Prevalence, Distribution, and Molecular Record of Four Hard Ticks from Livestock in the United Arab Emirates
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Approval
2.2. Study Site
2.3. Tick Sampling
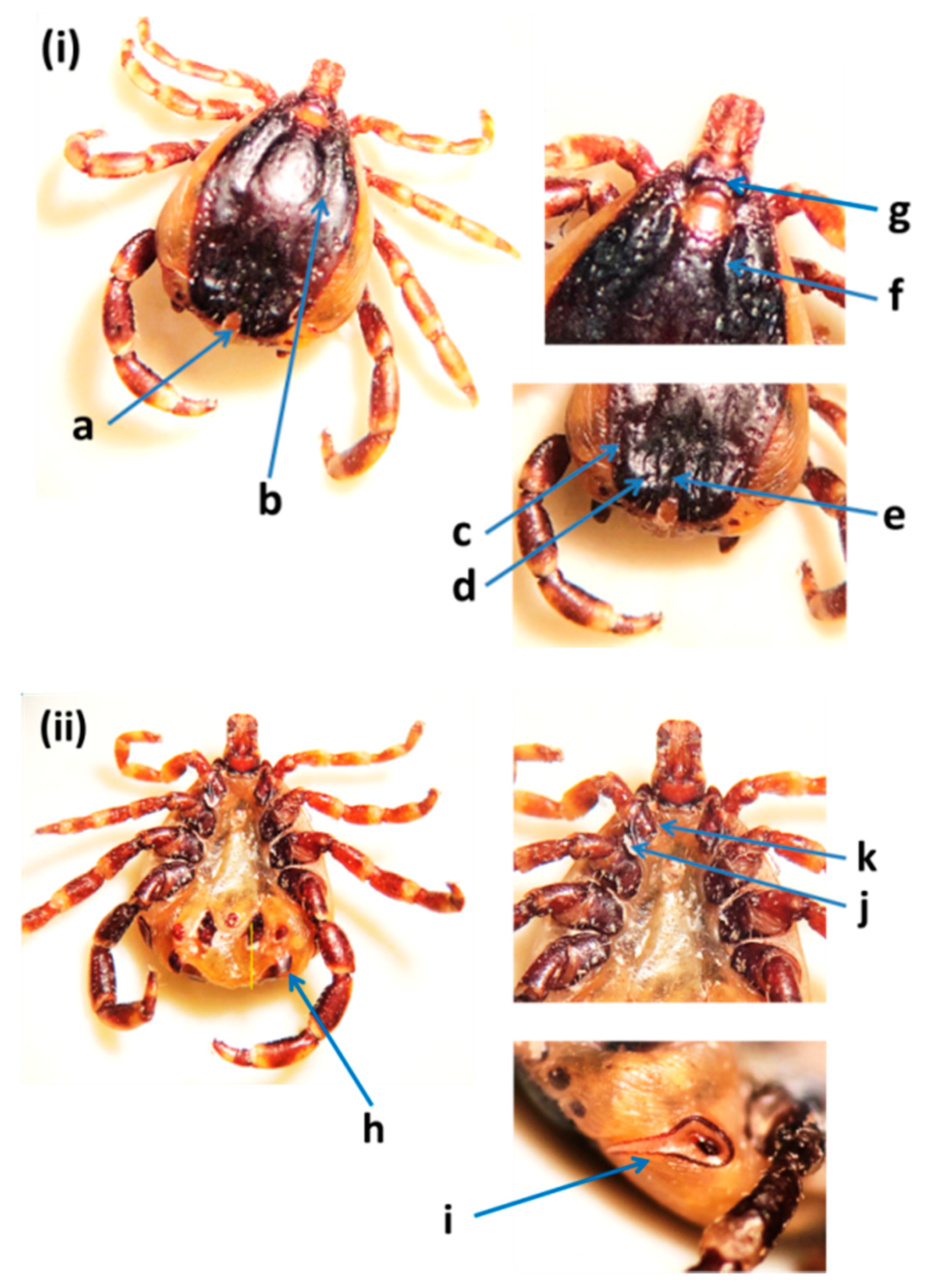
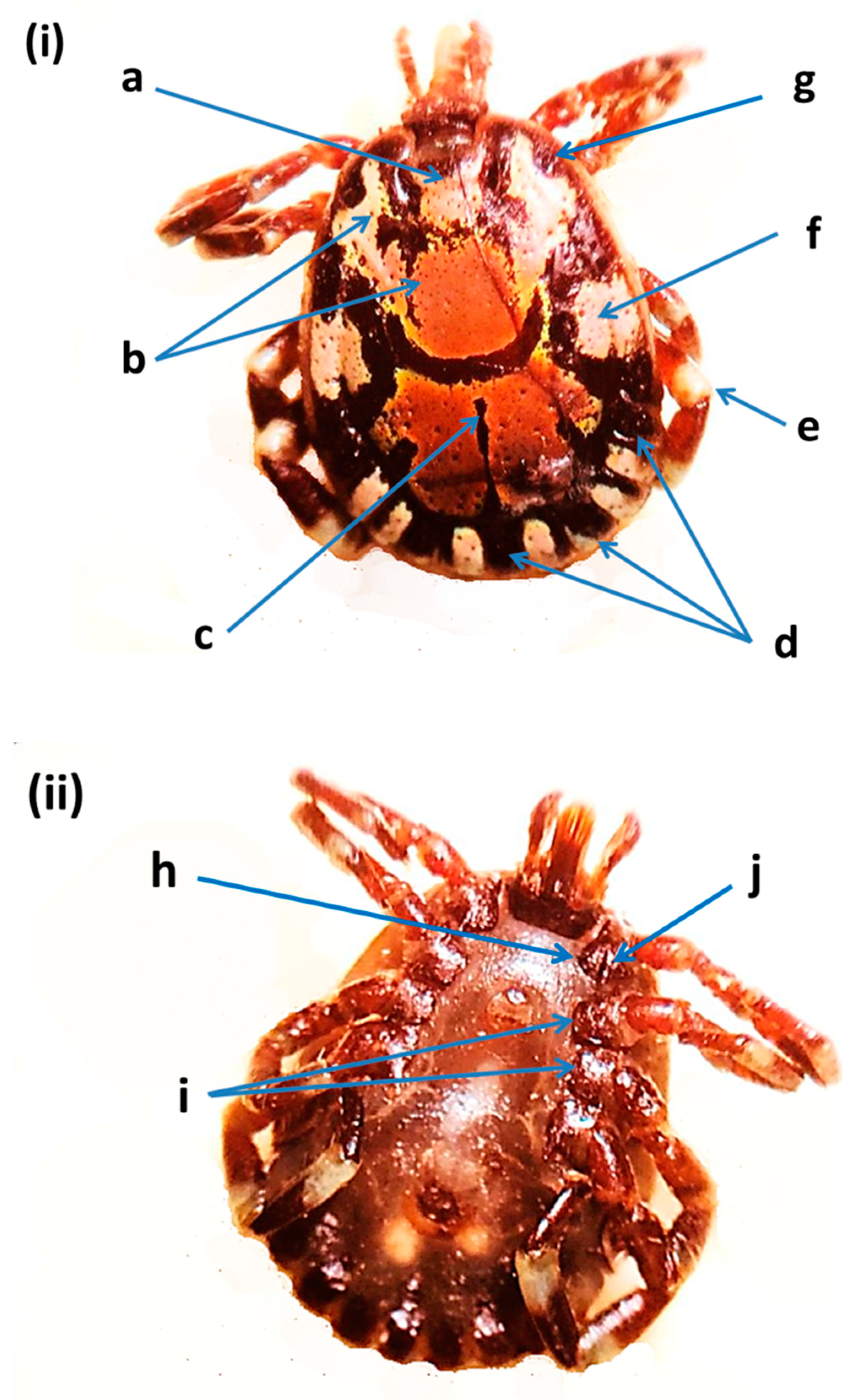
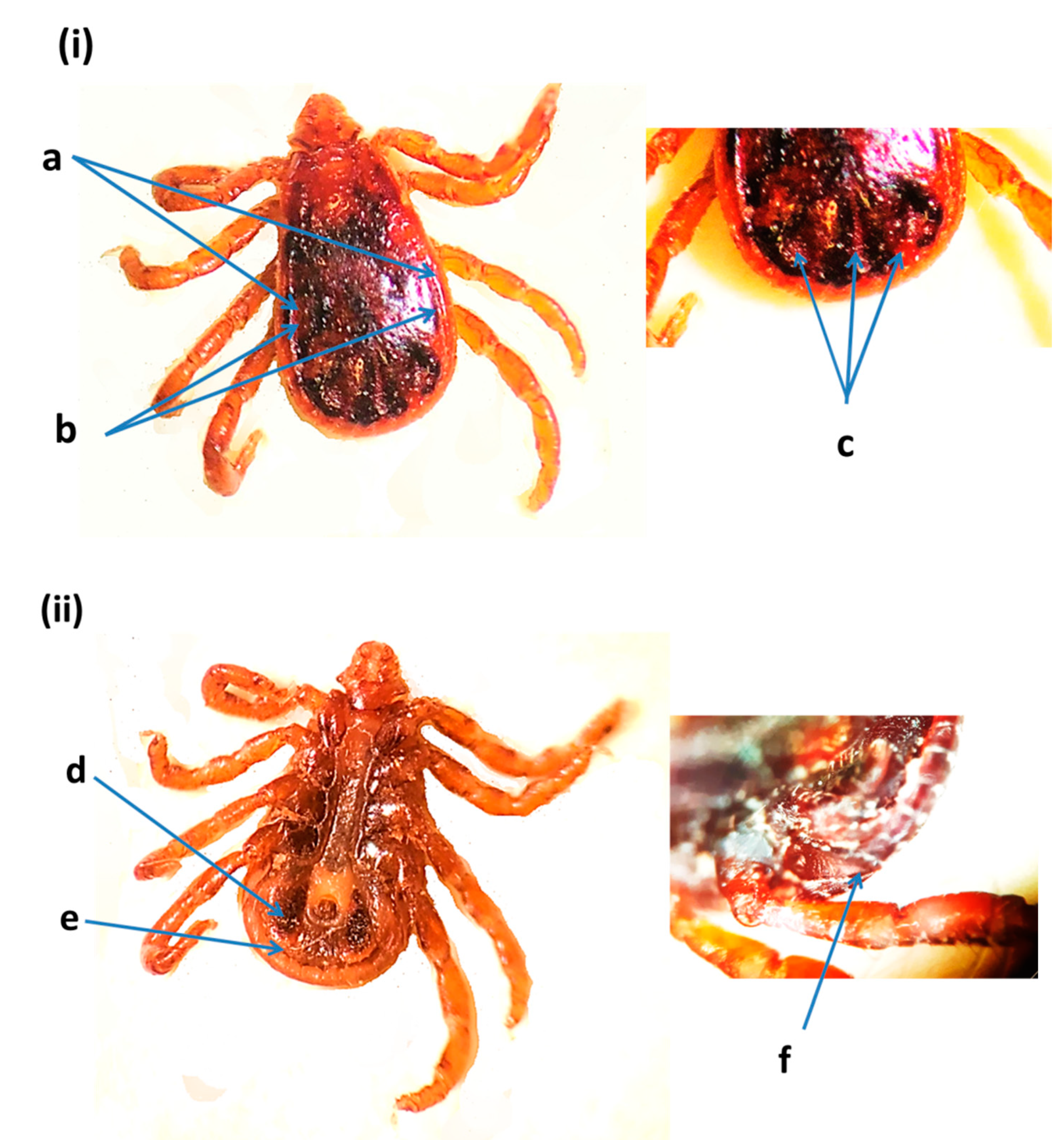
2.4. Morphological Identification of Ticks
2.5. Molecular Characterization of Ticks
2.5.1. DNA Extraction
2.5.2. Polymerase Chain Reaction Amplification
2.5.3. Agarose Gel Analysis, Amplicon Purification and Sequencing
2.5.4. DNA Sequence Analysis
2.5.5. Statistical Analysis
3. Results
3.1. Tick Identification
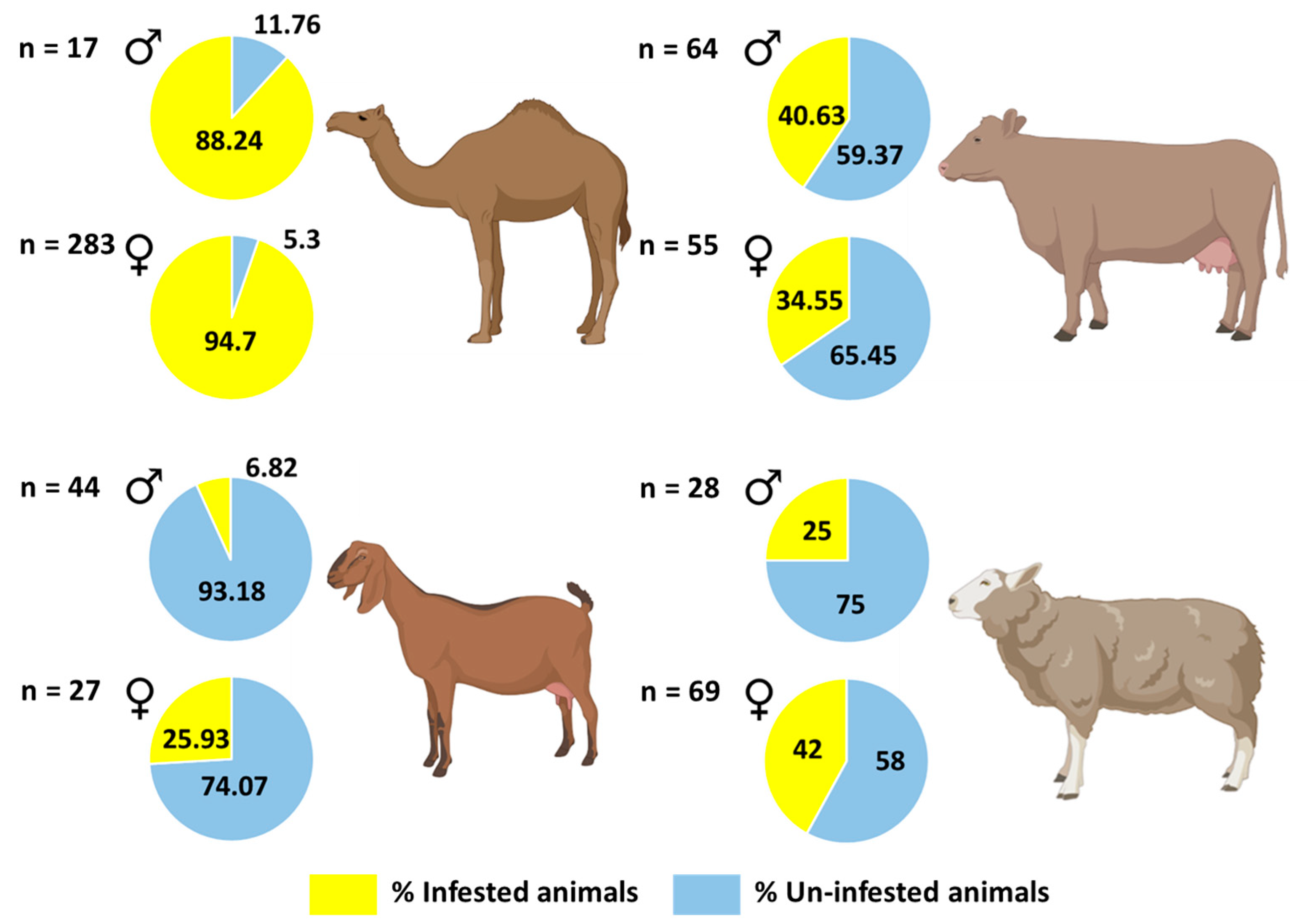
3.2. Tick Prevalence
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Uilenberg, G. Veterinary Significance of Ticks and Tick-Borne Diseases; Springer: Berlin/Heidelberg, Germany, 1992. [Google Scholar]
- Perveen, N.; Muzaffar, S.B.; Al-Deeb, M.A. Ticks and tick-borne diseases of livestock in the Middle East and North Africa: A review. Insects 2021, 12, 83. [Google Scholar] [CrossRef] [PubMed]
- de la Fuente, J.; Estrada-Pena, A.; Venzal, J.M.; Kocan, K.M.; Sonenshine, D.E. Overview: Ticks as vectors of pathogens that cause disease in humans and animals. Front. Biosci. 2008, 13, 6938–6946. [Google Scholar] [CrossRef]
- Yu, Z.; Wang, H.; Wang, T.; Sun, W.; Yang, X.; Liu, J. Tick-borne pathogens and the vector potential of ticks in China. Parasites Vectors 2015, 8, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.; Hussain, A.; Ho, J.; Li, J.; George, D.; Rehman, A.; Zeb, J.; Sparagano, O. An epidemiological survey regarding ticks and tick-borne diseases among livestock owners in Punjab, Pakistan: A one health context. Pathogens 2021, 10, 361. [Google Scholar] [CrossRef] [PubMed]
- Alanazi, A.D.; Al-Mohammed, H.I.; Alyousif, M.S.; Puschendorf, R.; Abdel-Shafy, S. Ticks (Acari: Ixodidae) Infesting domestic and wild mammalians on the Riyadh Province, Saudi Arabia. J. Entomol. 2018, 15, 75–82. [Google Scholar] [CrossRef][Green Version]
- Khan, A.S.; Maupin, G.O.; Rollin, P.E.; Noor, A.M.; Shurie, H.H.M.; Shalabi, A.G.A.; Wasef, S.; Haddad, Y.M.A.; Sadek, R.; Ijaz, K.; et al. An outbreak of Crimean-Congo hemorrhagic fever in the United Arab Emirates, 1994–1995. Am. J. Trop. Med. Hyg. 1997, 57, 519–525. [Google Scholar] [CrossRef]
- Bakheit, M.; Latif, A.; Vatansever, Z.; Seitzer, U.; Ahmed, J. The huge risks due to Hyalomma ticks. In Arthropods as Vectors of Emerging Diseases; Mehlhorn, H., Ed.; Springer: Berlin/Heidelberg, Germany, 2012. [Google Scholar]
- Doğan, M.; Devge, C.; Pata, Y.T.; Sönmezoğlu, M. Case report: Facial nerve paralysis due to intra-aural Hyalomma tick infestation. Turiye Parazitol. Derg. 2012, 36, 254–257. [Google Scholar] [CrossRef]
- Abdullah, H.H.A.M.; El-shanawany, E.E.; Abdel-shafy, S.; Abou-zeina, H.A.A.; Abdel-rahman, E.H. Molecular and immunological characterization of Hyalomma dromedarii and Hyalomma excavatum (Acari: Ixodidae) vectors of Q fever in camels. Vet. World 2018, 11, 1109–1119. [Google Scholar] [CrossRef]
- Walker, A.R.; Bouattour, A.; Camicas, J.L.; Estrada-Peña, A.; Horak, I.G.; Latif, A.A.; Pegram, R.G.; Preston, P.M. Ticks of Domestic Animals in Africa: A Guide to Identification of Species; Bioscience Reports: Edinburgh, UK, 2003. [Google Scholar]
- Perveen, N.; Muzaffar, S.B.; Al-Deeb, M.A. Population dynamics of Hyalomma dromedarii on camels in the United Arab Emirates. Insects 2020, 11, 320. [Google Scholar] [CrossRef]
- Alanazi, A.D.; Nguyen, V.L.; Alyousif, M.S.; Manoj, R.R.S.; Alouffi, A.S.; Donato, R.; Sazmand, A.; Mendoza-Roldan, J.A.; Dantas-Torres, F.; Otranto, D. Ticks and associated pathogens in camels (Camelus dromedarius) from Riyadh Province, Saudi Arabia. Parasites Vectors 2020, 13, 1–9. [Google Scholar] [CrossRef]
- Abdullah, H.H.A.M.; El-molla, A.; Salib, F.A.; Allam, N.A.T.; Ghazy, A.A.; Abdel-shafy, S. Morphological and molecular identification of the brown dog tick Rhipicephalus sanguineus and the camel tick Hyalomma dromedarii (Acari: Ixodidae) vectors of Rickettsioses in Egypt. Vet. World 2016, 9, 1087–1101. [Google Scholar] [CrossRef] [PubMed]
- Dantas-torres, F. Biology and ecology of the brown dog tick, Rhipicephalus sanguineus. Parasit. Vectors 2010, 3, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Cruickshank, R. Molecular markers for the phylogenetics of the mites and ticks. Syst. Appl. Acarol. 2002, 7, 3–14. [Google Scholar] [CrossRef]
- Dabert, M. PDNA markers in the phylogenetics of the Acari. Biol. Lett. 2006, 43, 97–107. [Google Scholar]
- Guglielmone, A.A.; Venzal, M.; Gonza’lez-Acuña, D.; Nava, S.; Hinojosa, A.; Mangold, A.J. The phylogenetic position of Ixodes stilesi Neumann, 1911 (Acari: Ixodidae): Morphological and preliminary molecular evidences from 16S rDNA sequences. Syst. Parasitol. 2006, 65, 1–11. [Google Scholar] [CrossRef]
- Apanaskevich, D.A.; Horak, I.G. The genus Hyalomma koch, 1844: V. re-evaluation of the taxonomic rank of taxa comprising the H. (Euhyalomma) marginatum koch complex of species (acari: Ixodidae) with redescription of all parasitic stages and notes on biology. Int. J. Acarol. 2008, 34, 13–42. [Google Scholar] [CrossRef]
- Ganjali, M.; Haddadzadeh, H.; Shayan, P. Nucleotide sequence analysis of the Second Internal Transcribed Spacer (ITS2) in Hyalomma anatolicum anatolicum in Iran. Int. J. Vet. Res. 2011, 5, 89–94. [Google Scholar]
- Jizhou, L.; Shaoqiang, W.; Yongning, Z.; Tianyi, Z.; Chunyan, F.; Guangle, J.; Xiangmei, L. Development of a DNA barcoding system for the Ixodida (Acari: Ixodida). Mitochondrial DNA 2014, 25, 142–149. [Google Scholar]
- Lv, J.; Wu, S.; Zhang, Y.; Chen, Y.; Feng, C.; Yuan, X.; Jia, G.; Deng, J.; Wang, C.; Wang, Q.; et al. Assessment of four DNA fragments (COI, 16S rDNA, ITS2, 12S rDNA) for species identification of the Ixodida (Acari: Ixodida). Parasit. Vectors 2014, 7, 1–11. [Google Scholar] [CrossRef]
- Dobson, S.J.; Barker, S.C. Phylogeny of the hard ticks (Ixodidae) inferred from 18S rRNA indicates that the genus Aponomma is paraphyletic. Mol. Phylogenet. Evol. 1999, 11, 288–295. [Google Scholar] [CrossRef]
- Norris, D.E.; Klompen, J.S.H.; Black, W.C. Comparison of the Mitochondrial 12S and 16S Ribosomal Dna genes in resolving phylogenetic relationships among hard ticks (Acari: Ixodidae). Ann. Entomol. Soc. Am. 1999, 92, 117–129. [Google Scholar] [CrossRef]
- Al-Deeb, M.A.; Muzaffar, S.B.; Abu-Zeid, Y.A.; Enan, M.R.; Karim, S. First record of a spotted fever group Rickettsia sp. and Theileria annulata in Hyalomma dromedarii (Acari: Ixodidae) ticks in the United Arab Emirates. Fla. Entomol. 2015, 98, 135–139. [Google Scholar] [CrossRef]
- Al-Deeb, M.A.; Enan, M.R. Genetic diversity in the camel tick Hyalomma dromedarii (Acari: Ixodidae) based on Mitochondrial Cytochrome c Oxidase Subunit I (COI) and Randomly Amplified Polymorphic DNA Polymerase Chain Reaction (RAPD-PCR). Adv. Entomol. 2018, 6, 285–294. [Google Scholar] [CrossRef]
- Suleiman, S.Y.A. Geophysical and Hydrogeological Studies of Al- Foah Area, North Al Ain, United Arab Emirates (UAE). Master’s Thesis, United Arab Emirates University, Abu Dhabi, United Arab Emirates, 2007. [Google Scholar]
- Carpi, G.; Cagnacci, F.; Wittekindt, N.E.; Zhao, F.; Qi, J.; Lynn, P.; Drautz, D.I.; Rizzoli, A.; Schuster, S.C. Metagenomic profile of the bacterial communities associated with Ixodes ricinus ticks. PLoS ONE 2011, 6, e25604. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, V.C. Ixodid Ticks (Acarina, Ixodidae) of West Pakistan. Ph.D. Thesis, University of Maryland, Maryland, MD, USA, 1967. [Google Scholar]
- Robinson, L.E. The Genus Amblyomma; Cambridge University Press: Cambridge, UK, 1962. [Google Scholar]
- Apanaskevich, D.A. Differentiation of closely related species Hyalomma anatolicum and Hyalomma excavatum (Acari, Ixodidae) based on a study of all life cycle stages, throughout entire geographical range. Parazitologiia 2003, 37, 259–280. [Google Scholar] [PubMed]
- Apanaskevich, D.A.; Schuster, A.L.; Horak, I.G. The Genus Hyalomma: VII. redescription of all parasitic stages of H. (Euhyalomma) dromedarii and H. (E.) schulzei (Acari: Ixodidae). J. Med. Entomol. 2008, 45, 817–831. [Google Scholar] [CrossRef] [PubMed]
- Black, W.C.; Piesman, J. Phylogeny of hard- and soft-tick taxa (Acari: Ixodida) based on mitochondrial 16S rDNA sequences. Proc. Natl. Acad. Sci. USA 1994, 91, 10034–10038. [Google Scholar] [CrossRef] [PubMed]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar]
- Rózsa, L.; Reiczigel, J.; Majoros, G. Quantifying parasites in samples of hosts. J. Parasitol. 2000, 86, 228–232. [Google Scholar] [CrossRef]
- Hasson, R.H. Tick distribution and infestation among sheep and cattle in Baghdad’s south suburb. Kufa J. Vet. Med. Sci. 2012, 3, 77–90. [Google Scholar]
- Sajid, M.S.; Iqbal, Z.; Khan, M.N.; Muhammad, G.; Khan, M.K. Prevalence and associated risk factors for bovine tick infestation in two districts of lower Punjab, Pakistan. Prev. Vet. Med. 2009, 92, 386–391. [Google Scholar] [CrossRef]
- Atif, F.A.; Khan, M.S.; Iqbal, H.J.; Ali, Z.; Ullah, S. Prevalence of cattle tick infestation in three districts of the Punjab, Pakistan. Pak. J. Sci. 2012, 64, 49. [Google Scholar]
- Al-Deeb, M.A.; Muzaffar, S.B. Prevalence, distribution on host’s body, and chemical control of camel ticks Hyalomma dromedarii in the United Arab Emirates. Vet. World 2020, 13, 114–120. [Google Scholar] [CrossRef] [PubMed]
- Perveen, N.; Muzaffar, S.B.; Vijayan, R.; Al-Deeb, M.A. Microbial communities associated with the camel tick, Hyalomma dromedarii: 16S rRNA gene-based analysis. Sci. Rep. 2020, 10, 1–11. [Google Scholar]
- Elbir, H.; Almathen, F.; Alhumam, N.A. A glimpse of the bacteriome of Hyalomma dromedarii ticks infesting camels reveals human Helicobacter pylori pathogen. J. Infect. Dev. Ctries. 2019, 13, 1001–1012. [Google Scholar] [CrossRef] [PubMed]
- Alreshidi, M.M. Description of microbial diversity associated with ticks Hyalomma dromedarii (Acari: Ixodidae) isolated from camels in Hail region (Saudi Arabia) using massive sequencing of 16S rDNA. Bioinformation 2020, 16, 602–610. [Google Scholar] [CrossRef]
- Musa, H.I.; Jajere, S.M.; Adamu, N.B.; Atsanda, N.N.; Lawal, J.R.; Adamu, S.G.; Lawal, E.K. Prevalence of tick infestation in different breeds of cattle in Maiduguri, Northeastern Nigeria. Bangl. J. Vet. Med. 2014, 12, 161–166. [Google Scholar] [CrossRef]
- Ghafar, A.; Gasser, R.B.; Rashid, I.; Ghafoor, A.; Jabbar, A. Exploring the prevalence and diversity of bovine ticks in five agro-ecological zones of Pakistan using phenetic and genetic tools. Ticks Tick. Borne. Dis. 2020, 11, 101472. [Google Scholar] [CrossRef]
- Malla, M.E.; Payne, V.K.; Cedric, Y. Prevalence of tick infestation of sheep and goats in Bui and Donga-Mantung Divisions of the North West Region of Cameroon. J. Anim. Sci. Vet. Med. 2021, 6, 20–29. [Google Scholar] [CrossRef]
- Damian, D.; Damas, M.; Wensman, J.J.; Berg, M. Molecular diversity of hard tick species from selected areas of a wildlife-livestock interface ecosystem at mikumi national park, Morogoro Region, Tanzania. Vet. Sci. 2021, 8, 36. [Google Scholar] [CrossRef]
- Aktas, M.; Dumanli, N.; Angin, M. Cattle infestation by Hyalomma ticks and prevalence of Theileria in Hyalomma species in the east of Turkey. Vet. Parasitol. 2004, 119, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Biglari, P.; Bakhshi, H.; Chinikar, S.; Belqeiszadeh, H.; Ghaffari, M.; Javaherizadeh, S.; Faghihi, F.; Telmadarraiy, Z. Hyalomma anatolicum as the main infesting tick in an important livestock rearing region, central area of Iran. Iran. J. Public Health 2018, 47, 742. [Google Scholar] [PubMed]
- Choubdar, N.; Karimian, F.; Koosha, M.; Oshaghi, M.A. An integrated overview of the bacterial flora composition of Hyalomma anatolicum, the main vector of cchf. PLoS Negl. Trop. Dis. 2021, 15, e0009480. [Google Scholar] [CrossRef] [PubMed]
- Perveen, N.; Muzaffar, S.B.; Al-Deeb, M.A. Four tick-borne microorganisms and their prevalence in Hyalomma ticks collected from livestock in United Arab Emirates. Pathogens 2021, 10, 1005. [Google Scholar] [CrossRef] [PubMed]
- Dinkisa, G. Review on control of cowdriosis in ruminants. Int. J. Vet. Sci. Technol. 2018, 3, 1–6. [Google Scholar]


| Target Gene | Primer | Sequence (5′–3′) | Cycle Conditions | Amplicon Size (bp) | Reference |
|---|---|---|---|---|---|
| 16S rRNA | 16S + 1 16S − 1 | CTGCTCAATGATTTTTTAAATTGCTGTGG CCGGTCTGAACTCAGATCAAGT | 94 °C 5 min 32 cycles: 94 °C 1 min 52.9 °C 1 min 72 °C 1 min 72 °C 15 min | 460 | [33] |
| cox1 | LCO1490 HCO2198 | GGTCAACAAATCATAAAGATATTGG TAAACTTCAGGGTGACCAAAAAATCA | 95 °C 5 min 30 cycles: 94 °C 1 min 54 °C 1 min 72 °C 1 min 30 s 72 °C 10 min | 710 | [34] |
| Sample Accession | Host | Location | GenBank Reference | Identity% | Species | |
|---|---|---|---|---|---|---|
| 16S | cox1 | |||||
| MZ976772 | Camel | Abu Dhabi | L34306.1 | - | 99.27 | H. dromedarii |
| OK017169 | Cow | Dubai | - | MT800311.1 | 99.70 | H. anatolicum |
| MZ976771 | Cow | Dubai | MK829042.1 | - | 99.28 | H. anatolicum |
| MZ976770 | Sheep | Dubai | MK829042.1 | - | 99.52 | H. anatolicum |
| MZ976780 | Goat | Dubai | KC203338.1 | - | 99.51 | H. anatolicum |
| MZ976769 | Cow | Sharjah | MG066692.1 | - | 99.03 | R. sanguineus |
| OK001821 | Cow | Dubai | - | KP987775.1 | 99.84 | A. lepidum |
| Hosts | Examined Animals | Infested with Ticks | Prevalence (95% Confidence Level) | Mean Intensity (95% Confidence Level) | Mean Abundance (95% Confidence Level) |
|---|---|---|---|---|---|
| Camels | 300 | 283 | 0.94 (0.91–0.97) | 17 (15.23–19.52) | 16 (14.39–18.63) |
| Cows | 119 | 45 | 0.38 (0.29–0.47) | 14.47 (11.18–18.87) | 5.47 (3.99–7.87) |
| Sheep | 97 | 36 | 0.37 (0.28–0.48) | 7.69 (5.69–10.67) | 2.85 (1.96–4.36) |
| Goats | 71 | 10 | 0.14 (0.07–0.24) | 21.9 (10.50–55) | 3.08 (1.14–9.08) |
| Hosts | Camels | Cows | Sheep | Goats | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P (%) | MI (M ± SE) | MA (M ± SE) | P (%) | MI (M ± SE) | MA (M ± SE) | P (%) | MI (M ± SE) | MA (M ± SE) | P (%) | MI (M ± SE) | MA (M ± SE) | |
| H. dromedarii | 94.3 | 16.52 ± 1.05 | 15.58 ± 1.01 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| H. anatolicum | 3.7 | 1.18 ± 0.12 | 0.04 ± 0.01 | 32.8 | 8.51 ± 1.34 | 2.79 ± 0.57 | 14.4 | 10.36 ± 3.25 | 1.5 ± 0.59 | 9.9 | 15.14 ± 10.38 | 1.49 ± 1.10 |
| R. sanguineus | 0 | 0 | 0 | 0.8 | 5 | 0.04 ± 0.04 | 0 | 0 | 0 | 0 | 0 | 0 |
| A. lepidum | 0 | 0 | 0 | 0.8 | 2 | 0.02 ± 0.02 | 0 | 0 | 0 | 0 | 0 | 0 |
| Others | 8 | 4.83 ± 0.56 | 0.39 ± 0.09 | 36.1 | 7.26 ± 0.96 | 2.62 ± 0.47 | 34 | 4 ± 1.01 | 1.4 ± 0.39 | 14.1 | 11.7 ± 3.53 | 1.65 ± 0.68 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Perveen, N.; Muzaffar, S.B.; Al-Deeb, M.A. Prevalence, Distribution, and Molecular Record of Four Hard Ticks from Livestock in the United Arab Emirates. Insects 2021, 12, 1016. https://doi.org/10.3390/insects12111016
Perveen N, Muzaffar SB, Al-Deeb MA. Prevalence, Distribution, and Molecular Record of Four Hard Ticks from Livestock in the United Arab Emirates. Insects. 2021; 12(11):1016. https://doi.org/10.3390/insects12111016
Chicago/Turabian StylePerveen, Nighat, Sabir Bin Muzaffar, and Mohammad Ali Al-Deeb. 2021. "Prevalence, Distribution, and Molecular Record of Four Hard Ticks from Livestock in the United Arab Emirates" Insects 12, no. 11: 1016. https://doi.org/10.3390/insects12111016
APA StylePerveen, N., Muzaffar, S. B., & Al-Deeb, M. A. (2021). Prevalence, Distribution, and Molecular Record of Four Hard Ticks from Livestock in the United Arab Emirates. Insects, 12(11), 1016. https://doi.org/10.3390/insects12111016








