RNA Interference of Genes Encoding the Vacuolar-ATPase in Liriomyza trifolii
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Insects
2.2. Cloning, Sequence Alignment, and Expression of V-ATPase Genes
2.3. Synthesis of dsRNA and Delivery by Microinjection
2.4. Analysis of Silencing Efficiency
2.5. Statistical Analyses
3. Results
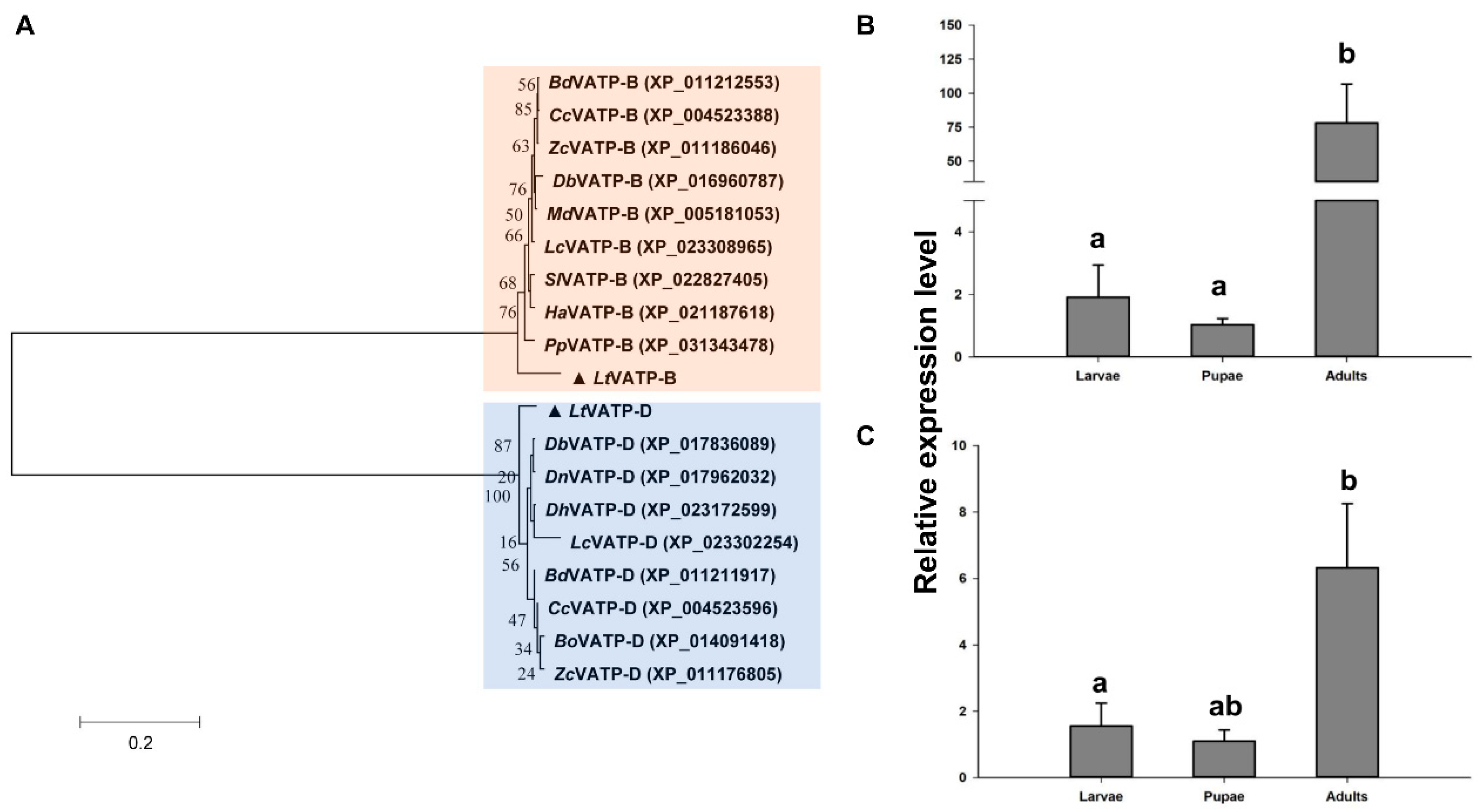
3.1. Sequence Analysis and Expression Patterns of V-ATPase Genes in L. trifolii
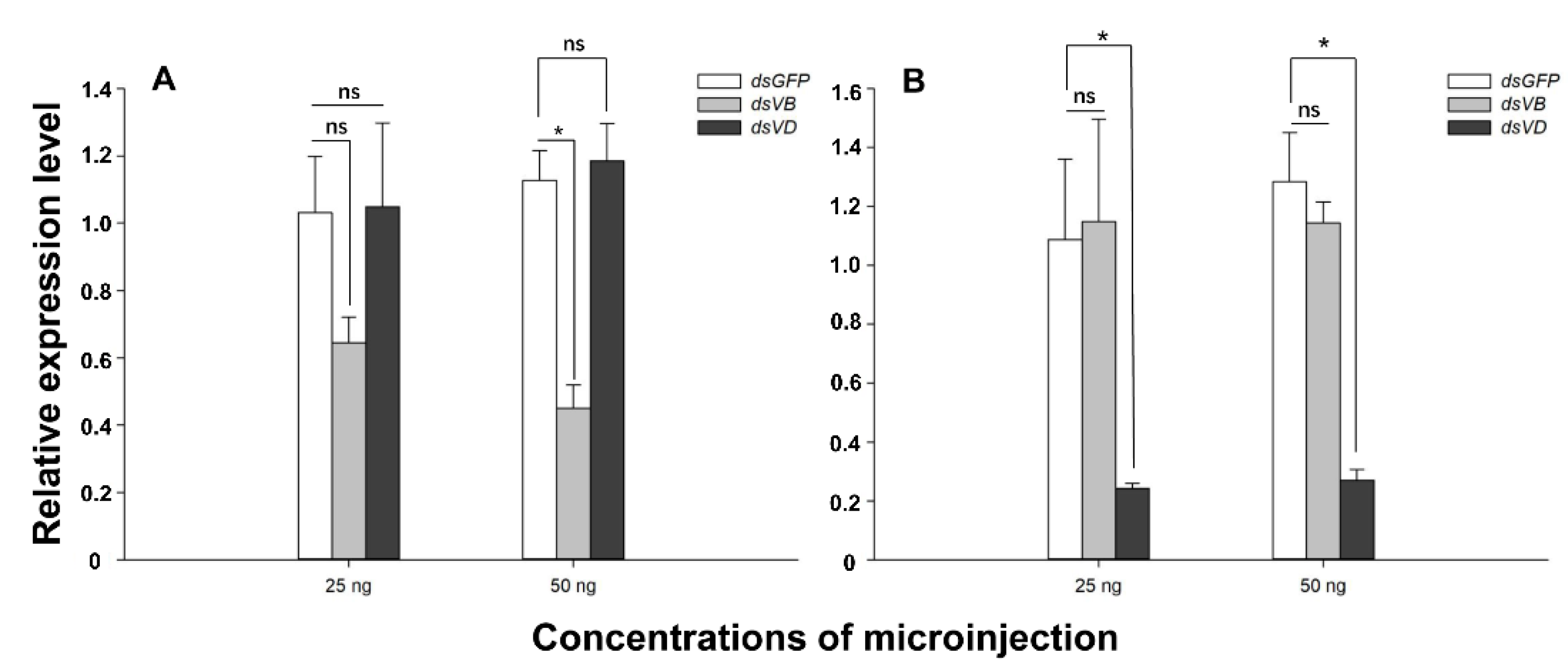
3.2. Knockdown of V-ATPase B and V-ATPase D Expression
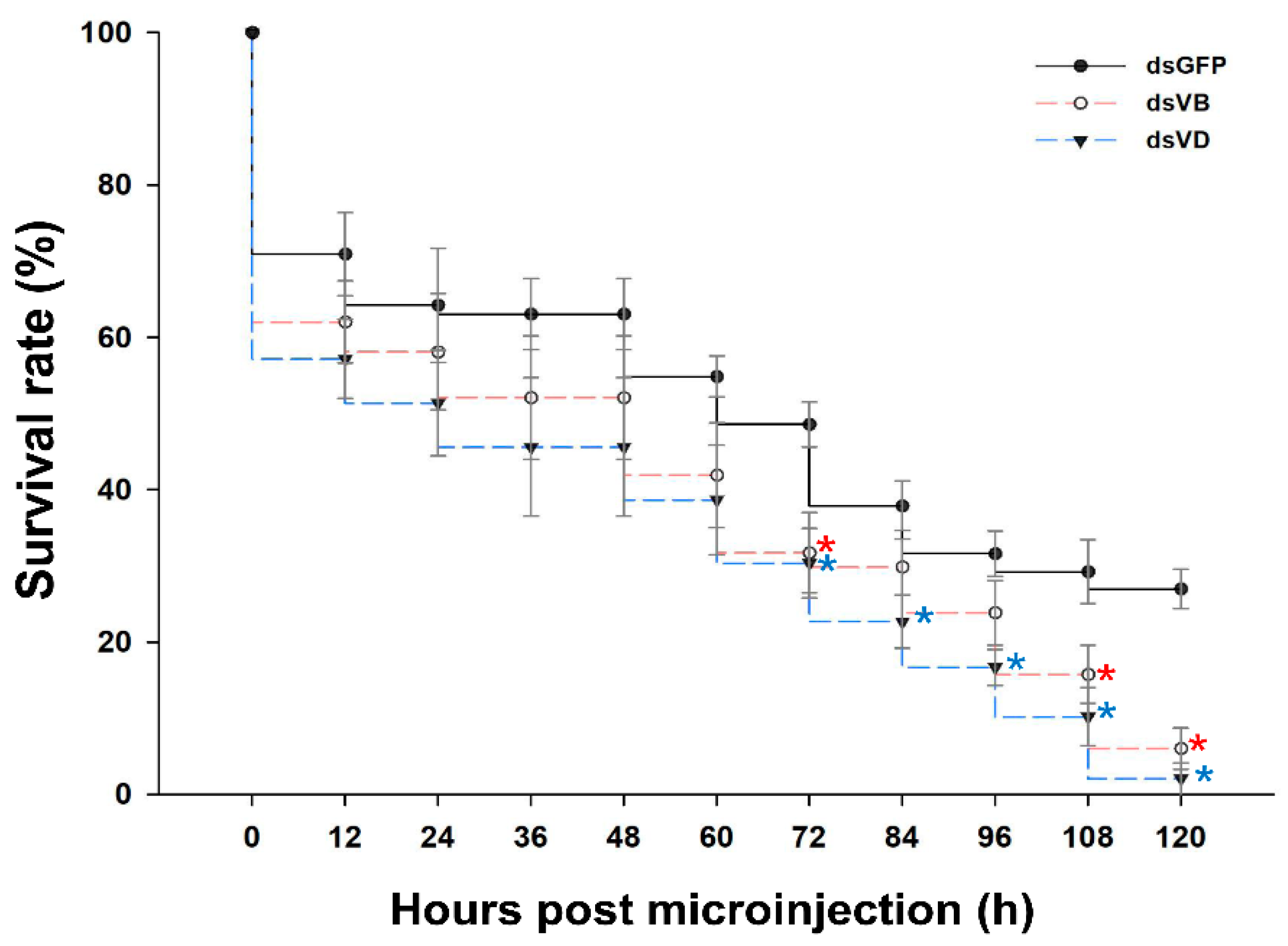
3.3. Mortality of L. trifolii Injected with dsV-ATPase-B and dsV-ATPase-D
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Spencer, K.A. Agromyzidae (Diptera) of Economic Importance. 9: Series Entomologica; The Hague Publishers: Bath, UK, 1973; pp. 19–28. [Google Scholar]
- Johnson, M.W.; Welter, S.C.; Toscano, N.C.; Tingi, P.; Trumble, J.T. Reduction of tomato leaflet photosynthesis rates by mining activity of Liriomyza sativae (Diptera: Agromyzidae). J. Econ. Entomol. 1983, 76, 1061–1063. [Google Scholar] [CrossRef]
- Parrella, M.P.; Jones, V.P.; Youngman, R.R.; Lebeck, L.M. Effect of leaf mining and leaf stippling of Liriomyza spp. on photosynthetic rates of chrysanthemum. Ann. Entomol. Soc. A 1985, 78, 90–93. [Google Scholar] [CrossRef]
- Reitz, S.R.; Kund, G.S.; Carson, W.G.; Phillips, P.A.; Trumble, J.T. Economics of reducing insecticide use on celery through low-input pest management strategies. Agric. Ecosyst. Environ. 1999, 73, 185–197. [Google Scholar] [CrossRef]
- Scheffer, S.J.; Lewis, M.L. Mitochondrial phylogeography of the vegetable pest Liriomyza trifolii (Diptera: Agromyzidae): Diverged clades and invasive populations. Ann. Entomol. Soc. Am. 2006, 99, 991–998. [Google Scholar] [CrossRef]
- Wang, Z.G.; Guan, W.; Chen, D.H. Preliminary report of the Liriomyza trifolii in Zhongshan area. Plant Quar. 2007, 21, 19–20. [Google Scholar]
- Gao, Y.L.; Reitz, S.; Xing, Z.L.; Ferguson, S.; Lei, Z.R. A decade of a leafminer invasion in China: Lessons learned. Pest Manag. Sci. 2017, 73, 1775–1779. [Google Scholar] [CrossRef]
- Kang, L.; Chen, B.; Wei, J.N.; Liu, T.X. Roles of thermal adaptation and chemical ecology in Liriomyza distribution and control. Annu. Rev. Entomol. 2009, 54, 127–145. [Google Scholar] [CrossRef]
- Xiang, J.C.; Lei, Z.R.; Wang, H.H. Interspecific competition among three invasive Liriomyza species. Acta Ecol. Sin. 2012, 32, 1616–1622. [Google Scholar] [CrossRef]
- Wan, F.H.; Yang, N.W. Invasion and management of agricultural alien insects in China. Annu. Rev. Entomol. 2016, 61, 77–98. [Google Scholar] [CrossRef]
- Wen, J.Z.; Wang, Y.; Lei, Z.R. New record of Liriomyza sativae Blanchard (Diptera: Agromyzidae) from China. Entomotaxonomia 1996, 18, 311–312. [Google Scholar]
- Wen, J.Z.; Lei, Z.R.; Wang, Y. Survey of Liriomyza huidobrensis in Yunnan Province and Guizhou Province, China. Plant Prot. 1998, 24, 18–20. [Google Scholar]
- Gao, Y.L.; Lei, Z.R.; Abe, Y.; Reitz, S.R. Species displacements are common to two invasive species of leafminer fly in China, Japan, and the United States. J. Econ. Entomol. 2011, 104, 1771–1773. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.L.; Reitz, S.R.; Wei, Q.B.; Yu, W.Y.; Lei, Z.R. Insecticide-mediated apparent displacement between two invasive species of leafminer fly. PLoS ONE 2012, 7, e36622. [Google Scholar] [CrossRef] [PubMed]
- Wei, Q.B.; Lei, Z.R.; Nauen, R.; Cai, D.C.; Gao, Y.L. Abamectin resistance in strains of vegetable leafminer, Liriomyza sativae (Diptera: Agromyzidae) is linked to elevated glutathione S-transferase activity. Insect Sci. 2015, 22, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.H.; Reitz, S.R.; Xiang, J.C.; Smagghe, G.; Lei, Z.R. Does Temperature-Mediated Reproductive Success Drive the Direction of Species Displacement in Two Invasive Species of Leafminer Fly? PLoS ONE 2014, 9, e98761. [Google Scholar] [CrossRef]
- Chang, Y.W.; Chen, J.Y.; Lu, M.X.; Gao, Y.; Tian, Z.H.; Gong, W.R.; Dong, C.S.; Du, Y.Z. Cloning and expression of genes encoding heat shock proteins in Liriomyza trifolii and comparison with two congener leafminer species. PLoS ONE 2017, 12, e0181355. [Google Scholar] [CrossRef]
- Reitz, S.R.; Trumble, J.T. Competitive displacement among insects and arachnids. Annu. Rev. Entomol. 2002, 47, 435–465. [Google Scholar] [CrossRef]
- Gao, Y.L.; Reitz, S.R. Emerging themes in our understanding of species displacements. Annu. Rev. Entomol. 2017, 62, 165–183. [Google Scholar] [CrossRef]
- Abe, Y.; Tokumaru, S. Displacement in two invasive species of leafminer fly in different localities. Biol. Invasions 2008, 10, 951–995. [Google Scholar] [CrossRef]
- Palumbo, J.C.; Mullis, C.H.; Reyes, F.J., Jr. Composition, seasonal abundance, and parasitism of Liriomyza (Diptera: Agromyzidae) species on lettuce in Arizona. J. Econ. Entomol. 1994, 87, 1070–1077. [Google Scholar] [CrossRef]
- Tokumaru, S.; Kurita, H.; Fukui, M.; Abe, Y. Insecticide susceptibility of Liriomyza sativae, L. trifolii, and L. bryoniae (Diptera: Agromyzidae). Jpn. J. Appl. Entomol. Z. 2005, 49, 1–10. [Google Scholar] [CrossRef]
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef] [PubMed]
- Joga, M.R.; Zotti, M.J.; Smagghe, G.; Christiaens, O. RNAi Efficiency, systemic properties, and novel delivery methods for pest insect control: What we know so far. Front. Physiol. 2016, 7, 553. [Google Scholar] [CrossRef] [PubMed]
- Bellés, X. Beyond Drosophila: RNAi in vivo and functional genomics in insects. Annu. Rev. Entomol. 2010, 55, 111–128. [Google Scholar] [CrossRef]
- Gordon, K.H.; Waterhouse, P.M. RNAi for insect-proof plants. Nat. Biotechnol. 2007, 25, 1231–1232. [Google Scholar] [CrossRef]
- Burand, J.P.; Hunter, W.B. RNAi: Future in insect management. J. Invertebr. Pathol. 2013, 112, S68–S74. [Google Scholar] [CrossRef]
- Palli, S.R. RNA interference in Colorado potato beetle: Steps toward development of dsRNA as a commercial insecticide. Curr. Opin. Insect Sci. 2014, 6, 1–8. [Google Scholar] [CrossRef]
- Baum, J.A.; Bogaert, T.; Clinton, W.; Heck, G.R.; Feldmann, P.; Ilagan, O.; Johnson, S.; Plaetinck, G.; Munyikwa, T.; Pleau, M.; et al. Control of coleopteran insect pests through RNA interference. Nat. Biotechnol. 2007, 25, 1322–1326. [Google Scholar] [CrossRef]
- Yao, J.; Rotenberg, D.; Afsharifar, A.; Barandoc-Alviar, K.; Whitfield, A.E. Development of RNAi methods for Peregrinus maidis, the corn planthopper. PLoS ONE 2013, 8, e70243. [Google Scholar] [CrossRef]
- Nelson, N.; Perzov, N.; Cohen, A.; Hagai, K.; Padler, V.; Nelson, H. The cellular biology of proton-motive force generation by V-ATPases. J. Exp. Biol. 2000, 203, 89–95. [Google Scholar]
- Badillo-Vargas, I.E.; Rotenberg, D.; Schneweis, B.A.; Whitfield, A.E. RNA interference tools for the western flower thrips, Frankliniella occidentalis. J. Insect Physiol. 2015, 76, 36–46. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Kang, L. Cold hardiness and supercooling capacity in the pea leafminer Liriomyza huidobrensis. Cryo Lett. 2002, 23, 173–182. [Google Scholar]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The Clustal-X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA 6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.W.; Chen, J.Y.; Lu, M.X.; Gao, Y.; Tian, Z.H.; Gong, W.R.; Zhu, W.; Du, Y.Z. Selection and validation of reference genes for quantitative real time PCR analysis under different experimental conditions in the leafminer Liriomyza trifolii (Diptera: Agromyzidae). PLoS ONE 2017, 12, e0181862. [Google Scholar] [CrossRef]
- Meyering-Vos, M.; Muller, A. RNA interference suggests sulfakinins as satiety effectors in the cricket Gryllus bimaculatus. J. Insect Physiol. 2007, 53, 840–848. [Google Scholar] [CrossRef]
- Shakesby, A.J.; Wallace, I.S.; Pritchard, H.V.; Roberts, D.M.; Douglas, A.E. A water-specific aquaporin involved in aphid osmoregulation. Insect Biochem. Mol. Biol. 2009, 39, 1–10. [Google Scholar] [CrossRef]
- Wang, K.; Peng, Y.; Pu, J.; Fu, W.; Wang, J. Variation in RNAi efficacy among insect species is attributable to dsRNA degradation in vivo. Insect Biochem. Mol. Biol. 2016, 77, 1–9. [Google Scholar] [CrossRef]
- Li, J.; Chen, Q.; Lin, Y.; Jiang, T.; Wu, G.; Hua, H. RNA interference in Nilaparvata lugens (Homoptera: Delphacidae) based on dsRNA ingestion. Pest Manag. Sci. 2011, 67, 852–859. [Google Scholar] [CrossRef]
- Wieczorek, H.; Beyenbach, K.W.; Huss, M.; Vitavska, O. Vacuolar-type proton pumps in insect epithelia. J. Exp. Biol. 2009, 212, 1611–1619. [Google Scholar] [CrossRef]
- Davies, S.; Goodwin, S.; Kelly, D.; Wang, Z.; Sozen, M.; Kaiser, K.; Dow, J. Analysis and inactivation of vha55, the gene encoding the vacuolar ATPase B subunit in Drosophila melanogaster reveals a larval lethal phenotype. J. Biol. Chem. 1996, 271, 30677–30684. [Google Scholar] [CrossRef] [PubMed]
- Whyard, S.; Singh, A.D.; Wong, S. Ingested double-stranded RNAs can act as species-specific insecticides. Insect Biochem. Mol. Biol. 2009, 39, 824–832. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Xu, J.; Palli, R.; Ferguson, J.; Palli, S.R. Ingested RNA interference for managing the populations of the Colorado potato beetle, Leptinotarsa decemlineata. Pest Manag. Sci. 2011, 67, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Wuriyanghan, H.; Rosa, C.; Falk, B.W. Oral delivery of double-stranded RNAs and siRNAs induces RNAi effects in the potato/tomato psyllid, Bactericerca cockerelli. PLoS ONE 2011, 6, e27736. [Google Scholar] [CrossRef] [PubMed]
- Nunes, F.M.; Simões, Z.L. A non-invasive method for silencing gene transcription in honeybees maintained under natural conditions. Insect Biochem. Mol. Biol. 2009, 39, 157–160. [Google Scholar] [CrossRef] [PubMed]
- Walshe, D.P.; Lehane, S.M.; Lehane, M.J.; Haines, L.R. Prolonged gene knockdown in the tsetse fly Glossina by feeding double stranded RNA. Insect Mol. Biol. 2009, 18, 11–19. [Google Scholar] [CrossRef]
- Rangasamy, M.; Siegfried, B.D. Validation of RNA interference in western corn rootworm Diabrotica virgifera virgifera LeConte (Coleoptera: Chrysomelidae) adults. Pest Manag. Sci. 2012, 68, 587–591. [Google Scholar] [CrossRef]
- Basnet, S.; Kamble, S.T. Knockdown of the chromatin remodeling gene brahma by RNA interference reduces reproductive fitness and lifespan in common Bed Bug (Hemiptera: Cimicidae). J. Med. Entomol. 2017, 55, 534–539. [Google Scholar] [CrossRef]
- Basnet, S.; Kamble, S.T. RNAi-mediated knockdown of V-ATPase subunits affects survival and reproduction in bed bugs (Hemiptera: Cimicidae). J. Med. Entomol. 2018, 55, 540–546. [Google Scholar] [CrossRef]
- Parrella, M.P. Biology of Liriomyza. Annu. Rev. Entomol. 1987, 32, 201–224. [Google Scholar] [CrossRef]


| Gene | Primer Sequences (5′→3′) | Fragment Length (bp) | |
|---|---|---|---|
| Primers for cDNA Cloning and Full-Length cDNA Amplification | |||
| V-ATPase B | F | ACAAAACAGTGACAGGCGTGAAT | 669 |
| R | TTGTAGGGTCATTAGCCAGGTTT | ||
| 5′ | GCCAGCACTTGTTCGTAGTAGCCTTT | 200 | |
| 3′ | GGTCGTCGTGGTTTCCCTGGTTACAT | 920 | |
| V-ATPase D | F | TCTGCCCGTGTTTGAGTCCTATC | 365 |
| R | CCTGTTGGAGCAGTTCAGCGTTA | ||
| 5′ | ACACGGGCAGAGTGACACCAGCAAC | 398 | |
| 3′ | ACGCTGAACTGCTCCAACAGGGTA | 196 | |
| Primers for dsRNA synthesis | |||
| dsV-ATPB | F | TAATACGACTCACTATAGGGAG AACTTTCAATGGGTCTGGTA | 412 |
| R | TAATACGACTCACTATAGGGAG GTCATTAGCCAGGTTTAGAA | ||
| dsV-ATPD | F | TAATACGACTCACTATAGGGAG TGAGTCCTATCAAGATGGTT | 284 |
| R | TAATACGACTCACTATAGGGAG TCTTCAAACGATAGAACTCC | ||
| dsGFP | F | TAATACGACTCACTATAGGGAGA CCTCGTGACCACCCTGACCTAC | 314 |
| R | TAATACGACTCACTATAGGGAGA CACCTTGATGCCGTTCTTCTGC | ||
| Primers for qRT-PCR | |||
| V-ATPase B | F | ACAAAACAGTGACAGGCGTGAATG | 92 |
| R | CGGCAAAGTCAAGTAAACTATCTC | ||
| V-ATPase D | F | GGCGGAAGCCAAGTTCACTAC | 115 |
| R | GGGCAGAGTGACACCAGCAAC | ||
| ACTIN | F | TTGTATTGGACTCTGGTGACGG | 73 |
| R | GATAGCGTGAGGCAAAGCATAA | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, Y.-W.; Wang, Y.-C.; Zhang, X.-X.; Iqbal, J.; Du, Y.-Z. RNA Interference of Genes Encoding the Vacuolar-ATPase in Liriomyza trifolii. Insects 2021, 12, 41. https://doi.org/10.3390/insects12010041
Chang Y-W, Wang Y-C, Zhang X-X, Iqbal J, Du Y-Z. RNA Interference of Genes Encoding the Vacuolar-ATPase in Liriomyza trifolii. Insects. 2021; 12(1):41. https://doi.org/10.3390/insects12010041
Chicago/Turabian StyleChang, Ya-Wen, Yu-Cheng Wang, Xiao-Xiang Zhang, Junaid Iqbal, and Yu-Zhou Du. 2021. "RNA Interference of Genes Encoding the Vacuolar-ATPase in Liriomyza trifolii" Insects 12, no. 1: 41. https://doi.org/10.3390/insects12010041
APA StyleChang, Y.-W., Wang, Y.-C., Zhang, X.-X., Iqbal, J., & Du, Y.-Z. (2021). RNA Interference of Genes Encoding the Vacuolar-ATPase in Liriomyza trifolii. Insects, 12(1), 41. https://doi.org/10.3390/insects12010041





