Selection and Evaluation of Reference Genes for qRT-PCR in Spodoptera frugiperda (Lepidoptera: Noctuidae)
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Insect Rearing
2.2. Experimental Treatments and Sample Collection
2.2.1. Different Developmental Stages
2.2.2. Various Tissues
2.2.3. Mating Status
2.2.4. Hormone Treatment
2.2.5. Diet Treatment
2.2.6. Temperature Treatment
2.3. RNA Isolation and cDNA Synthesis
2.4. Reference Genes Selection and Primer Design
2.5. qRT-PCR Analysis
2.6. Stability Analysis
2.7. Stability Verification of Candidate Reference Genes
3. Results
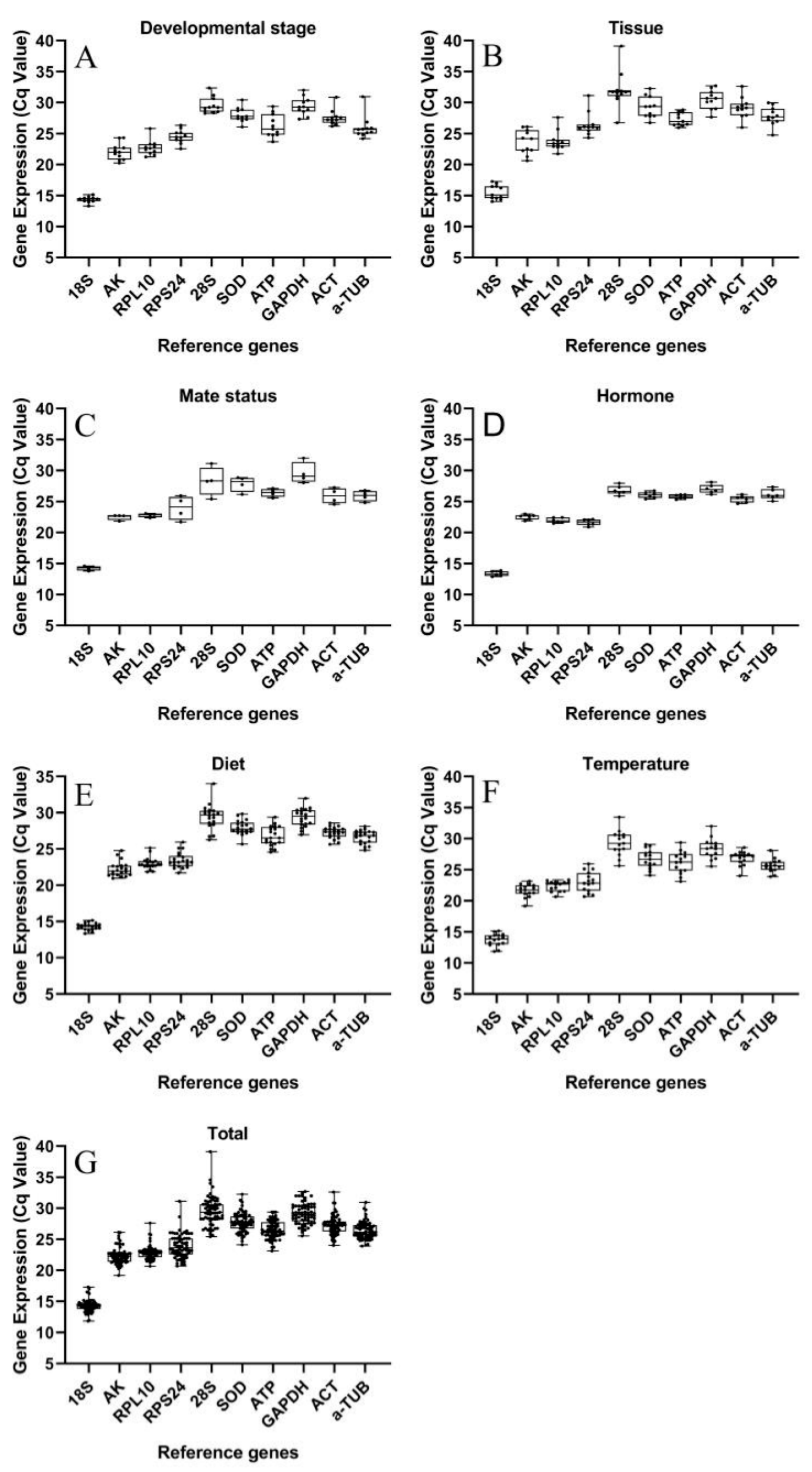
3.1. qRT-PCR Analysis
3.2. Expression Stability Analysis of Candidate Reference Genes
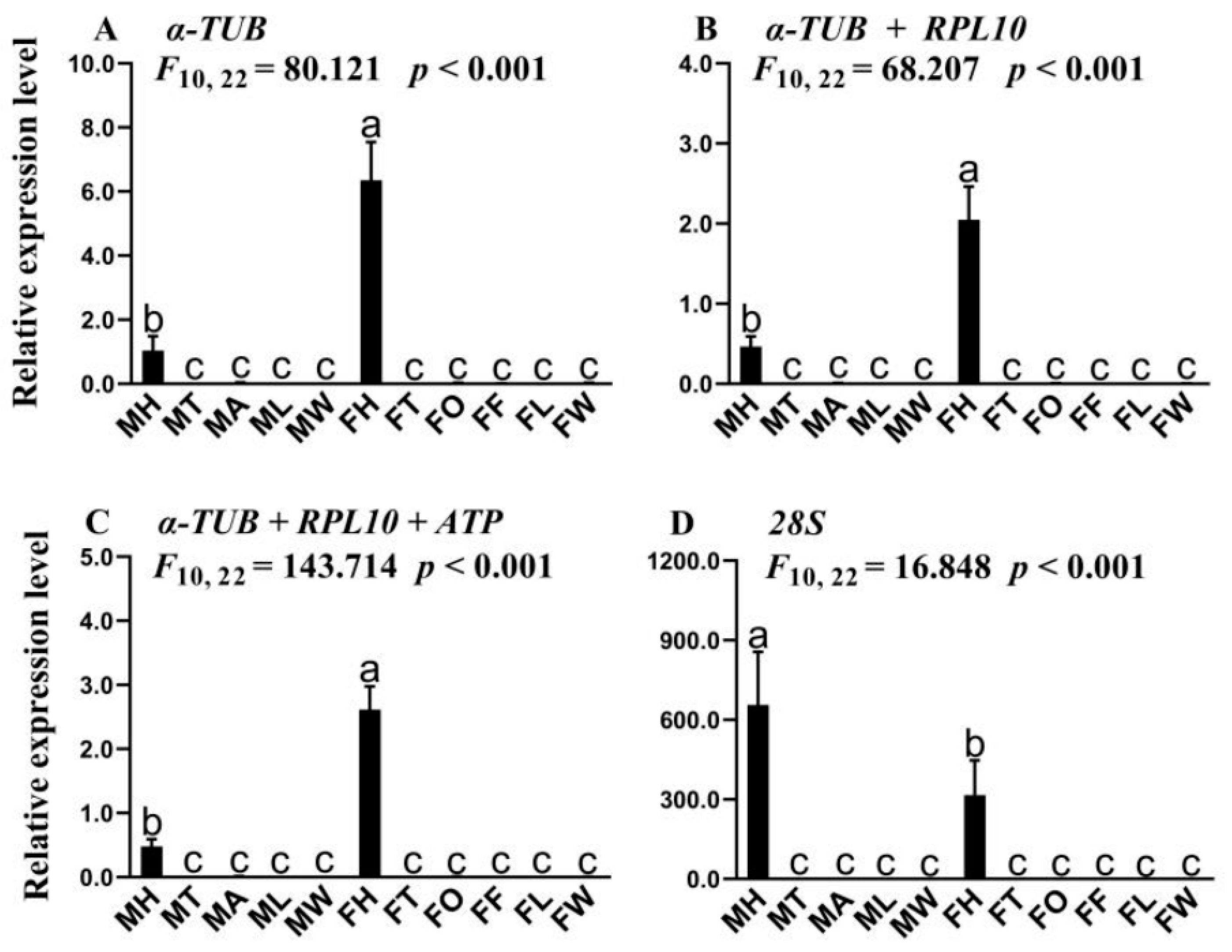
3.3. Verification of Candidate Reference Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Todd, E.L.; Poole, R.W. Keys and Illustrations for the Armyworm Moths of the Noctuid Genus Spodoptera Guenée from the Western Hemisphere. Ann. Entomol. Soc. Am. 1980, 73, 722–738. [Google Scholar] [CrossRef]
- Jing, D.P.; Guo, J.F.; Jiang, Y.Y.; Zhao, J.Z.; Sethi, A.; He, K.L.; Wang, Z.Y. Initial detections and spread of invasive Spodoptera frugiperda in China and comparisons with other noctuid larvae in cornfields using molecular techniques. Insect Sci. 2020, 27, 780–790. [Google Scholar] [CrossRef]
- Montezano, D.; Specht, A.; Sosa-Gómez, D.; Roque-Specht, V.; Sousa-Silva, J.; Paula-Moraes, S.; Peterson, J.; Hunt, T. Host Plants of Spodoptera frugiperda (Lepidoptera: Noctuidae) in the Americas. Afr. Entomol. 2018, 26, 286–300. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; He, P.; Zhang, Y.; Liu, T.; Jing, X.; Zhang, S. The Population Growth of Spodoptera frugiperda on Six Cash Crop Species and Implications for Its Occurrence and Damage Potential in China. Insects 2020, 11, 639. [Google Scholar] [CrossRef] [PubMed]
- Sharanabasappa; Kalleshwaraswamy, C.M.; Asokan, R.; Swamy, H.M.; Maruthi, M.S.; Pavithra, H.B.; Kavita, H.; Shivaray, N.; Prabhu, S.T.; Goergen, G. First report of the Fall armyworm, Spodoptera frugiperda (J E Smith) (Lepidoptera: Noctuidae), an alien invasive pest on maize in India. J. Entomol. Zool. 2018, 6, 432–436. [Google Scholar]
- Wan, J.; Huang, C.; Li, C.-Y.; Zhou, H.-X.; Ren, Y.-L.; Li, Z.-Y.; Xing, L.-S.; Zhang, B.; Qiao, X.; Liu, B.; et al. Biology, invasion and management of the agricultural invader: Fall armyworm, Spodoptera frugiperda (Lepidoptera: Noctuidae). J. Integr. Agric. 2021, 20, 646–663. [Google Scholar] [CrossRef]
- Goergen, G.; Kumar, P.L.; Sankung, S.B.; Togola, A.; Tamò, M. First Report of Outbreaks of the Fall Armyworm Spodoptera frugiperda (J E Smith) (Lepidoptera, Noctuidae), a New Alien Invasive Pest in West and Central Africa. PLoS ONE 2016, 11, e0165632. [Google Scholar] [CrossRef] [Green Version]
- Sun, X.; Hu, C.; Jia, H.; Wu, Q.; Shen, X.; Zhao, S.; Jiang, Y.; Wu, K. Case study on the first immigration of fall armyworm Spodoptera frugiperda invading into China. J. Integr. Agric. 2019, 18, 2–10. [Google Scholar]
- Pinheiro, D.H.; Siegfried, B.D. Selection of reference genes for normalization of RT-qPCR data in gene expression studies in Anthonomus eugenii Cano (Coleoptera: Curculionidae). Sci. Rep. 2020, 10, 5070. [Google Scholar] [CrossRef] [Green Version]
- Xie, J.; Liu, T.; Khashaveh, A.; Yi, C.; Liu, X.; Zhang, Y. Identification and Evaluation of Suitable Reference Genes for RT-qPCR Analysis in Hippodamia variegata (Coleoptera: Coccinellidae) Under Different Biotic and Abiotic Conditions. Front. Physiol. 2021, 12, 669510. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Wong, M.L.; Medrano, J.F. Real-time PCR for mRNA quantitation. Biotechniques 2005, 39, 75–85. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bustin, S.A. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): Trends and problems. J. Mol. Endocrinol. 2002, 29, 23–39. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, 34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kozera, B.; Rapacz, M. Reference genes in real-time PCR. J. Appl. Genet. 2013, 54, 391–406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shu, B.; Zhang, J.; Cui, G.; Sun, R.; Sethuraman, V.; Yi, X.; Zhong, G. Evaluation of Reference Genes for Real-Time Quantitative PCR Analysis in Larvae of Spodoptera litura Exposed to Azadirachtin Stress Conditions. Front. Physiol. 2018, 9, 372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Meng, Q.; Zhu, X.; Sun, S.; Gao, S.; Gou, Y.; Liu, A. Evaluation and Validation of Reference Genes for Quantitative Real-Time PCR in Helopeltis theivora Waterhouse (Hemiptera: Miridae). Sci. Rep. 2019, 9, 13291. [Google Scholar] [CrossRef] [Green Version]
- Chapman, J.R.; Waldenstrom, J. With Reference to Reference Genes: A Systematic Review of Endogenous Controls in Gene Expression Studies. PLoS ONE 2015, 10, e141853. [Google Scholar] [CrossRef]
- Pan, H.; Yang, X.; Bidne, K.; Hellmich, R.L.; Siegfried, B.D.; Zhou, X. Selection of Reference Genes for RT-qPCR Analysis in the Monarch Butterfly, Danaus plexippus (L.), a Migrating Bio-Indicator. PLoS ONE 2015, 10, e129482. [Google Scholar] [CrossRef]
- Guo, H.; Jiang, L.; Xia, Q. Selection of reference genes for analysis of stress-responsive genes after challenge with viruses and temperature changes in the silkworm Bombyx mori. Mol. Genet. Genom. 2016, 291, 999–1004. [Google Scholar] [CrossRef]
- Li, K.; Xu, N.; Yang, Y.J.; Zhang, J.H.; Yin, H. Identification and Validation of Reference Genes for RT-qPCR Normalization in Mythimna separata (Lepidoptera: Noctuidae). Biomed. Res. Int. 2018, 2018, 1828253. [Google Scholar] [CrossRef]
- Piron Prunier, F.; Chouteau, M.; Whibley, A.; Joron, M.; Llaurens, V. Selection of Valid Reference Genes for Reverse Transcription Quantitative PCR Analysis in Heliconius numata (Lepidoptera: Nymphalidae). J. Insect Sci. 2016, 16, 50. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Lu, M.; Cui, Y.; Du, Y. Selection and Evaluation of Reference Genes for Expression Analysis Using qRT-PCR in Chilo suppressalis (Lepidoptera: Pyralidae). J. Econ. Entomol. 2017, 2, w297. [Google Scholar] [CrossRef]
- Zhang, S.; An, S.; Li, Z.; Wu, F.; Yang, Q.; Liu, Y.; Cao, J.; Zhang, H.; Zhang, Q.; Liu, X. Identification and validation of reference genes for normalization of gene expression analysis using qRT-PCR in Helicoverpa armigera (Lepidoptera: Noctuidae). Gene 2015, 555, 393–402. [Google Scholar] [CrossRef]
- Zhu, X.; Yuan, M.; Shakeel, M.; Zhang, Y.; Wang, S.; Wang, X.; Zhan, S.; Kang, T.; Li, J. Selection and evaluation of reference genes for expression analysis using qRT-PCR in the beet armyworm Spodoptera exigua (Hubner) (Lepidoptera: Noctuidae). PLoS ONE 2014, 9, e84730. [Google Scholar] [CrossRef]
- Boaventura, D.; Ulrich, J.; Lueke, B.; Bolzan, A.; Okuma, D.; Gutbrod, O.; Geibel, S.; Zeng, Q.; Dourado, P.M.; Martinelli, S.; et al. Molecular characterization of Cry1F resistance in fall armyworm, Spodoptera frugiperda from Brazil. Insect Biochem. Mol. Biol. 2020, 116, 103280. [Google Scholar] [CrossRef] [PubMed]
- Andersen, C.L.; Jensen, J.L.; Orntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper--Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Lü, J.; Chen, S.; Guo, M.; Ye, C.; Qiu, B.; Yang, C.; Pan, H. Selection of appropriate reference genes for RT-qPCR analysis in Propylea japonica (Coleoptera: Coccinellidae). PLoS ONE 2018, 13, e208027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, Y.; Zhao, H.; Yang, S.; Zhang, L.; Zhang, L.; Hou, C. Screening and Validation of Reference Genes for RT-qPCR Under Different Honey Bee Viral Infections and dsRNA Treatment. Front. Microbiol. 2020, 11, 1715. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Wang, A.; Cheng, Y.; Rong, H.; Guo, L.; Peng, Y.; Xu, L. Selection and Validation of Suitable Reference Genes for RT-qPCR Analysis in Apolygus lucorum (Hemiptera: Miridae). J. Econ. Entomol. 2020, 113, 451–460. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Zheng, H.; Liu, Y.; Li, H.; Jiang, Y.; Lin, L.; Deng, X.; Zhang, Q. Selection of Reference Genes for Quantitative Real-Time PCR in Aquatica leii (Coleoptera: Lampyridae) Under Five Different Experimental Conditions. Front. Physiol. 2020, 11, 555233. [Google Scholar] [CrossRef]
- Zheng, C.; Zhao, D.; Xu, Y.; Shi, F.; Zong, S.; Tao, J. Reference Gene Selection for Expression Analyses by qRT-PCR in Dendroctonus valens. Insects 2020, 11, 328. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Pandher, S.; Gupta, M.; Kaur, G.; Rathore, P. Reference Gene Selection in Phenacoccus solenopsis Tinsley (Hemiptera: Pseudococcidae) and Their Normalization Impact on Gene Expression in RNAi Studies. J. Econ. Entomol. 2019, 112, 371–381. [Google Scholar] [CrossRef]
- Yang, Q.; Li, Z.; Cao, J.; Zhang, S.; Zhang, H.; Wu, X.; Zhang, Q.; Liu, X. Selection and Assessment of Reference Genes for Quantitative PCR Normalization in Migratory Locust Locusta migratoria (Orthoptera: Acrididae). PLoS ONE 2014, 9, e98164. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.; Liao, W.J.; Liao, J.M.; Liao, P.; Hua, L. Ribosomal proteins: Functions beyond the ribosome. J. Mol. Cell Biol. 2015, 7, 92–104. [Google Scholar] [CrossRef] [Green Version]
- Ladror, D.T.; Frey, B.L.; Scalf, M.; Levenstein, M.E.; Artymiuk, J.M.; Smith, L.M. Methylation of yeast ribosomal protein S2 is elevated during stationary phase growth conditions. Biochem. Biophys. Res. Commun. 2014, 445, 535–541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bin, S.; Pu, X.; Shu, B.; Kang, C.; Luo, S.; Tang, Y.; Wu, Z.; Lin, J. Selection of Reference Genes for Optimal Normalization of Quantitative Real-Time Polymerase Chain Reaction Results for Diaphorina citri Adults. J. Econ. Entomol. 2019, 112, 355–363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, Y.; Zhou, X.R.; Pang, B.P. Reference gene selection and evaluation for expression analysis using qRT-PCR in Galeruca daurica (Joannis). Bull. Entomol. Res. 2017, 107, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Fu, W.; Xie, W.; Zhang, Z.; Wang, S.; Wu, Q.; Liu, Y.; Zhou, X.; Zhou, X.; Zhang, Y. Exploring Valid Reference Genes for Quantitative Real-time PCR Analysis in Plutella xylostella (Lepidoptera: Plutellidae). Int. J. Biol. Sci. 2013, 9, 792–802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, J. Odorant-binding proteins in insects. Vitam. Horm. 2010, 83, 241–272. [Google Scholar] [PubMed]


| Gene | GenBank Accession Number | Primer Sequences (5′-3′) | Length (bp) | Amplification Efficiency (%) | Regression Coefficients |
|---|---|---|---|---|---|
| 18S | KY554596 | F: GACTCAACACGGGAAATCTC R: CCACGCACACCTAAATGAC | 185 | 102.6 | 0.995 |
| AK | KC262642 | F: TCTACCACAACGAGAACAAGAC R: AAAGTGAGGAAACCAAGCC | 180 | 97.88 | 0.992 |
| RPL10 | XM_035580073 | F: TGGGTAAGAAGAAGGCTACG R: TGTTGATGCGGATGACAT | 194 | 104.2 | 0.994 |
| RPS24 | XM_035577015 | F: CACTGGCTTTGCTCTCATC R: TCATCCTGTTCTTGCGTTC | 136 | 102.5 | 0.995 |
| 28S | XM_035580587 | F: GACCAGATTCCGATTTCACT R: CTTTGTTTACCGCTTGCC | 122 | 94.1 | 0.996 |
| SOD | XM_035587671 | F: TCGGCACAATCATCAGTC R: AGTCCTTCTCAATAGCCTGC | 137 | 93.6 | 0.995 |
| ATP | XM_035575197 | F: AAAGTAGTTCCGTGGAGTGAG R: AAAGAAGGGTCCGTAGATTG | 115 | 107.1 | 0.994 |
| GAPDH | XM_035587881 | F: AGAAGACTGTTGACGGACC R: AGGAATGACTTTGCCGAC | 112 | 103.9 | 0.984 |
| ACT | KT218672 | F: TTCTTCCACCCTGAGTTCTC R: AGTCCTCTTGATGTCACGC | 182 | 90.6 | 0.999 |
| a-TUB | XM_035600854 | F: AGGGCTGTGTTTGTTGACT R: TCCTTACCGATGGTGTAGTG | 148 | 93.6 | 0.999 |
| OBP1 | XM_035578840 | F: GTGGGTGTGAGAGAGATAGAAC R: GAACAGGTCTGCTATGATGTG | 125 | 100.0 | 0.994 |
| Treatments | Genes | Delta Ct | geNorm | NormFinder | BestKeeper | ||||
|---|---|---|---|---|---|---|---|---|---|
| Stability | Ranking | Stability | Ranking | Stability | Ranking | Stability | Ranking | ||
| Developmental stages | 18S | 1.282 | 9 | 0.954 | 6 | 0.680 | 7 | 0.352 | 1 |
| AK | 1.236 | 7 | 1.081 | 8 | 0.683 | 8 | 1.053 | 7 | |
| RPL10 | 0.885 | 2 | 0.607 | 3 | 0.174 | 1 | 0.822 | 3 | |
| RPS24 | 1.061 | 3 | 0.444 | 1 | 0.453 | 3 | 0.825 | 4 | |
| 28S | 1.117 | 5 | 0.873 | 5 | 0.625 | 5 | 1.010 | 6 | |
| SOD | 0.813 | 1 | 0.444 | 1 | 0.189 | 2 | 0.904 | 5 | |
| ATP | 1.255 | 8 | 1.143 | 9 | 0.803 | 9 | 1.498 | 10 | |
| GAPDH | 1.207 | 6 | 1.037 | 7 | 0.677 | 6 | 1.104 | 9 | |
| ACT | 1.065 | 4 | 0.797 | 4 | 0.557 | 4 | 0.807 | 2 | |
| a-TUB | 1.283 | 10 | 1.197 | 10 | 0.820 | 10 | 1.079 | 8 | |
| Tissues | 18S | 1.649 | 9 | 1.168 | 8 | 0.961 | 9 | 1.043 | 2 |
| AK | 1.563 | 8 | 1.251 | 9 | 0.937 | 8 | 1.694 | 9 | |
| RPL10 | 1.058 | 1 | 0.995 | 5 | 0.318 | 1 | 1.084 | 4 | |
| RPS24 | 1.209 | 6 | 1.073 | 7 | 0.580 | 5 | 1.246 | 7 | |
| 28S | 1.964 | 10 | 1.405 | 10 | 1.284 | 10 | 1.752 | 10 | |
| SOD | 1.142 | 3 | 0.802 | 3 | 0.502 | 4 | 1.374 | 8 | |
| ATP | 1.153 | 5 | 0.914 | 4 | 0.587 | 6 | 0.919 | 1 | |
| GAPDH | 1.228 | 7 | 0.625 | 1 | 0.593 | 7 | 1.176 | 6 | |
| ACT | 1.146 | 4 | 1.036 | 6 | 0.499 | 3 | 1.135 | 5 | |
| a-TUB | 1.092 | 2 | 0.625 | 1 | 0.483 | 2 | 1.080 | 3 | |
| Mating status | 18S | 1.182 | 4 | 0.408 | 3 | 0.388 | 3 | 0.268 | 2 |
| AK | 1.062 | 1 | 0.190 | 1 | 0.066 | 1 | 0.338 | 3 | |
| RPL10 | 1.067 | 2 | 0.190 | 1 | 0.181 | 2 | 0.212 | 1 | |
| RPS24 | 1.703 | 8 | 1.177 | 8 | 1.069 | 8 | 1.568 | 10 | |
| 28S | 1.968 | 10 | 1.516 | 10 | 1.496 | 10 | 1.459 | 9 | |
| SOD | 1.182 | 4 | 0.770 | 5 | 0.416 | 4 | 0.913 | 6 | |
| ATP | 1.098 | 3 | 0.556 | 4 | 0.644 | 6 | 0.485 | 4 | |
| GAPDH | 1.765 | 9 | 1.312 | 9 | 1.204 | 9 | 1.220 | 8 | |
| ACT | 1.185 | 6 | 0.896 | 6 | 0.549 | 5 | 1.028 | 7 | |
| a-TUB | 1.401 | 7 | 0.999 | 7 | 0.846 | 7 | 0.633 | 5 | |
| Treatments | Genes | Delta Ct | geNorm | NormFinder | BestKeeper | ||||
|---|---|---|---|---|---|---|---|---|---|
| Stability | Ranking | Stability | Ranking | Stability | Ranking | Stability | Ranking | ||
| Hormone treatment | 18S | 0.545 | 7 | 0.210 | 1 | 0.216 | 5 | 0.316 | 3 |
| AK | 0.484 | 2 | 0.210 | 1 | 0.119 | 2 | 0.315 | 2 | |
| RPL10 | 0.502 | 4 | 0.341 | 5 | 0.119 | 2 | 0.338 | 4 | |
| RPS24 | 0.506 | 5 | 0.316 | 4 | 0.162 | 4 | 0.436 | 7 | |
| 28S | 0.675 | 8 | 0.486 | 8 | 0.499 | 8 | 0.554 | 9 | |
| SOD | 0.509 | 6 | 0.409 | 7 | 0.271 | 6 | 0.413 | 5 | |
| ATP | 0.432 | 1 | 0.222 | 3 | 0.106 | 1 | 0.267 | 1 | |
| GAPDH | 0.799 | 10 | 0.631 | 10 | 0.557 | 9 | 0.541 | 8 | |
| ACT | 0.499 | 3 | 0.582 | 6 | 0.271 | 6 | 0.419 | 6 | |
| a-TUB | 0.703 | 9 | 0.563 | 9 | 0.565 | 10 | 0.629 | 10 | |
| Diet treatment | 18S | 1.041 | 4 | 0.808 | 3 | 0.224 | 1 | 0.385 | 1 |
| AK | 1.230 | 6 | 0.694 | 5 | 0.654 | 6 | 0.787 | 6 | |
| RPL10 | 1.034 | 3 | 0.636 | 1 | 0.363 | 2 | 0.562 | 2 | |
| RPS24 | 1.254 | 8 | 0.957 | 5 | 0.652 | 5 | 0.812 | 7 | |
| 28S | 1.713 | 10 | 1.319 | 10 | 1.221 | 10 | 1.239 | 10 | |
| SOD | 0.958 | 1 | 0.636 | 1 | 0.441 | 4 | 0.751 | 4 | |
| ATP | 1.249 | 7 | 1.165 | 9 | 0.745 | 8 | 1.149 | 9 | |
| GAPDH | 1.256 | 9 | 1.024 | 7 | 0.715 | 7 | 1.055 | 8 | |
| ACT | 1.003 | 2 | 0.870 | 4 | 0.416 | 3 | 0.596 | 3 | |
| a-TUB | 1.214 | 5 | 1.100 | 8 | 0.793 | 9 | 0.763 | 5 | |
| Temperature treatment | 18S | 1.127 | 3 | 0.950 | 3 | 0.393 | 2 | 0.810 | 3 |
| AK | 1.294 | 9 | 1.143 | 7 | 0.666 | 5 | 0.820 | 4 | |
| RPL10 | 1.070 | 1 | 1.056 | 5 | 0.331 | 1 | 0.687 | 1 | |
| RPS24 | 1.210 | 5 | 0.871 | 1 | 0.599 | 4 | 1.307 | 8 | |
| 28S | 1.722 | 10 | 1.345 | 10 | 1.109 | 10 | 1.554 | 10 | |
| SOD | 1.190 | 4 | 0.871 | 1 | 0.743 | 9 | 1.270 | 7 | |
| ATP | 1.261 | 8 | 1.229 | 9 | 0.720 | 8 | 1.422 | 9 | |
| GAPDH | 1.244 | 7 | 1.004 | 4 | 0.699 | 6 | 1.202 | 6 | |
| ACT | 1.096 | 2 | 1.100 | 6 | 0.527 | 3 | 0.833 | 5 | |
| a-TUB | 1.224 | 6 | 1.180 | 8 | 0.709 | 7 | 0.752 | 2 | |
| Total | 18S | 1.269 | 5 | 1.025 | 3 | 0.524 | 3 | 0.673 | 1 |
| AK | 1.369 | 8 | 1.173 | 6 | 0.745 | 7 | 0.993 | 3 | |
| RPL10 | 1.096 | 1 | 0.977 | 1 | 0.359 | 1 | 0.715 | 2 | |
| RPS24 | 1.369 | 8 | 1.291 | 9 | 0.752 | 8 | 1.491 | 9 | |
| 28S | 1.750 | 10 | 1.414 | 10 | 1.168 | 10 | 1.722 | 10 | |
| SOD | 1.099 | 2 | 0.977 | 1 | 0.512 | 2 | 1.147 | 6 | |
| ATP | 1.349 | 7 | 1.250 | 8 | 0.732 | 6 | 1.168 | 7 | |
| GAPDH | 1.317 | 6 | 1.217 | 7 | 0.720 | 5 | 1.325 | 8 | |
| ACT | 1.128 | 3 | 1.064 | 4 | 0.532 | 4 | 1.065 | 4 | |
| a-TUB | 1.262 | 4 | 1.136 | 5 | 0.757 | 9 | 1.097 | 5 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, S.; Qin, Q.; Wang, D.; Zhou, Y.; He, Y. Selection and Evaluation of Reference Genes for qRT-PCR in Spodoptera frugiperda (Lepidoptera: Noctuidae). Insects 2021, 12, 902. https://doi.org/10.3390/insects12100902
Han S, Qin Q, Wang D, Zhou Y, He Y. Selection and Evaluation of Reference Genes for qRT-PCR in Spodoptera frugiperda (Lepidoptera: Noctuidae). Insects. 2021; 12(10):902. https://doi.org/10.3390/insects12100902
Chicago/Turabian StyleHan, Shipeng, Qiuju Qin, Da Wang, Yayuan Zhou, and Yunzhuan He. 2021. "Selection and Evaluation of Reference Genes for qRT-PCR in Spodoptera frugiperda (Lepidoptera: Noctuidae)" Insects 12, no. 10: 902. https://doi.org/10.3390/insects12100902
APA StyleHan, S., Qin, Q., Wang, D., Zhou, Y., & He, Y. (2021). Selection and Evaluation of Reference Genes for qRT-PCR in Spodoptera frugiperda (Lepidoptera: Noctuidae). Insects, 12(10), 902. https://doi.org/10.3390/insects12100902





