The Transcriptomic Landscape of Molecular Effects after Sublethal Exposure to Dinotefuran on Apis mellifera
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Honey Bees
2.2. Exposure to Sublethal Concentrations of Dinotefuran
2.3. RNA Preparation and Library Construction
2.4. RNA Sequencing and Analysis
2.5. Real-Time Quantitative PCR
2.6. Statistical Analysis
3. Results
3.1. Acute (48 h) Oral Toxicity of Dinotefuran
3.2. Mapping RNA-seq Reads to the Apis Mellifera Genome
3.3. Differential Gene Expression Analysis
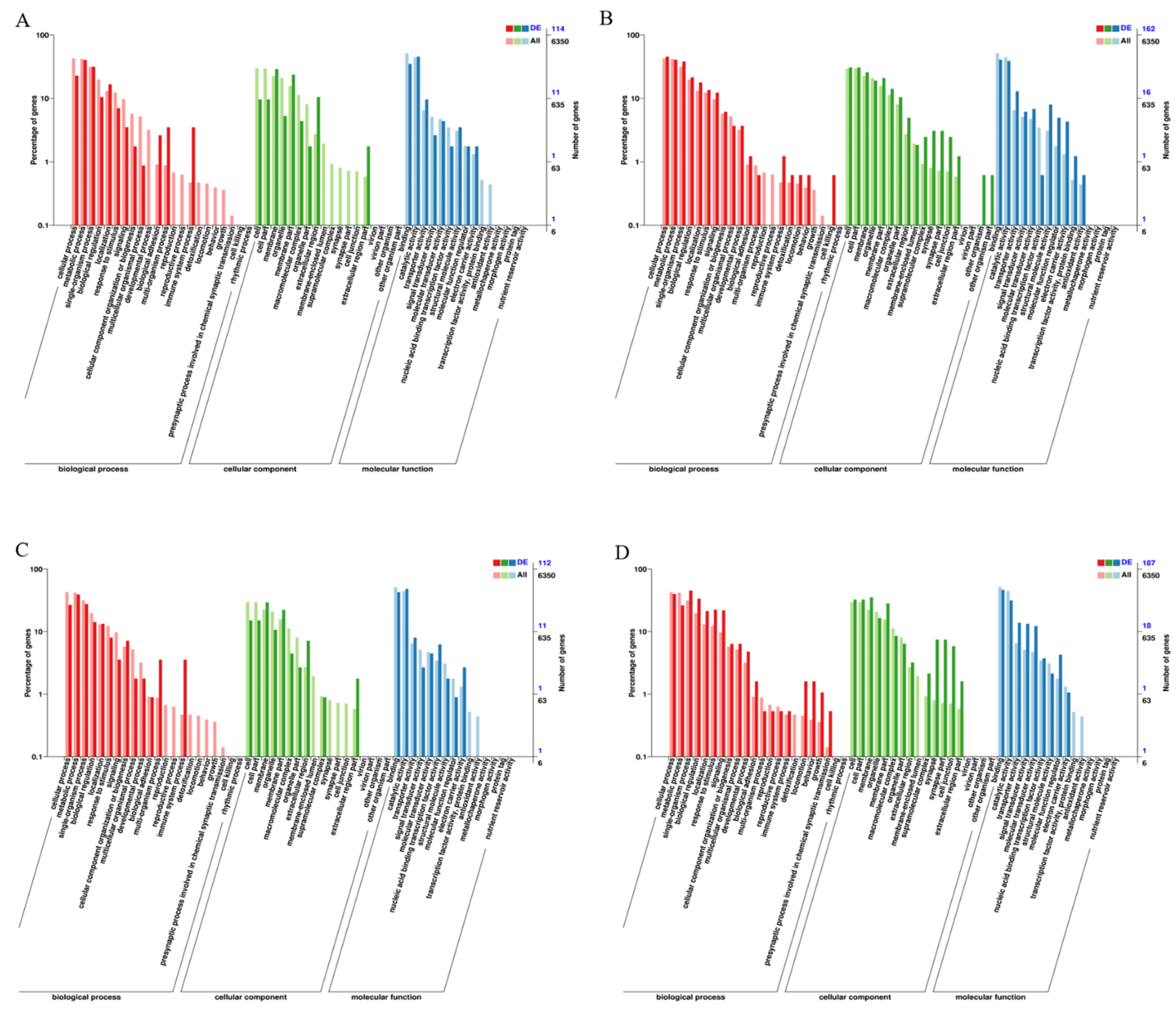
3.4. Analysis of GO Enrichment and KEGG Enrichment of DEGs
3.5. Validation of DEGs by qPCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Moritz, R.F.A.; de Miranda, J.; Fries, I.; Le Conte, Y.; Neumann, P.; Paxton, R.J. Research strategies to improve honeybee health in Europe. Apidologie 2010, 41, 227–242. [Google Scholar] [CrossRef] [Green Version]
- Klein, A.-M.; Vaissiere, B.E.; Cane, J.H.; Steffan-Dewenter, I.; Cunningham, S.A.; Kremen, C.; Tscharntke, T. Importance of pollinators in changing landscapes for world crops. Proc. R. Soc. B Boil. Sci. 2007, 274, 303–313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Zhu, C.; Wang, F.; Huang, D.; Zhang, Y.; Ding, L.; Huang, H. Current research on the status of wild bees and their pollination roles. Biodivers. Sci. 2007, 15, 687. [Google Scholar] [CrossRef]
- Vanengelsdorp, D.; Evans, J.; Saegerman, C.; Mullin, C.; Haubruge, E.; Nguyen, B.K.; Frazier, M.; Frazier, J.; Cox-Foster, D.; Chen, Y.; et al. Colony Collapse Disorder: A Descriptive Study. PLoS ONE 2009, 4, e6481. [Google Scholar] [CrossRef] [PubMed]
- Farooqui, T. A potential link among biogenic amines-based pesticides, learning and memory, and colony collapse disorder: A unique hypothesis. Neurochem. Int. 2013, 62, 122–136. [Google Scholar] [CrossRef]
- Klein, M. Scientific Opinion on the science behind the development of a risk assessment of Plant Protection Products on bees (Apis mellifera, Bombusspp. and solitary bees). EFSA J. 2012, 10. [Google Scholar] [CrossRef]
- Nauen, R.; Denholm, I. Resistance of insect pests to neonicotinoid insecticides: Current status and future prospects. Arch. Insect Biochem. Physiol. 2005, 58, 200–215. [Google Scholar] [CrossRef]
- Claudianos, C.; Ranson, H.; Johnson, R.M.; Biswas, S.; Schuler, M.A.; Berenbaum, M.R.; Feyereisen, R.; Oakeshott, J.G. A deficit of detoxification enzymes: Pesticide sensitivity and environmental response in the honeybee. Insect Mol. Biol. 2006, 15, 615–636. [Google Scholar] [CrossRef] [Green Version]
- Kessler, S.C.; Tiedeken, E.J.; Simcock, K.L.; Derveau, S.; Mitchell, J.; Softley, S.; Radcliffe, A.; Stout, J.C.; Wright, G.A. Bees prefer foods containing neonicotinoid pesticides. Nature 2015, 521, 74–76. [Google Scholar] [CrossRef]
- Ma, C.; Zhang, Y.; Sun, J.; Imran, M.; Yang, H.; Wu, J.; Zou, Y.; Li-Byarlay, H.; Luo, S. Impact of acute oral exposure to thiamethoxam on the homing, flight, learning acquisition and short-term retention of Apis cerana. Pest Manag. Sci. 2019, 75, 2975–2980. [Google Scholar] [CrossRef]
- Williams, G.R.; Troxler, A.; Retschnig, G.; Roth, K.; Yañez, O.; Shutler, D.; Neumann, P.; Gauthier, L. Neonicotinoid pesticides severely affect honey bee queens. Sci. Rep. 2015, 5, 14621. [Google Scholar] [CrossRef] [Green Version]
- Yang, E.-C.; Chang, H.-C.; Wu, W.-Y.; Chen, Y.-W. Impaired Olfactory Associative Behavior of Honeybee Workers Due to Contamination of Imidacloprid in the Larval Stage. PLoS ONE 2012, 7, e49472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wakita, T. Molecular Design of Dinotefuran with Unique Insecticidal Properties. J. Agric. Food Chem. 2011, 59, 2938–2942. [Google Scholar] [CrossRef]
- Tan, L.; Cheng, Y.; Zhu, Y.; Bu, Y.; Zhou, J.; Shan, Z. Safety Evaluation of Nicotinic Insecticide Dinotefuran on Honeybee (Apis mellifera L.). Asian J. Ecotoxicol. 2017, 227–233. [Google Scholar] [CrossRef]
- Huang, M.; Dong, J.; Guo, H.; Wang, D. Effects of Dinotefuran on Brain miRNA Expression Profiles in Young Adult Honey Bees (Hymenopptera: Apidae). J. Insect Sci. 2021, 21, 3. [Google Scholar] [CrossRef]
- Badawy, M.E.I.; Nasr, H.M.; Rabea, E.I. Toxicity and biochemical changes in the honey bee Apis mellifera exposed to four insecticides under laboratory conditions. Apidologie 2015, 46, 177–193. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Liu, Y.M.; He, F.M.; Zhang, H.; Li, X.; Tan, H. Enantioselective Olfactory Effects of the Neonicotinoid Dinotefuran on Honey Bees (Apis mellifera L.). J. Agric. Food Chem. 2019, 67, 12105–12116. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Dong, J.; Guo, H.; Xiao, M.; Wang, D. Identification of long noncoding RNAs reveals the effects of dinotefuran on the brain in Apis mellifera (Hymenopptera: Apidae). BMC Genom. 2021, 22, 502. [Google Scholar] [CrossRef]
- Christen, V.; Mittner, F.; Fent, K. Molecular Effects of Neonicotinoids in Honey Bees (Apis mellifera). Environ. Sci. Technol. 2016, 50, 4071–4081. [Google Scholar] [CrossRef] [PubMed]
- Jia, C. Calculatiang the LC50 of insecticides with software SPSS. Chin. J. Appl. Entomol. 2006, 43, 414–417. [Google Scholar] [CrossRef]
- Chmiel, J.A.; Daisley, B.A.; Burton, J.P.; Reid, G. Deleterious Effects of Neonicotinoid Pesticides on Drosophila melanogaster Immune Pathways. mBio 2019, 10, e01395-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Prisco, G.; Cavaliere, V.; Annoscia, D.; Varricchio, P.; Caprio, E.; Nazzi, F.; Gargiulo, G.; Pennacchio, F. Neonicotinoid clothianidin adversely affects insect immunity and promotes replication of a viral pathogen in honey bees. Proc. Natl. Acad. Sci. USA 2013, 110, 18466–18471. [Google Scholar] [CrossRef] [Green Version]
- James, R.; Xu, J. Mechanisms by which pesticides affect insect immunity. J. Invertebr. Pathol. 2012, 109, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Amdam, G.V.; Simões, Z.L.; Hagen, A.; Norberg, K.; Schrøder, K.; Mikkelsen, Ø.; Kirkwood, T.B.L.; Omholt, S.W. Hormonal control of the yolk precursor vitellogenin regulates immune function and longevity in honeybees. Exp. Gerontol. 2004, 39, 767–773. [Google Scholar] [CrossRef] [PubMed]
- Seehuus, S.-C.; Norberg, K.; Gimsa, U.; Krekling, T.; Amdam, G.V. Reproductive protein protects functionally sterile honey bee workers from oxidative stress. Proc. Natl. Acad. Sci. USA 2006, 103, 962–967. [Google Scholar] [CrossRef] [Green Version]
- Johnson, R.M.; Wen, Z.; Schuler, M.A.; Berenbaum, M.R. Mediation of Pyrethroid Insecticide Toxicity to Honey Bees (Hymenoptera: Apidae) by Cytochrome P450 Monooxygenases. J. Econ. Èntomol. 2006, 99, 1046–1050. [Google Scholar] [CrossRef] [PubMed]
- Alptekin, S.; Bass, C.; Nicholls, C.; Paine, M.J.I.; Clark, S.J.; Field, L.; Moores, G.D. Induced thiacloprid insensitivity in honeybees (Apis melliferaL.) is associated with up-regulation of detoxification genes. Insect Mol. Biol. 2016, 25, 171–180. [Google Scholar] [CrossRef] [Green Version]
- Kostaropoulos, I.; Papadopoulos, A.I.; Metaxakis, A.; Boukouvala, E.; Papadopoulou-Mourkidou, E. Glutathione S–transferase in the defence against pyrethroids in insects. Insect Biochem. Mol. Biol. 2001, 31, 313–319. [Google Scholar] [CrossRef]
- Hemingway, J.; Hawkes, N.J.; McCarroll, L.; Ranson, H. The molecular basis of insecticide resistance in mosquitoes. Insect Biochem. Mol. Biol. 2004, 34, 653–665. [Google Scholar] [CrossRef]
- Nauen, R.; Ebbinghaus-Kintscher, U.; Schmuck, R. Toxicity and nicotinic acetylcholine receptor interaction of imidacloprid and its metabolites in Apis mellifera (Hymenoptera: Apidae). Pest Manag. Sci. 2001, 57, 577–586. [Google Scholar] [CrossRef]
- Györi, J.; Farkas, A.; Stolyar, O.; Székács, A.; Mörtl, M.; Vehovszky, Á. Inhibitory effects of four neonicotinoid active ingredients on acetylcholine esterase activity. Acta Biol. Hung. 2017, 68, 345–357. [Google Scholar] [CrossRef] [PubMed]
- Lämsä, J.; Kuusela, E.; Tuomi, J.; Juntunen, S.; Watts, P.C. Low dose of neonicotinoid insecticide reduces foraging motivation of bumblebees. Proc. R. Soc. B Boil. Sci. 2018, 285, 20180506. [Google Scholar] [CrossRef]
- Williamson, S.M.; Wright, G.A. Exposure to multiple cholinergic pesticides impairs olfactory learning and memory in honeybees. J. Exp. Biol. 2013, 216, 1799–1807. [Google Scholar] [CrossRef] [Green Version]
- Chang, H.; Liu, Y.; Yang, T.; Pelosi, P.; Dong, S.; Wang, G. Pheromone binding proteins enhance the sensitivity of olfactory receptors to sex pheromones in Chilo suppressalis. Sci. Rep. 2015, 5, 13093. [Google Scholar] [CrossRef] [Green Version]
- Li, R.; Zhang, L.; Fang, Y.; Han, B.; Lu, X.; Zhou, T.; Feng, M.; Li, J. Proteome and phosphoproteome analysis of honeybee (Apis mellifera) venom collected from electrical stimulation and manual extraction of the venom gland. BMC Genom. 2013, 14, 766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, L.A.; Ihara, M.; Buckingham, S.D.; Matsuda, K.; Sattelle, D.B. Neonicotinoid insecticides display partial and super agonist actions on native insect nicotinic acetylcholine receptors. J. Neurochem. 2006, 99, 608–615. [Google Scholar] [CrossRef]
- Woitecki, A.M.; Müller, J.A.; Van Loo, K.M.; Sowade, R.F.; Becker, A.J.; Schoch, S. Identification of Synaptotagmin 10 as Effector of NPAS4-Mediated Protection from Excitotoxic Neurodegeneration. J. Neurosci. 2016, 36, 2561–2570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huo, X.; Wu, B.; Feng, M.; Han, B.; Fang, Y.; Hao, Y.; Meng, L.; Wubie, A.J.; Fan, P.; Hu, H.; et al. Proteomic Analysis Reveals the Molecular Underpinnings of Mandibular Gland Development and Lipid Metabolism in Two Lines of Honeybees (Apis mellifera ligustica). J. Proteome Res. 2016, 15, 3342–3357. [Google Scholar] [CrossRef]
- Pankiw, T. Cued in: Honey bee pheromones as information flow and collective decision-making. Apidologie 2004, 35, 217–226. [Google Scholar] [CrossRef] [Green Version]





| Simple Name | Clean Reads | Q30 (%) | Total Mapped | Mapped Reads | Unique Mapped Reads |
|---|---|---|---|---|---|
| CK-1 | 27,658,539 | 94.85% | 55,317,078 | 50,192,862 (90.74%) | 48,804,054 (88.23%) |
| CK-2 | 20,392,556 | 91.24% | 40,785,112 | 33,658,507 (82.53%) | 32,785,064 (80.38%) |
| CK-3 | 22,237,202 | 91.09% | 44,474,404 | 38,962,393 (87.61%) | 37,942,202 (85.31%) |
| LC05-1 | 20,561,037 | 94.72% | 41,122,074 | 33,264,911 (80.89%) | 32,254,135 (78.44%) |
| LC05-2 | 23,028,809 | 95.07% | 46,057,618 | 37,855,789 (82.19%) | 36,641,503 (79.56%) |
| LC05-3 | 20,344,696 | 94.87% | 40,689,392 | 36,729,260 (90.27%) | 35,098,847 (86.26%) |
| LC20-1 | 21,104,445 | 94.95% | 42,208,890 | 37,238,021 (88.22%) | 36,368,799 (86.16%) |
| LC20-2 | 19,639,136 | 95.09% | 39,278,272 | 34,726,789 (88.41%) | 34,104,678 (86.83%) |
| LC20-3 | 20,027,148 | 95.21% | 40,054,296 | 35,496,091 (88.62%) | 34,376,252 (85.82%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Du, Y.; Ma, W.; Liu, J.; Jiang, Y. The Transcriptomic Landscape of Molecular Effects after Sublethal Exposure to Dinotefuran on Apis mellifera. Insects 2021, 12, 898. https://doi.org/10.3390/insects12100898
Zhang Y, Du Y, Ma W, Liu J, Jiang Y. The Transcriptomic Landscape of Molecular Effects after Sublethal Exposure to Dinotefuran on Apis mellifera. Insects. 2021; 12(10):898. https://doi.org/10.3390/insects12100898
Chicago/Turabian StyleZhang, Yuhao, Yali Du, Weihua Ma, Jinjia Liu, and Yusuo Jiang. 2021. "The Transcriptomic Landscape of Molecular Effects after Sublethal Exposure to Dinotefuran on Apis mellifera" Insects 12, no. 10: 898. https://doi.org/10.3390/insects12100898






