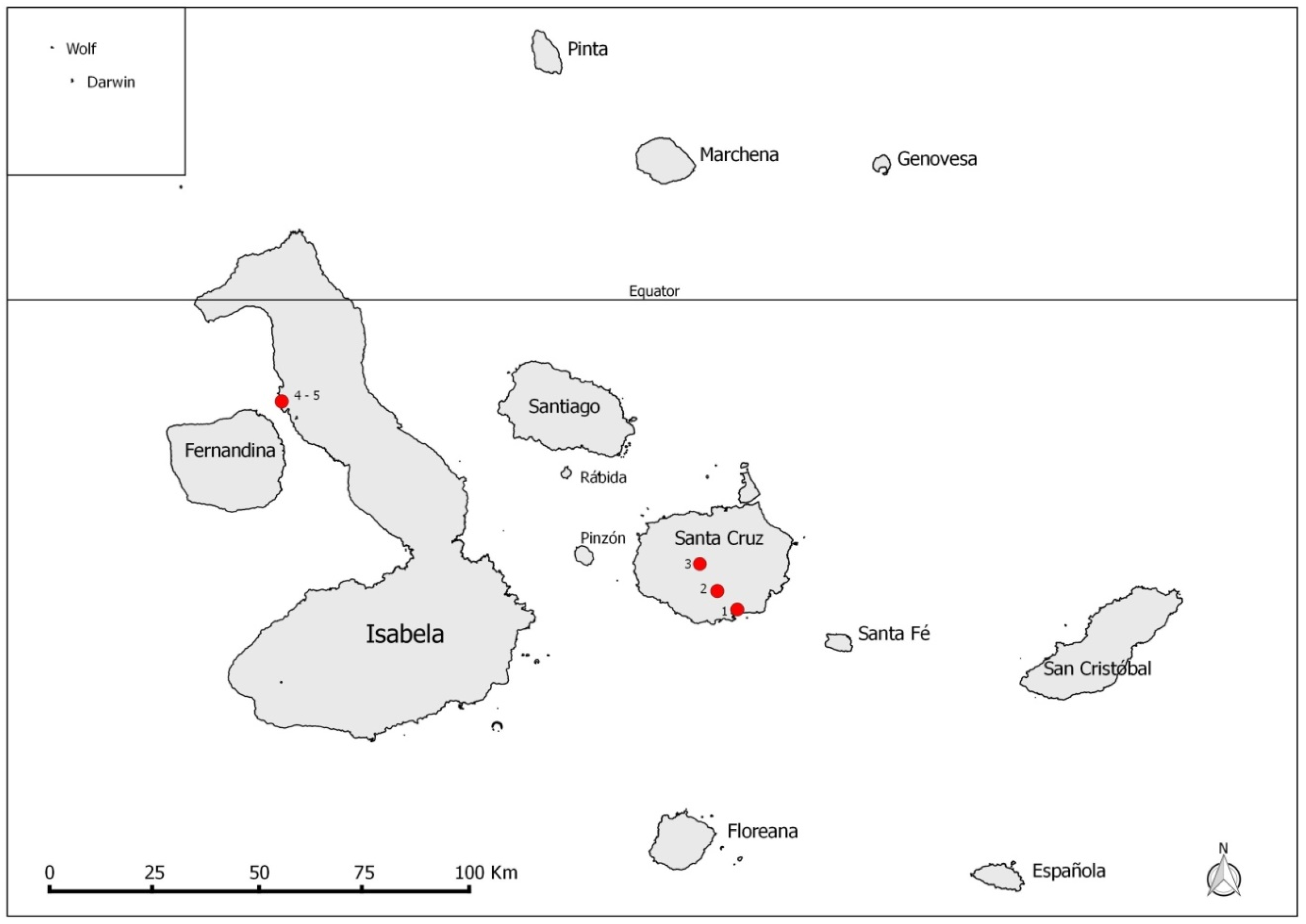
2. Materials and Methods
3. Results
3.1. Molecular Testing
3.2. Phylogenetic Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Friend, M.; McLean, R.G.; Dein, J. Disease emergence in birds: Challenges for the twenty-first century. Auk 2001, 118, 290–303. [Google Scholar] [CrossRef]
- Parker, P.G.; Whiteman, N.K.; Miller, R.E. Conservation medicine on the Galápagos Islands: Partnerships among behavioral, population, and veterinary scientists. Auk 2006, 123, 625–638. [Google Scholar] [CrossRef] [PubMed]
- Aguirre, A.A. Parasitic diseases of wildlife and domestic animals: New trends of disease emergence. In Infectious and Parasitic Diseases of Livestock 1: General Considerations; Lefevre, P.C., Blancou, J., Chermette, R., Uilenberg, G., Eds.; Lavoisier: Paris, France, 2010; Volume 1, pp. 73–79. [Google Scholar]
- Thompson, R.C.A.; Lymbery, A.J.; Smith, A. Parasites, emerging disease and wildlife conservation. Int. J. Parasitol. 2010, 40, 1163–1170. [Google Scholar] [CrossRef] [Green Version]
- Loope, L.L.; Howarth, F.G.; Kraus, F.; Pratt, T.K. Newly emergent and future threats of alien species to pacific birds and ecosystems. Stud. Avian Biol. 2001, 22, 291–304. [Google Scholar]
- Wilkelski, M.; Foufopoulos, J.; Vargas, H.; Snell, H. Galapagos birds and diseases: Invasive pathogens as threats for island species. Ecol. Soc. 2004, 9, 5. [Google Scholar] [CrossRef]
- Simberloff, D. Invasive Species. In Conservation Biology for All; Sodhi, N.S., Ehrlich, P.R., Eds.; Oxford University Press: Oxford, UK, 2010; pp. 131–152. [Google Scholar]
- Blackburn, T.M.; Ewen, J.G. Parasites as drivers and passengers of human-mediated biological invasions. EcoHealth 2016, 14, 61–73. [Google Scholar] [CrossRef] [Green Version]
- Hatcher, M.J.; Dick, J.T.A.; Dunn, A.M. Disease emergence and invasions. Funct. Ecol. 2012, 26, 1275–1287. [Google Scholar] [CrossRef]
- Wilfert, L.; Long, G.; Leggett, H.C.; Schmid-Hempel, P.; Butlin, R.; Martin, S.J.; Boots, M. Deformed wing virus is a recent global epidemic in honeybees driven by Varroa mites. Science 2016, 5, 594–597. [Google Scholar] [CrossRef] [Green Version]
- Wyatt, K.B.; Campos, P.F.; Gilbert, M.T.P.; Kolokotronis, S.-O.; Hynes, W.H.; DeSalle, R.; Daszak, P.; MacPhee, R.D.E.; Greenwood, A.D. Historical mammal extinction on Christmas Island (Indian Ocean) correlates with introduced infectious disease. PLoS ONE 2008, 3, e3602. [Google Scholar] [CrossRef] [Green Version]
- McNew, S.M.; Clayton, D.H. Alien invasion: Biology of Philornis flies highlighting Philornis downsi, an introduced parasite of Galapagos birds. Ann. Rev. Entomol. 2018, 63, 369–387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Causton, C.E.; Peck, S.B.; Sinclair, B.J.; Roque-Albelo, L.; Hodgson, C.J.; Landry, B. Alien insects: Threats and implications for conservation of Galápagos Islands. Ann. Entomol. Soc. Am. 2006, 99, 121–143. [Google Scholar] [CrossRef]
- O’Connor, J.A.; Sulloway, F.J.; Robertson, J.; Kleindorfer, S. Philornis downsi parasitism is the primary cause of nestling mortality in the critically endangered Darwin’s medium tree finch (Camarhynchus pauper). Biodivers. Conserv. 2010, 19, 853–866. [Google Scholar] [CrossRef]
- Fessl, B.; Young, G.H.; Young, R.P.; Rodríguez-Matamoros, J.; Dvorak, M.; Tebbich, S.; Fa, J.E. How to save the rarest Darwin’s finch from extinction: The mangrove finch on Isabela Island. Philos. Trans. R. Soc. B 2010, 365, 1019–1030. [Google Scholar] [CrossRef] [PubMed]
- Fessl, B.; Heimpel, G.E.; Causton, C.E. Invasion of an avian nest parasite, Philornis downsi, to the Galapagos Islands: Colonization history, adaptations to novel ecosystems, and conservation challenges. In Disease Ecology: Galapagos Birds and Their Parasites; Parker, P.G., Ed.; Springer International Publishing: Cham, Switzerland, 2018; pp. 213–268. [Google Scholar]
- Koop, J.A.H.; Huber, S.K.; Laverty, S.M.; Clayton, D.H. Experimental demonstration of the fitness consequences of an introduced parasite of Darwin’s finches. PLoS ONE 2011, 6, e19706. [Google Scholar] [CrossRef]
- Knutie, S.A.; McNew, S.M.; Bartlow, A.W.; Vargas, D.A.; Clayton, D.H. Darwin’s finches combat introduced nest parasites with fumigated cotton. Curr. Biol. 2014, 24, R355–R356. [Google Scholar] [CrossRef] [Green Version]
- Ben-Yosef, M.; Zaada, D.S.Y.; Dudaniec, R.Y.; Pasternak, Z.; Jurkevitch, E.; Smith, R.J.; Causton, C.E.; Lincango, M.P.; Tobe, S.S.; Mitchell, J.G.; et al. Host-specific associations affect the microbiome of Philornis downsi, an introduced parasite to the Galápagos Islands. Mol. Ecol. 2017, 26, 4644–4656. [Google Scholar] [CrossRef]
- Podlipaev, S. The more insect trypanosomatids under study - the more diverse Trypanosomatidae appears. Int. J. Parasitol. 2001, 31, 648–652. [Google Scholar] [CrossRef]
- Podlipaev, S.A. Insect trypanosomatids: The need to know more. Mem. Inst. Oswaldo Cruz 2000, 95, 517–522. [Google Scholar] [CrossRef]
- Undeen, A.H.; Vávra, J. Research methods for entomopathogenic protozoa. In Manual of Techniques in Insect Pathology, 1st ed.; Lacey, L., Ed.; Academic Press: London, UK, 1997; pp. 117–151. [Google Scholar]
- Lange, C.E.; Lord, J.C. Protistan entomopathogens. In Insect Pathology, 2nd ed.; Vega, F.E., Kaya, H.K., Eds.; Academic Press: London, UK, 2012; pp. 367–394. [Google Scholar]
- Erler, S.; Popp, M.; Wolf, S.; Lattorff, H.M.G. Sex, horizontal transmission, and multiple hosts prevent local adaptation of Crithidia bombi, a parasite of bumblebees (Bombus spp.). Ecol. Evol. 2012, 2, 930–940. [Google Scholar] [CrossRef]
- Olsen, O.W. Animal Parasites: Their Life Cycles and Ecology, 3rd ed.; Dover Publications: Mineola, NY, USA, 1986; pp. 21–24. [Google Scholar]
- Dias, F.A.; Vasconcellos, L.R.C.; Romeiro, A.; Attias, M.; Souto-Padrón, T.C.; Lopes, A.H. Transovum transmission of trypanosomatid cysts in the Milkweed Bug, Oncopeltus fasciatus. PLoS ONE 2014, 9, e108746. [Google Scholar] [CrossRef] [Green Version]
- Frolov, A.O.; Malysheva, M.N.; Ganyukova, A.I.; Yurchenko, V.; Kostygov, A.Y. Life cycle of Blastocrithidia papi sp. n. (Kinetoplastea, Trypanosomatidae) in Pyrrhocoris apterus (Hemiptera, Pyrrhocoridae). Eur. J. Protistol. 2017, 57, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Conchon, I.; Campaner, M.; Sbravate, C.; Camargo, E.P. Trypanosomatids, other than Phytomonas spp. isolated and cultured from fruit. J. Protozool. 1989, 36, 412–414. [Google Scholar] [CrossRef]
- Schaub, G.A.; Jensen, C. Developmental time and mortality of the reduviid bug Triatoma infestans with differential exposure to coprophagic infections with Blastocrithidia triatomae (Trypanosomatidae). J. Invertebr. Pathol. 1990, 55, 17–27. [Google Scholar] [CrossRef]
- Brown, M.J.F.; Schmid-Hempel, R.; Schmid-Hempel, P. Strong context-dependent virulence in a host-parasite system: Reconciling genetic evidence with theory. J. Anim. Ecol. 2003, 72, 994–1002. [Google Scholar] [CrossRef] [Green Version]
- Ibraham, E.A.; Molyneux, D.H. Pathogenicity of Crithidia fasciculata in the haemocoele of Glossina. Acta Trop. 1987, 44, 13–22. [Google Scholar]
- Arnquist, G.; Mäki, M. Infection rates and pathogenicity of trypanosomatid gut parasites in the water strider Gerris odontogaster (Zett.) (Heteroptera: Gerridae). Oecologia 1990, 84, 194–198. [Google Scholar] [CrossRef]
- Tanada, Y.; Kaya, H.K. Protozoan infections: Zoomastigina, rhizopoda, and ciliophoran. In Insect Pathology, 1st ed.; Tanada, Y., Kaya, H., Eds.; Academic Press: San Diego, CA, USA, 1993; pp. 388–397. [Google Scholar]
- Lipa, J.J.; Carl, K.P.; Valentine, E.W. Blastocrithidia caliroae sp. n., a flagellate parasite of Caliroa cerasi (L.) (Hymenoptera: Tenthredinidae) and notes on its epizootics in host field populations. Acta Protozool. 1977, 16, 121–129. [Google Scholar]
- Bartlett-Healy, K.; Crans, W.; Gaugler, R. Vertebrate hosts and phylogenetic relationships of amphibian trypanosomes from a potential invertebrate vector, Culex territans Walker (Diptera: Culicidae). J. Parasitol. 2009, 95, 381–387. [Google Scholar] [CrossRef]
- Bennett, G.F. On the specificity and transmission of some avian trypanosomes. Can. J. Zool. 1961, 39, 17–33. [Google Scholar] [CrossRef]
- Ramos, B.; Urdaneta-Morales, S. Hematophagous insects as vectors for frog trypanosomes. Rev. Biol. Trop. 1978, 25, 209–217. [Google Scholar]
- Wamwiri, F.N.; Changasi, R.E. Tsetse flies (Glossina) as vectors of human African trypanosomiasis: A review. Biomed. Res. Int. 2016, 2016, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hartman, D. Galapagos Mosquitoes as Avian Disease Vectors. Master’s Thesis, University of Missouri-St. Louis, St. Louis, MO, USA, 2014. [Google Scholar]
- Wallace, F.G. The trypanosomatid parasites of insects and arachnids. Exp. Parasitol. 1966, 18, 124–193. [Google Scholar] [CrossRef]
- Bulat, S.A.; Mokrousov, I.V.; Podlipaev, S.A. Classification of trypanosomatids from insects and plants by the UP-PCR (Universally Primed PCR) technique and cross dot blot hybridization of PCR products. Eur. J. Protistol. 1999, 35, 319–326. [Google Scholar] [CrossRef]
- Podlipaev, S.A.; Sturm, N.R.; Fiala, I.; Fernandes, O.; Westenberger, S.J.; Dollet, M.; Campbell, D.A.; Lukeš, J. Diversity of insect trypanosomatids assessed from the spliced leader RNA and 5S rRNA genes and intergenic regions. J. Eukaryot. Microbiol. 2004, 51, 283–290. [Google Scholar] [CrossRef]
- Causton, C.E.; Moon, R.D.; Cimadom, A.; Boulton, R.A.; Cedeño, D.; Lincango, M.P.; Tebbich, S.; Ulloa, A. Population dynamics of an invasive bird parasite, Philornis downsi (Diptera: Muscidae), in the Galapagos Islands. PLoS ONE 2019, 14, e0224125. [Google Scholar] [CrossRef] [Green Version]
- Maslov, D.A.; Lukeš, J.; Jirku, M.; Simpson, L. Phylogeny of trypanosomes as inferred from the small and large subunit rRNAs: Implication for the evolution of parasitism in the trypanosomatid protozoa. Mol. Biochem. Parasitol. 1996, 75, 197–205. [Google Scholar] [CrossRef]
- Votýpka, J.; Szabová, J.; Rádrová, J.; Zídková, L.; Svobodová, M. Trypanosoma culicavium sp. nov., an avian trypanosome transmitted by Culex mosquitoes. Int. J. Syst. Evol. Microbiol. 2012, 62, 745–754. [Google Scholar] [CrossRef] [Green Version]
- Sehgal, R.M.N.; Jones, H.I.; Smith, T.B. Host specificity and incidence of Trypanosoma in some African rainforest birds: A molecular approach. Mol. Ecol. 2001, 10, 2319–2327. [Google Scholar] [CrossRef]
- Nzelu, C.O.; Kato, H.; Puplampu, N.; Desewu, K.; Odoom, K.; Wilson, M.D.; Sakurai, T.; Katakura, K.; Boakye, D. First detection of Leishmania tropica DNA and Trypanosoma species in Sergentomyia sand flies (Diptera: Psychodidae) from an outbreak area of cutaneous leishmaniasis in Ghana. PLoS Negl. Trop. Dis. 2014, 8, e2630. [Google Scholar] [CrossRef] [Green Version]
- Van Dyken, M.; Bolling, B.G.; Moore, C.G.; Blair, C.D.; Beaty, B.J.; Black, W.C., IV; Foy, B.D. Molecular evidence for trypanosomatids in Culex mosquitoes collected during a West Nile virus survey. Int. J. Parasitol. 2006, 36, 1015–1023. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kimura, M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Suchard, M.A.; Lemey, P.; Baele, G.; Ayres, D.L.; Drummond, A.J.; Rambaut, A. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 2018, 4, vey016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bouckaert, R.R.; Drummond, A.J. Bmodeltest: Bayesian phylogenetic site model averaging and model comparison. BMC Evol. Biol. 2017, 17, 42. [Google Scholar] [CrossRef] [Green Version]
- Stöver, B.C.; Müller, K.F. TreeGraph 2: Combining and visualizing evidence from different phylogenetic analyses. BMC Bioinform. 2010, 11, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pike, C.; Lincango, P.; Causton, C.; Parker, P. Sequence Alignment Data for Trypanosomatidae Parasites Found in Philornis downsi from the Galapagos Islands; Version 2; Mendeley Repository: Amsterdam, The Netherlands, 2020; Available online: http://dx.doi.org/10.17632/c6wsxd2jz5.2 (accessed on 19 March 2020).
- Jukes, T.H.; Cantor, C.R. Evolution of protein molecules. In Mammalian Protein Metabolism; Munro, H.N., Ed.; Academic Press: New York, NY, USA, 1969; pp. 21–132. [Google Scholar]
- Solter, L.F.; Becnel, J.J.; Vavra, J. Research methods for entomopathogenic microsporidia and other protists. In Manual of Techniques in Invertebrate Pathology, 2nd ed.; Lacey, L.A., Ed.; Elsevier: San Diego, CA, USA, 2012; pp. 329–371. [Google Scholar]

| Island | Site | Total Sample Pools (# Total Flies) | Molecular Tests (# Positive Pools/# Total Tested Pools) | MIR (%) | |
|---|---|---|---|---|---|
| [44] 1 | [45] 1 | ||||
| Santa Cruz | Lowland, dry (El Barranco) | 165 (376) | 138/165 | 19/40 | 36.7 |
| Agricultural zone (Los Guayabillos) | 2 (2) | 2/2 | 0/0 | 100.0 | |
| Highland, humid (Los Gemelos) | 110 (267) | 109/110 | 18/43 | 40.8 | |
| Isabela | Mangrove forest (Playa Tortuga Negra) | 17 (45) | 14/17 | 3/9 | 31.1 |
| Lava field (Playa Tortuga Negra) | 3 (4) | 3/3 | 0/3 | 75.0 | |
| Total | 297 (694) | 267/297 | 40/95 | 38.5 | |
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Trypanosomatidae sp. P041 | 0.17 | 0.14 | 0.30 | 0.32 | 0.19 | 0.29 | 0.43 | 1.58 | 1.44 | 1.58 | 1.35 | 1.41 | 1.44 | 1.54 | 1.48 | 1.46 | |
| 2 | Trypanosomatidae sp. P034 | 0.32 | 0.11 | 0.32 | 0.34 | 0.26 | 0.34 | 0.49 | 1.53 | 1.42 | 1.53 | 1.34 | 1.41 | 1.41 | 1.51 | 1.44 | 1.43 | |
| 3 | Trypanosomatidae sp. P057 | 0.21 | 0.11 | 0.34 | 0.35 | 0.24 | 0.32 | 0.46 | 1.53 | 1.42 | 1.53 | 1.34 | 1.41 | 1.41 | 1.51 | 1.44 | 1.42 | |
| 4 | Trypanosomatidae sp. P120 | 0.86 | 0.97 | 1.08 | 0.21 | 0.37 | 0.41 | 0.50 | 1.57 | 1.40 | 1.57 | 1.32 | 1.37 | 1.45 | 1.52 | 1.48 | 1.47 | |
| 5 | Blastocrithidia miridarum | 0.97 | 1.08 | 1.19 | 0.43 | 0.39 | 0.42 | 0.54 | 1.57 | 1.40 | 1.57 | 1.31 | 1.38 | 1.43 | 1.53 | 1.47 | 1.46 | |
| 6 | Crithidia bombi | 0.32 | 0.65 | 0.54 | 1.19 | 1.30 | 0.36 | 0.50 | 1.57 | 1.45 | 1.57 | 1.35 | 1.42 | 1.46 | 1.54 | 1.50 | 1.48 | |
| 7 | Crithidia confusa | 0.75 | 1.08 | 0.97 | 1.52 | 1.63 | 1.08 | 0.53 | 1.59 | 1.43 | 1.59 | 1.34 | 1.37 | 1.41 | 1.51 | 1.44 | 1.42 | |
| 8 | Crithidia dedva | 1.75 | 2.08 | 1.97 | 2.19 | 2.53 | 2.08 | 2.42 | 1.54 | 1.43 | 1.54 | 1.37 | 1.47 | 1.41 | 1.46 | 1.46 | 1.47 | |
| 9 | Leptomonas collosoma | 15.12 | 14.81 | 14.81 | 15.12 | 15.27 | 14.81 | 15.12 | 14.96 | 1.56 | 0.15 | 1.50 | 1.50 | 1.48 | 1.52 | 1.43 | 1.40 | |
| 10 | Leptomonas mirabilis | 14.20 | 14.05 | 14.05 | 13.75 | 14.05 | 14.50 | 14.20 | 14.35 | 16.52 | 1.55 | 1.10 | 1.11 | 1.42 | 1.40 | 1.42 | 1.41 | |
| 11 | Leptomonas rigidus | 15.12 | 14.81 | 14.81 | 15.12 | 15.27 | 14.81 | 15.12 | 14.81 | 0.21 | 16.36 | 1.49 | 1.49 | 1.48 | 1.52 | 1.43 | 1.40 | |
| 12 | Leptomonas samueli | 13.16 | 13.01 | 13.01 | 12.86 | 12.86 | 13.16 | 13.31 | 13.75 | 15.74 | 9.49 | 15.58 | 0.80 | 1.54 | 1.62 | 1.55 | 1.55 | |
| 13 | Leptomonas costoris | 14.50 | 14.50 | 14.50 | 13.90 | 14.20 | 14.50 | 14.20 | 15.27 | 15.89 | 9.49 | 15.74 | 5.61 | 1.49 | 1.53 | 1.47 | 1.46 | |
| 14 | Trypanosoma avium | 13.90 | 13.75 | 13.60 | 13.90 | 13.75 | 14.20 | 13.45 | 13.75 | 14.96 | 15.42 | 14.96 | 16.05 | 15.74 | 0.80 | 0.63 | 0.62 | |
| 15 | Trypanosoma bennetti | 15.12 | 14.96 | 14.96 | 14.81 | 14.96 | 15.12 | 14.66 | 14.66 | 15.58 | 14.96 | 15.58 | 16.84 | 16.52 | 5.37 | 0.82 | 0.84 | |
| 16 | Trypanosoma corvi | 14.50 | 14.35 | 14.20 | 14.50 | 14.35 | 14.81 | 13.90 | 14.35 | 14.50 | 15.74 | 14.50 | 16.52 | 16.36 | 3.93 | 5.49 | 0.18 | |
| 17 | Trypanosoma culicavium | 14.20 | 14.05 | 13.90 | 14.20 | 14.05 | 14.50 | 13.60 | 14.50 | 14.05 | 15.74 | 14.05 | 16.52 | 16.36 | 4.04 | 5.86 | 0.32 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pike, C.L.; Lincango, M.P.; Causton, C.E.; Parker, P.G. Trypanosomatids Detected in the Invasive Avian Parasite Philornis downsi (Diptera: Muscidae) in the Galapagos Islands. Insects 2020, 11, 422. https://doi.org/10.3390/insects11070422
Pike CL, Lincango MP, Causton CE, Parker PG. Trypanosomatids Detected in the Invasive Avian Parasite Philornis downsi (Diptera: Muscidae) in the Galapagos Islands. Insects. 2020; 11(7):422. https://doi.org/10.3390/insects11070422
Chicago/Turabian StylePike, Courtney L., María Piedad Lincango, Charlotte E. Causton, and Patricia G. Parker. 2020. "Trypanosomatids Detected in the Invasive Avian Parasite Philornis downsi (Diptera: Muscidae) in the Galapagos Islands" Insects 11, no. 7: 422. https://doi.org/10.3390/insects11070422





