Resistance of Common Bean Genotypes to the Broad Mite, Polyphagotarsonemus latus (Banks, 1904) (Acari: Tarsonemidae): Offspring Development and Biochemical Basis
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Screening Genotypes
2.2. Resistance of the Genotypes Previously Selected
2.3. Quantifying Leaf Chemical Compounds
2.4. Statistics
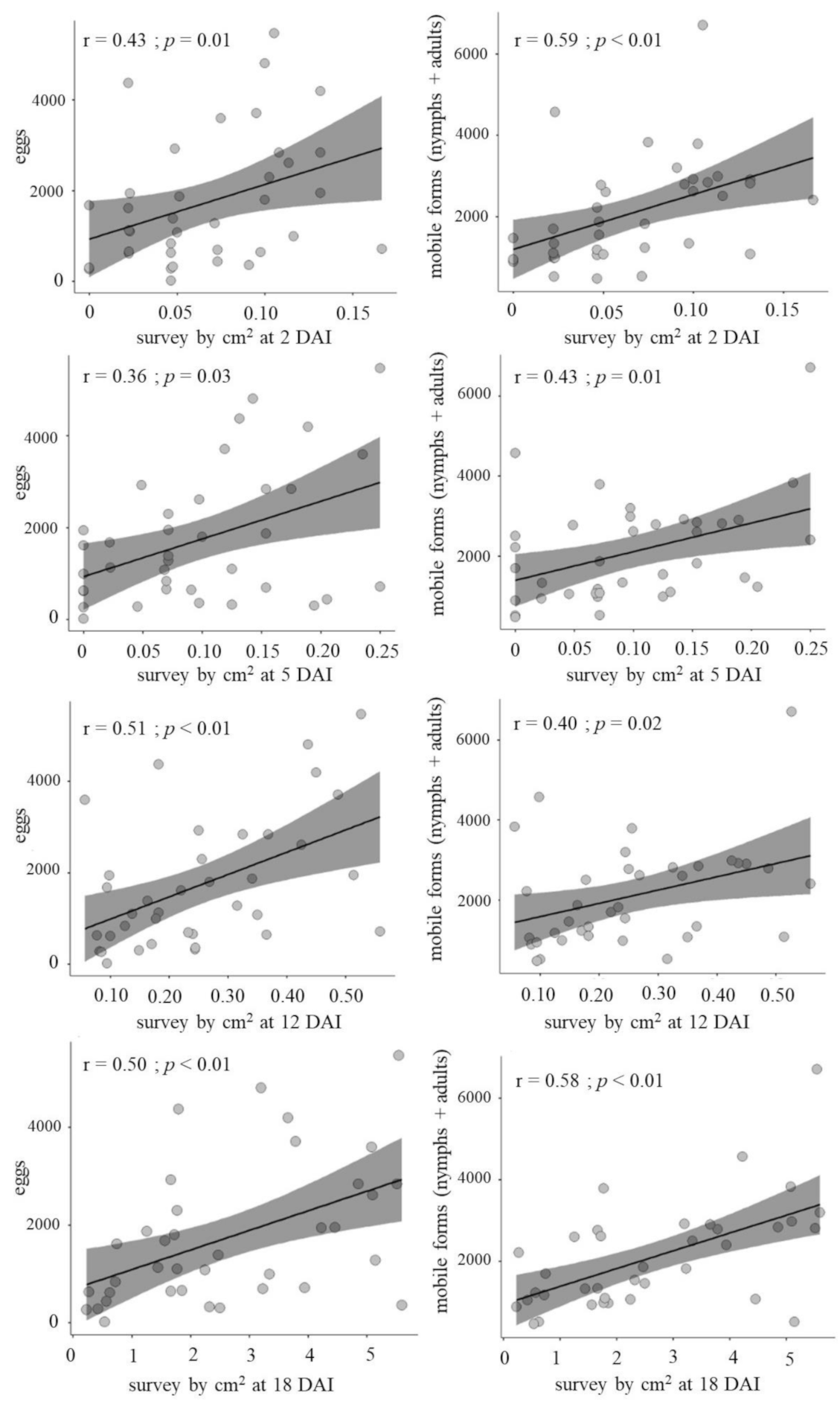
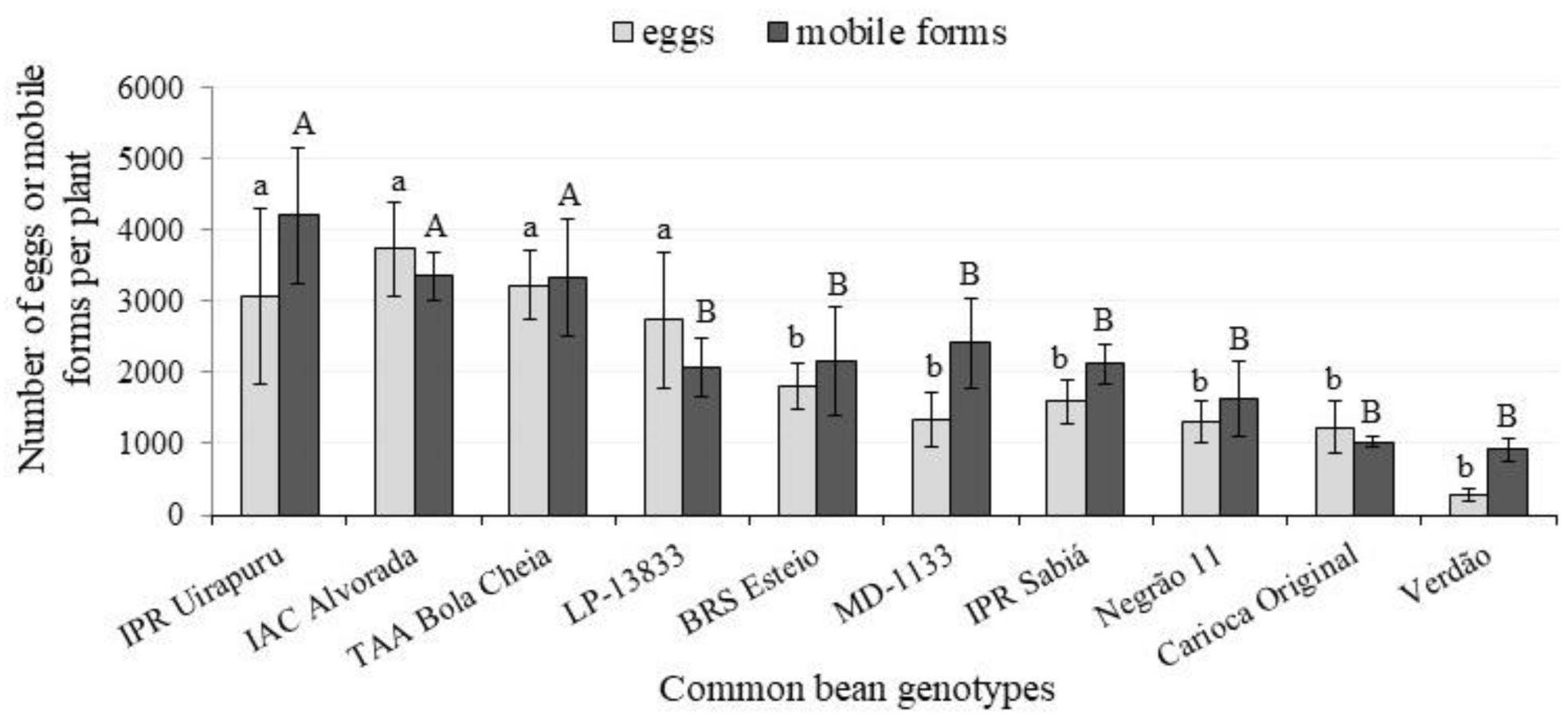
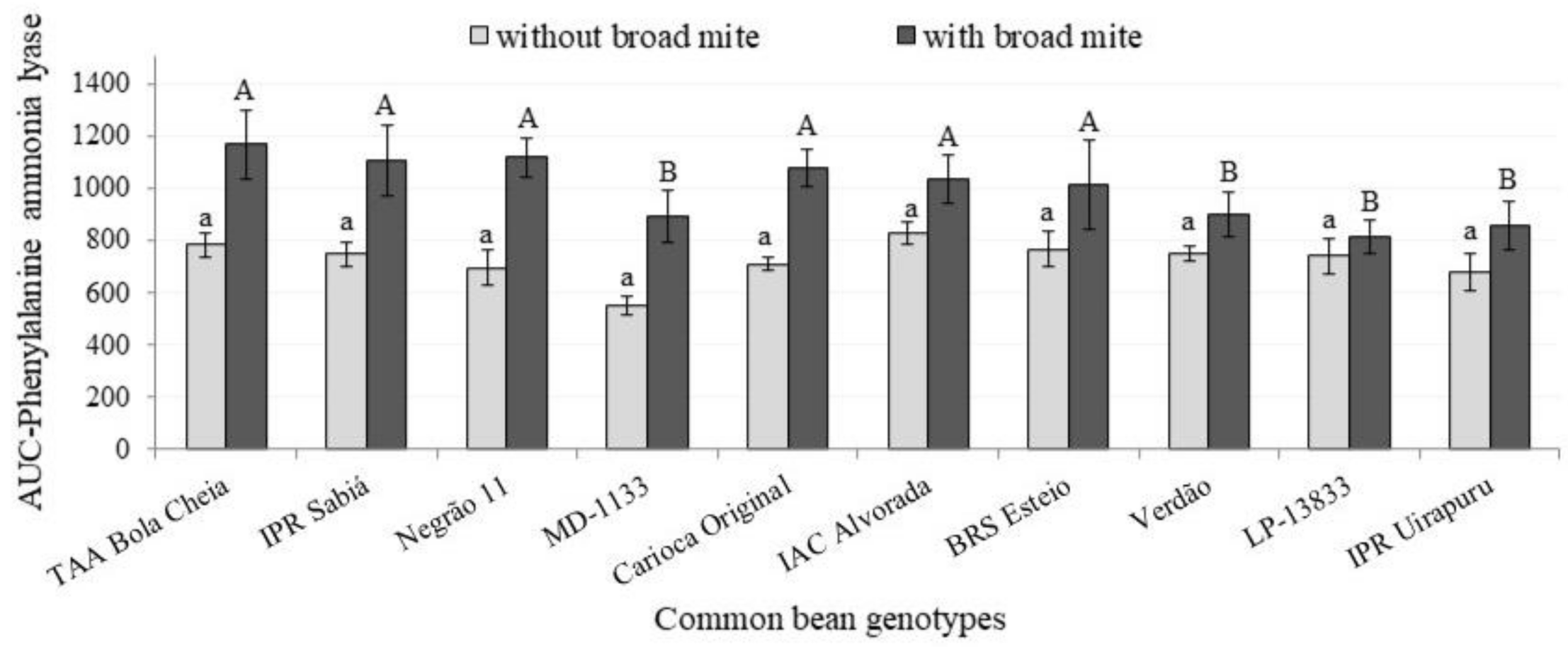
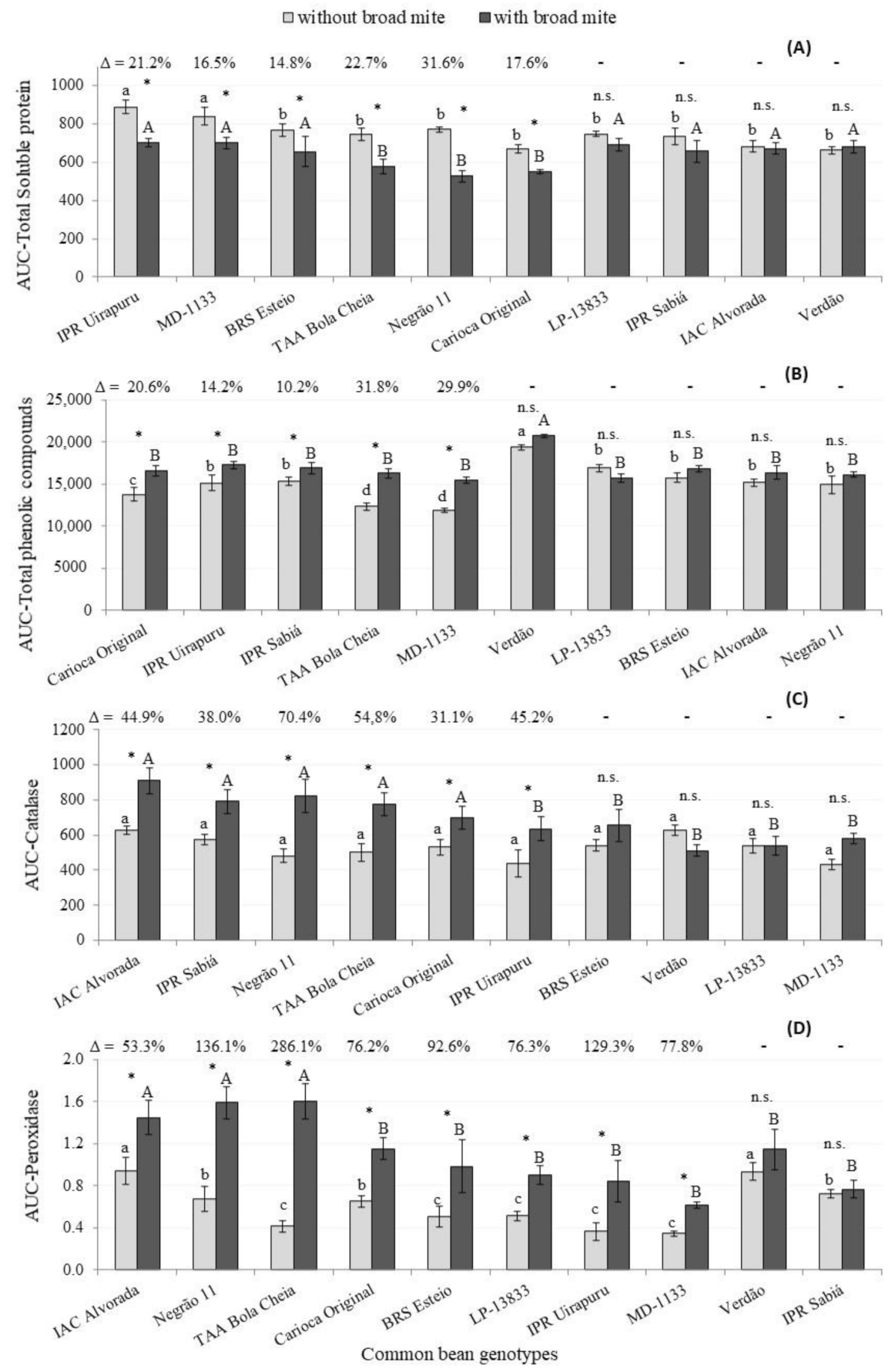
3. Results and Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fores-Sosa, Á.R.; Aquino-Bolaños, E.N.; Cardador-Martínez, A.; Chávez-Servia, J.L.; Vera-Guzmán, A.M.; Carrillo-Rodríguez, J.C.; Jiménez, J.E.A. Variation in protein and amino acids content among landraces of common bean (Phaseolus vulgaris L.). Emir. J. Food Agric. 2020, 32, 750–760. [Google Scholar] [CrossRef]
- Aquino-Bolaños, E.N.; Garzón-García, A.K.; Alba-Jiménez, J.E.; Chávez-Servia, J.L.; Vera-Guzmán, A.M.; Carrillo-Rodríguez, J.C.; Santos-Basurto, M.A. Physicochemical Characterization and Functional Potential of Phaseolus vulgaris L. and Phaseolus coccineus L. Landrace Green Beans. Agronomy 2021, 11, 803. [Google Scholar] [CrossRef]
- Kramer, P.J.; Boyers, J.S. Evolution and agriculture. In Water Relations of Plants and Soils; Kramer, P.J., Boyers, J.S., Eds.; Academic Press: San Diego, CA, USA, 1995; pp. 377–404. [Google Scholar]
- Gerson, U. Biology and control of the broad mite, Polyphagotarsonemus latus (Banks) (Acari: Tarsonemidae). Exp. Appl. Acarol. 1992, 13, 163–178. [Google Scholar] [CrossRef]
- Peña, J.E.; Bullock, R.C. Effects of feeding of broad mite (Acari: Tarsonemidae) on vegetative plant growth. Fla. Entomol. 1994, 77, 180–184. [Google Scholar] [CrossRef]
- Vacante, V. The Handbook of Mites of Economic Plants: Identification, Bio-Ecology and Control; CABI: Wallingford, UK, 2015; 872p. [Google Scholar]
- Jones, V.P.; Brown, R.D. Reproductive responses of the broad mite Polyphagotarsonemus latus (Acari: Tarsonemidae), to constant temperature-humidity regimes. Ann. Entomol. Soc. Am. 1983, 76, 466–469. [Google Scholar] [CrossRef]
- Cardona, C.; Flor, C.A.; Morales, F.J.; Pastor Corrale, M. Problemas de Campo em los Cultivos de Frijol em América Latina; Centro Internacional de Agricultura Tropical 2: Palmira, Colombia, 1982; 100p. [Google Scholar]
- Hill, M.P.; Macfadyen, S.; Nash, M.A. Broad spectrum pesticide application alters natural enemy communities and may facilitate secondary pest outbreaks. PeerJ 2017, 5, e4179. [Google Scholar] [CrossRef] [Green Version]
- Nagai, H.; Lourenção, A.L. Pimenta Cambuci “IAC UBATUBA”. O Agronômico 1987, 39, 120–121. [Google Scholar]
- Rameash, K.; Pandravada, S.R.; Sivaraj, N.; Babu, B.S.; Chakrabarty, S.K. Screening chilli (Capsicum annuum L.) genotypes for resistance to broad mite (Polyphagotarsonemus latus Banks) and analysing the geographic distribution of resistance. Electron. J. Plant. Breed. 2015, 6, 928–937. [Google Scholar]
- Latha, S.; Hunumanthraya, L. Screening of chilli genotypes against chilli thrips (Scirtothrips dorsalis Hood) and yellow mite [Polyphagotarsonemus latus (Banks)]. J. Entomol. Zool. Stud. 2018, 6, 2739–2744. [Google Scholar]
- Cabedo-López, M.; Cruz-Miralles, J.; Peris, D.; Ibáñez-Gual, M.V.; Flors, V.; Jaques, J.A. The response of citrus plants to the broad mite Polyphagotarsonemus latus (Banks) (Acari: Tarsonemidae). Agric. Forest Entomol. 2021. early view. [Google Scholar] [CrossRef]
- Santana, M.F.; de Oliveira, J.V.; Breda, M.O.; Silva Barbosa, D.R.; Filho, A.B.E.; de Oliveira, C.M. Host preference, acaricides effects and population growth of Polyphagotarsonemus latus (Banks) (Acari: Tarsonemidae) on white and colored cotton cultivars. Pest. Manag. Sci. 2021, 77, 217–223. [Google Scholar] [CrossRef]
- Kousik, C.S.; Shepard, B.M.; Hassell, R.; Levi, A.; Simmons, A.M. Potential sources of resistance to broad mites (Polyphagotarsonemus latus) in watermelon germplasm. HortScience 2007, 42, 1539–1544. [Google Scholar] [CrossRef] [Green Version]
- Bonaventure, G. Perception of insect feeding by plants. Plant. Biol. 2012, 14, 872–880. [Google Scholar] [CrossRef]
- Pearce, G.; Strydom, D.; Johnson, S.; Ryan, C.A. A polypeptide from tomato leaves induces wound inducible proteinase inhibitor proteins. Science 1991, 253, 895–897. [Google Scholar] [CrossRef]
- Schmelz, E.A.; Carrol, M.J.; Leclere, S.; Phipps, S.M.; Meredith, J.; Chourey, P.S.; Alborn, H.T.; Teal, P.E.A. Fragments of ATP synthase mediate plant perception of insect attack. Proc. Natl. Acad. Sci. USA 2006, 103, 8894–8899. [Google Scholar] [CrossRef] [Green Version]
- Yamaguchi, Y.; Huffaker, A. Endogenous peptide elicitors in higher plants. Curr. Opin. Plant Biol. 2011, 14, 351–357. [Google Scholar] [CrossRef]
- Halliwell, B.; Gutteridge, J.M.C. Free Radicals in Biology and Medicine; Oxford University Press: Oxford, UK, 2015; 905p. [Google Scholar]
- Hu, Z.; Shen, Y.; Shen, F.; Su, X. Effects of feeding Clostera anachoreta on hydrogen peroxide accumulation and activities of peroxidase, catalase, and ascorbate peroxidase in Populus simonii × P. pyramidalis ‘Opera 8277’ leaves. Acta. Physiol. Plant 2009, 31, 995–1002. [Google Scholar] [CrossRef]
- Panzarino, O.; Hyršl, P.; Dobeš, P.; Vojtek, L.; Vernile, P.; Bari, G.; Terzano, R.; Spagnuolo, M.; Lillo, E. Rank-based biomarker index to asses cadmium eco-toxicity on the earthworm Eisenia andrei. Chemosphere 2016, 145, 480–486. [Google Scholar] [CrossRef]
- Almagro, L.; Gómez-Ros, L.V.; Belchi-Navarro, S.; Bru, R.; Rosbarcelo, A.; Pedreño, M.A. Class III peroxidases in plant defence reactions. J. Exp. Bot. 2009, 60, 377–390. [Google Scholar] [CrossRef] [Green Version]
- Gulsen, O.; Eickhoff, T.; Heng-Moss, T.; Shearman, R.; Baxendale, F.; Sarath, G.; Lee, D. Characterization of peroxidase changes in resistant and susceptible warm-season turf grasses challenged by Blissus occiduus. Arthropod-Plant Interact. 2010, 4, 45–55. [Google Scholar] [CrossRef]
- War, A.R.; Paulraj, M.G.; Ignacimuthu, S.; Sharma, H.C. Defensive responses in groundnut against chewing and sap-sucking insects. J. Plant Growth Regul. 2013, 32, 259–272. [Google Scholar] [CrossRef] [Green Version]
- Appel, H.M. Phenolics in ecological interactions: The importance of oxidation. J. Chem. Ecol. 1993, 19, 1521–1552. [Google Scholar] [CrossRef]
- Kielkiewicz, M. Concentration of some phenylpropanoid compounds and the activity of oxidative enzymes within tomato plant (Lycopersicon esculentum Mill.) locally infested by the carmine spider mite (Tetranychus cinnabarinus Boisd.). Zesz. Nauk. Ochr. Srod. 1998, 214, 41–47. [Google Scholar]
- Kant, M.R.; Ament, K.; Sabelis, M.W.; Haring, M.A.; Scuurink, R.C. Diferential timing of spider mite induced direct and indirect defenses in tomato plants. Plant Physiol. 2004, 135, 483–495. [Google Scholar] [CrossRef] [Green Version]
- Grinberg, M.; Perl-Treves, R.; Palevsky, E.; Shomer, I.; Soroker, V. Interaction between cucumber plants and the broad mite, Polyphagotarsonemus latus: From damage to defense gene expression. Entomol. Exp. Appl. 2005, 115, 135–144. [Google Scholar] [CrossRef]
- Gagné, F. Biochemical Ecotoxicology, Principles and Methods; Academic Press: San Diego, CA, USA, 2014; p. 282. [Google Scholar]
- Resende, M.D.V.D. Software Selegen-REML/BLUP: A useful tool for plant breeding. Crop. Breed. Appl. Biotechnol. 2016, 16, 330–339. [Google Scholar] [CrossRef]
- Rao, C.R. Linear Statistical Inference and Its Applications, 2nd ed.; John Wiley & Sons: Hoboken, NJ, USA, 1973; pp. 220–308. [Google Scholar]
- Singleton, V.L.; Orthofer, R.; Lamuela-Raventos, R.M. Analysis of total phenols and other oxidation substrates and antioxidants by means of folin-ciocaulteau reagent. Methods Enzymol. 1999, 299, 152. [Google Scholar]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Anderson, G.P.; Wang, J.; Chetwynd, J.H. An update and recent validations against airborne high resolution interferometer measurements. In Annual JPL Airborne Earth Science Workshop, 5; Summaries, Jet Propulsion Laboratory: Pasadena, CA, USA, 1995; pp. 5–8. [Google Scholar]
- Lusso, M.F.G.; Pascholati, S.F. Activity and isoenzymatic pattern of soluble peroxidases in maize tissues after mechanical injury or fungal inoculation. Summa Phytopathol. 1999, 25, 244–249. [Google Scholar]
- Kuhn, O.J. Indução de Resistência em Feijoeiro (Phaseolus vulgaris) por Acibenzolar-S-Metil e Bacillus cereus: Aspectos Fisiológiocos, Bioquímicos e Parâmetros de Crescimento e Produção. Ph.D. Thesis, Escola Superior de Agricultura Luiz de Queiroz, Piracicaba, Brazil, 2007. [Google Scholar]
- Shimizu, G.D.; Marubayashi, R.Y.P.; Goncalves, L.S.A. Package AgroR version 1.2.0., Cran R. 2021. Available online: https://cran.r-project.org/web/packages/AgroR/index.html (accessed on 20 May 2021).
- Resende, M.D.V. Genética Biométrica e Estatística no Melhoramento de Plantas Perenes; Embrapa Informação Tecnológica: Brasίlia, Brazil; Embrapa Florestas: Colombo, Sri Lanka, 2002; 975p. [Google Scholar]
- Silva, E.A.; Oliveira, J.V.; Gondim Jr, M.G.; Menezes, D. Biologia de Polyphagotarsonemus latus (Banks) (Acari: Tarsonemidae) em pimentão. An. Soc. Entomol. Bras. 1998, 27, 223–228. [Google Scholar] [CrossRef]
- Ferreira, R.C.; Oliveira, J.V.D.; Haji, F.N.; Gondim Jr, M.G. Biologia, exigências térmicas e tabela de vida de fertilidade do ácaro-branco Polyphagotarsonemus latus (Banks) (Acari: Tarsonemidae) em videira (Vitis vinifera L.) cv. Itália. Neotrop. Entomol. 2006, 35, 126–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.-Q. Mites of Greenhouses: Identification, Biology and Control; CABI: Wallingford, UK, 2003; 244p. [Google Scholar]
- Bernards, M.A.; Båstrup-Spohr, L. Phenylpropanoid metabolism induced by wounding and insect herbivory. In Induced Plant Resistance to Herbivory; Springer: Dordrecht, The Netherlands, 2008; pp. 189–211. [Google Scholar]
- Weintraub, P.G.; Kleitman, S.; Mori, R.; Shapira, N.; Palevsky, E. Control of the broad mite (Polyphagotarsonemus latus (Banks)) on organic greenhouse sweet peppers (Capsicum annuum L.) with the predatory mite, Neoseiulus cucumeris (Oudemans). Biol. Control. 2003, 27, 300–309. [Google Scholar] [CrossRef]
- Qian, L.; He, S.; Liu, X.; Huang, Z.; Chen, F.; Gui, F. Effect of elevated CO2 on the interaction between invasive thrips, Frankliniella occidentalis, and its host kidney bean, Phaseolus vulgaris. Pest. Manag. Sci. 2018, 74, 2773–2782. [Google Scholar] [CrossRef] [PubMed]
- Gayler, S.; Leser, C.; Priesack, E.; Treutter, D. Modelling the effect of environmental factors on the “trade-off” between growth and defensive compounds in young apple trees. Trees 2004, 18, 363–371. [Google Scholar] [CrossRef]
- Kumar, S.; Abedin, M.M.; Singh, A.K.; Das, S. Role of phenolic compounds in plant-defensive mechanisms. In Plant Phenolics in Sustainable Agriculture; Springer: Singapore, 2020; pp. 517–532. [Google Scholar]
- Hata, F.T.; Ventura, M.U.; de Souza, M.S.D.J.; de Sousa, N.V.; Oliveira, B.G.; da Silva, J.B. Mineral and organic fertilization affects Tetranychus urticae, pseudofruit production and leaf nutrient content in strawberry. Phytoparasitica 2019, 47, 513–521. [Google Scholar] [CrossRef]
- Pandhair, V.; Sekhon, B.S. Reactive oxygen species and antioxidants in plants: An overview. J. Plant. Biochem. Biot. 2006, 15, 71–78. [Google Scholar] [CrossRef]
- Lu, F.; Liang, X.; Lu, H.; Li, Q.; Chen, Q.; Zhang, P.; Li, K.; Liu, G.; Yan, W.; Song, J.; et al. Overproduction of superoxide dismutase and catalase confers cassava resistance to Tetranychus cinnabarinus. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef]
- Taggar, G.K.; Gill, R.S.; Gupta, A.K.; Sandhu, J.S. Fluctuations in peroxidase and catalase activities of resistant and susceptible black gram (Vigna mungo (L.) Hepper) genotypes elicited by Bemisia tabaci (Gennadius) feeding. Plant. Signal. Behav. 2012, 7, 1321–1329. [Google Scholar] [CrossRef] [Green Version]
- Mitra, N.; Gotyal, B.S.; Selvaraj, K.; Satpathy, S.; Babu, V.R. Biochemical effects of cultivated and wild jute species on life stages of the broad mite, Polyphagotarsonemus latus (Prostigmata: Tarsonemidae). Fla. Entomol. 2015, 98, 1044–1049. [Google Scholar] [CrossRef]
- Hildebrand, D.F.; Rodriguez, J.G.; Brown, G.C.; Luu, K.T.; Volden, C.S. Peroxidative responses of leaves in two soybean genotypcs injured by twospotted spider mites (Acari: Tetranychidae). J. Econ. Entomol. 1986, 79, 1459–1465. [Google Scholar] [CrossRef]
- Montasser, A.A.; Hanafy, A.R.I.; Taha, A.M.; Hassan, G.M. Infestation of the pepper plant Capsicum annuum L. by the broad mite Polyphagotarsonemus latus (Banks)(Acari: Tarsonemidae) in relation to susceptibility, leaf chemical components, dispersal with phoretic insects and damage caused to the plant leaves. Egyptj J. Zool. 2011, 56, 123–138. [Google Scholar]
- Gotyal, B.S.; De, R.K.; Selvaraj, K.; Satpathy, S.; Kumar, M.; Meena, P.N. Effect of nitrogen fertilizer on yellow mite infestation in Corchorus spp. J. Environ. Biol. 2016, 37, 431–436. [Google Scholar]
- Mondal, C.K.; Acharyya, P.; Saha, U. Evaluation of chilli genotypes against chilli leaf curl complex based on phenol and isozymes study. Bioscan 2016, 11, 3011–3016. [Google Scholar]



| Effect | HT | MT | LT | WP | ||||
|---|---|---|---|---|---|---|---|---|
| Dev | LRT | Dev | LRT | Dev | LRT | Dev | LRT | |
| Genotype | 2164.70 | 2.67 ns | 1547.70 | 1.28 ns | 1341.44 | 0.01 ns | 2281.66 | 4.05 * |
| Complete model | 2162.03 | 1546.42 | 1341.43 | 2277.61 | ||||
| Components/Parameters | HT | MT | LT | WP |
|---|---|---|---|---|
| σ2g | 51.37 | 4.35 | 0.07 | 100.73 |
| σ2perm | 7.78 | 7.23 | 0.55 | 12.94 |
| σ2e | 836.01 | 83.23 | 43.74 | 1258.53 |
| σ2f | 895.15 | 94.81 | 44.36 | 1372.20 |
| h2g | 0.06 | 0.05 | 0.01 | 0.07 |
| h2mg | 0.32 | 0.26 | 0.01 | 0.38 |
| ρ | 0.07 | 0.12 | 0.01 | 0.08 |
| c2perm | 0.01 | 0.08 | 0.01 | 0.01 |
| Overall means | 18.03 | 8.43 | 5.11 | 31.57 |
| Genotypes | g | GV | f | Genotypes | g | GV | f |
|---|---|---|---|---|---|---|---|
| Carioca Original | −8.08 | 23.49 | 12.00 | Rio Tibagi | −1.12 | 30.45 | 31.57 |
| Verdão | −6.82 | 24.76 | 15.57 | LP 13-827 | −0.99 | 30.58 | 29.00 |
| IAPAR 57 | −5.95 | 25.62 | 16.13 | MD-1140 | 0.36 | 31.93 | 32.50 |
| IPR Curió | −5.81 | 25.76 | 16.50 | LP 12-420 | 0.50 | 32.07 | 32.88 |
| Negrão 11 | −5.67 | 25.91 | 16.88 | LP 14-55 | 0.89 | 32.46 | 33.88 |
| MD-1092 | −4.12 | 27.45 | 20.88 | IAPAR 139 | 1.08 | 32.65 | 34.38 |
| IPR Sabiá | −4.08 | 27.50 | 21.00 | RAZ-49 | 1.18 | 32.75 | 34.63 |
| IAPAR-65 | −3.79 | 27.78 | 21.75 | MD-1133 | 1.37 | 32.94 | 35.13 |
| LP 13-833 | −3.64 | 27.93 | 22.13 | LP 11-363 | 1.62 | 33.19 | 39.36 |
| LP 13-827 | −3.50 | 28.07 | 22.50 | H-Amarelo | 2.19 | 33.76 | 37.25 |
| IPR Urutau | −3.02 | 28.56 | 23.75 | IPR Corujinha | 2.86 | 34.44 | 39.00 |
| IPR Celeiro | −2.87 | 28.7 | 24.13 | RAZ-55 | 3.06 | 34.63 | 39.50 |
| IAPAR 81 | −2.58 | 28.99 | 24.88 | Capitão | 3.39 | 34.97 | 40.38 |
| MD-1140 | −2.40 | 29.17 | 28.00 | BRS Pontal | 3.59 | 35.16 | 40.88 |
| IPR Gralha | −2.15 | 29.42 | 26.00 | TAA Bola Cheia | 4.84 | 36.41 | 44.13 |
| RAZ-56 | −1.86 | 29.71 | 26.75 | IPR Tangará | 7.73 | 39.30 | 51.63 |
| IPR Saracura | −1.75 | 29.82 | 29.86 | MD-1092 | 9.08 | 40.65 | 55.13 |
| BRS Esteio | −1.62 | 29.95 | 27.38 | IAC Alvorada | 28.07 | 59.64 | 104.38 |
| Genotypes | Higher | Medium | Lower | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Eggs | |||||||||
| BRS Esteio | 1284.8 | AB | a | 256.8 | AB | b | 264.3 | A | b |
| Carioca Original | 453.8 | BC | a | 391.5 | AB | a | 381.5 | A | a |
| IAC-Alvorada | 2368.0 | A | a | 796.5 | AB | b | 571.8 | A | b |
| LP 13833 | 1286.7 | AB | a | 381.0 | AB | b | 160.8 | A | b |
| MD 1133 | 772.0 | ABC | a | 303.3 | AB | a | 261.8 | A | a |
| Negrão 11 | 464.3 | BC | a | 681.5 | AB | a | 163.0 | A | a |
| IPR Sabiá | 748.5 | ABC | a | 658.3 | AB | ab | 185.3 | A | b |
| TAA Bola Cheia | 1532.3 | AB | a | 1075.3 | A | ab | 621.3 | A | b |
| IPR Uirapuru | 1590.5 | AB | a | 834.0 | AB | ab | 651.3 | A | b |
| Verdão | 78.5 | C | a | 111.8 | B | a | 77.5 | A | a |
| Genotypes | Mobile Forms (Larvae and Adults) | ||||||||
| BRS Esteio | 931.3 | ABC | a | 735.3 | AB | a | 487.8 | A | a |
| Carioca Original | 404.5 | C | a | 315.5 | B | a | 299.3 | A | a |
| IAC-Alvorada | 2078.0 | A | a | 875.8 | AB | b | 407.8 | A | b |
| LP 13833 | 1223.8 | ABC | a | 462.0 | AB | b | 378.8 | A | b |
| MD 1133 | 1047.5 | ABC | a | 816.3 | AB | a | 551.3 | A | a |
| Negrão 11 | 535.5 | BC | a | 773.5 | AB | a | 311.8 | A | a |
| Sabiá | 1233.8 | ABC | a | 643.8 | AB | ab | 251.0 | A | b |
| TAA Bola Cheia | 1758.5 | AB | a | 859.5 | AB | b | 716.0 | A | b |
| Uirapuru | 1861.5 | A | a | 1439.8 | A | ab | 900.0 | A | b |
| Verdão | 383.0 | C | a | 219.3 | B | a | 314.5 | A | a |
| Correlation | AUC-Protein | AUC-TPC | AUC-Cat | AUC-Pox | AUC-PAL | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Eggs | La + Ad | Eggs | La + Ad | Eggs | La + Ad | Eggs | La + Ad | Eggs | La + Ad | |
| r | 0.27 | 0.45 | 0.07 | 0.22 | 0.18 | 0.09 | 0.03 | 0.19 | 0.22 | 0.35 |
| p-value | 0.45 | 0.19 | 0.86 | 0.54 | 0.63 | 0.81 | 0.95 | 0.61 | 0.54 | 0.33 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Androcioli, H.G.; Hoshino, A.T.; Ventura, M.U.; Hata, F.T.; Brugnerotto, M.d.R.; Constantino, L.V.; Marques, F.d.A. Resistance of Common Bean Genotypes to the Broad Mite, Polyphagotarsonemus latus (Banks, 1904) (Acari: Tarsonemidae): Offspring Development and Biochemical Basis. Insects 2021, 12, 910. https://doi.org/10.3390/insects12100910
Androcioli HG, Hoshino AT, Ventura MU, Hata FT, Brugnerotto MdR, Constantino LV, Marques FdA. Resistance of Common Bean Genotypes to the Broad Mite, Polyphagotarsonemus latus (Banks, 1904) (Acari: Tarsonemidae): Offspring Development and Biochemical Basis. Insects. 2021; 12(10):910. https://doi.org/10.3390/insects12100910
Chicago/Turabian StyleAndrocioli, Humberto Godoy, Adriano Thibes Hoshino, Maurício Ursi Ventura, Fernando Teruhiko Hata, Marco dos Reis Brugnerotto, Leonel Vinicius Constantino, and Francisco de Assis Marques. 2021. "Resistance of Common Bean Genotypes to the Broad Mite, Polyphagotarsonemus latus (Banks, 1904) (Acari: Tarsonemidae): Offspring Development and Biochemical Basis" Insects 12, no. 10: 910. https://doi.org/10.3390/insects12100910






