Hemolymph Ecdysteroid Titer Affects Maternal mRNAs during Bombyx mori Oogenesis
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Animals and 20-Hydroxyecdysone
2.2. Ligature and 20-Hydroxyecdysone Treatment
2.3. Sample Preparation
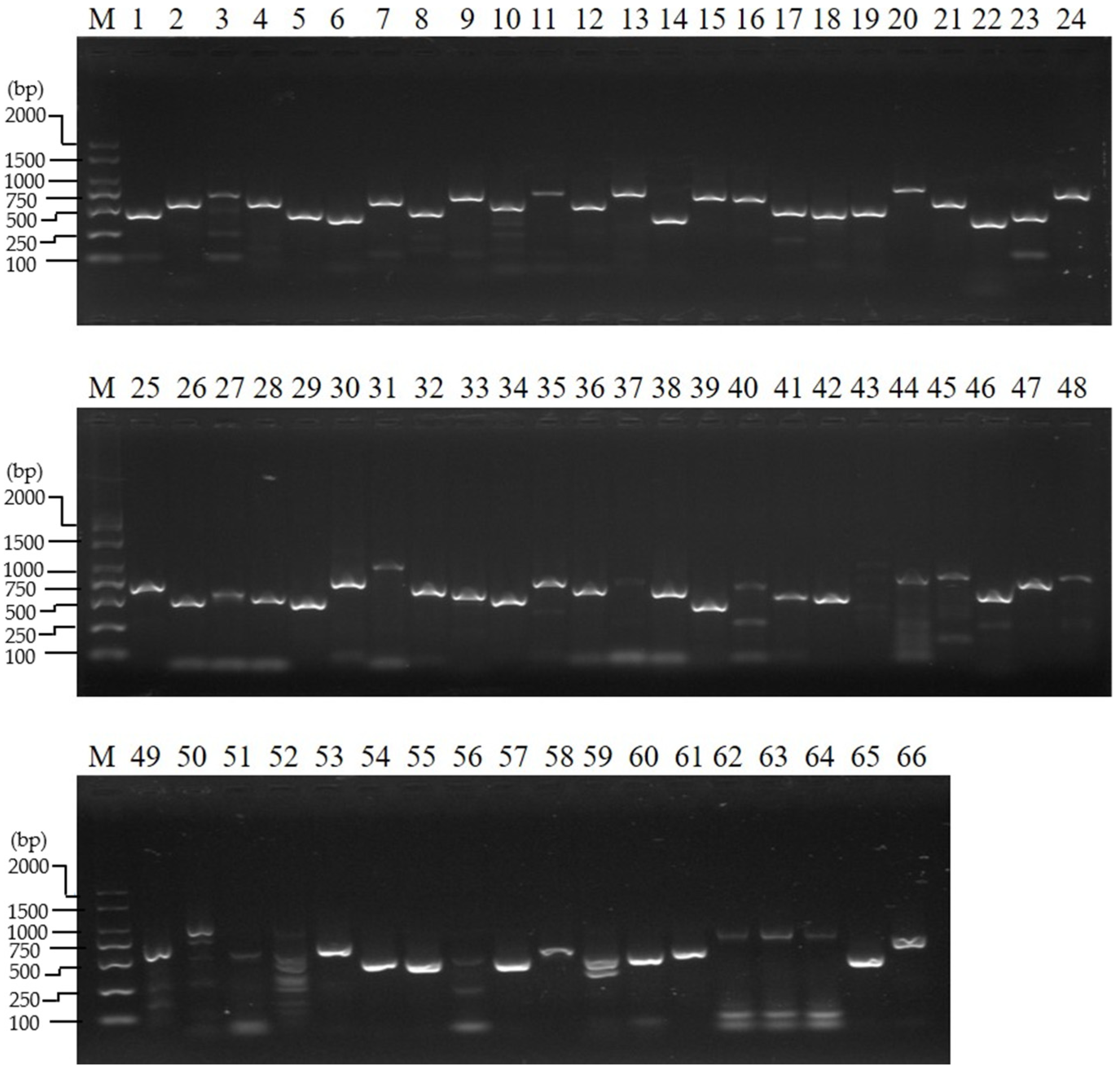
2.4. Reverse-Transcription PCR and Reverse Transcription–Quantitative PCR Analysis
2.5. Kyoto Encyclopedia of Genes and Genomes Annotation
3. Results
3.1. Gene Expression Analysis of the Ovary at the Wandering Stage
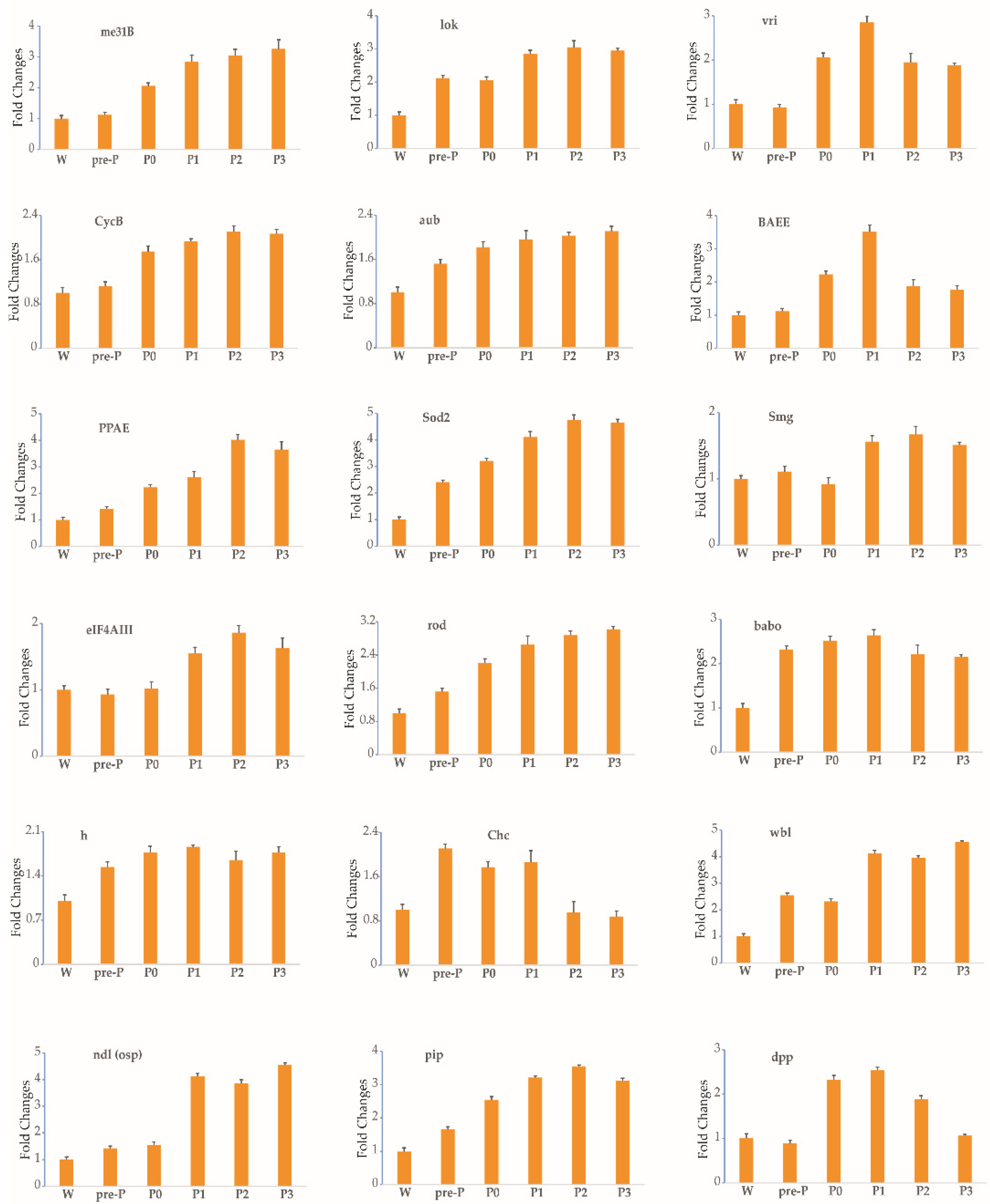
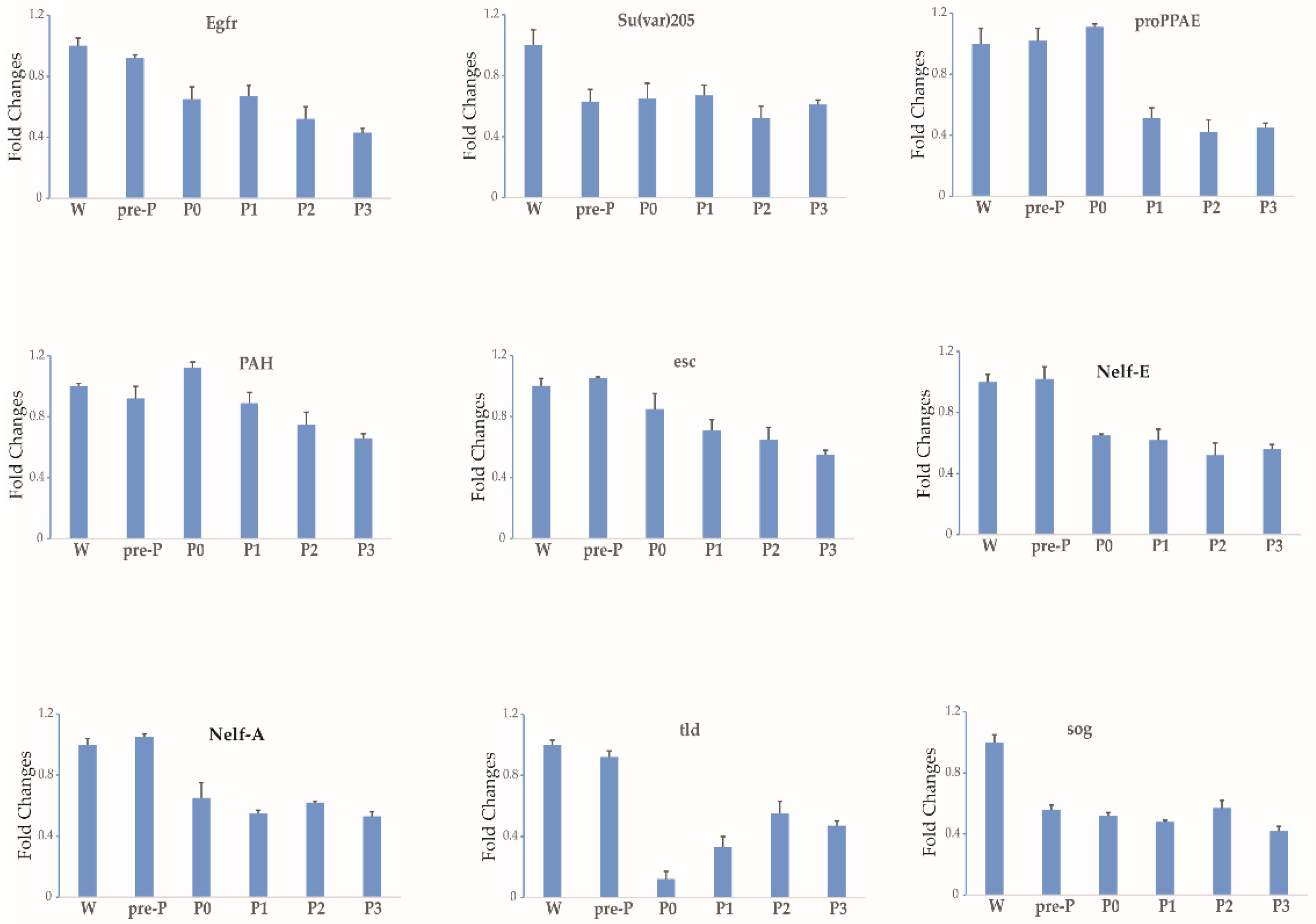
3.2. Expression Profile of Each Gene during Stages 3–5 of Oogenesis
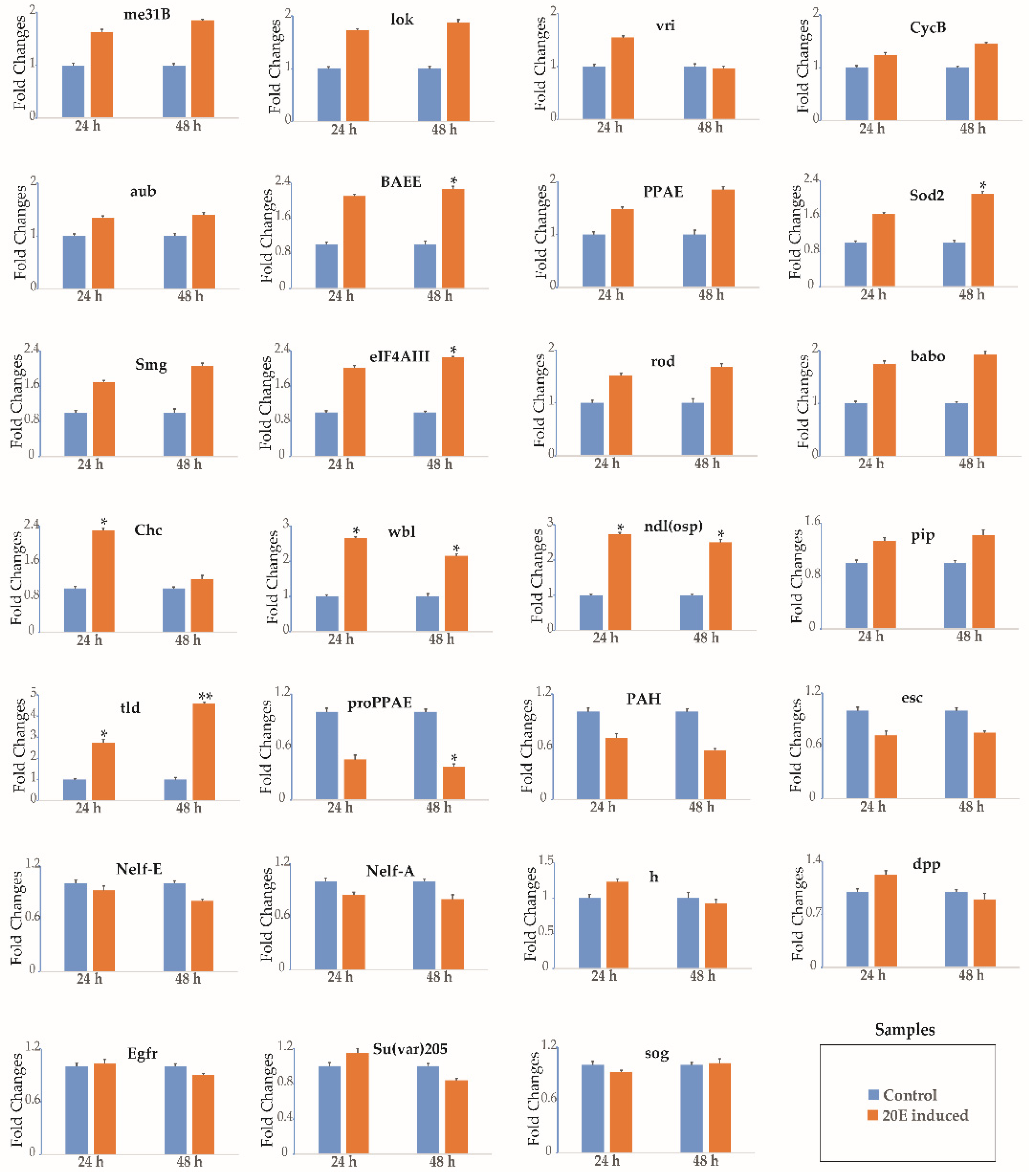
3.3. 20-Hydroxyecdysone Regulates the Expression of Select Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Langley, A.R.; Smith, J.C.; Stemple, D.L.; Harvey, S.A. New insights into the maternal to zygotic transition. Development 2014, 141, 3834–3841. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vastenhouw, N.L.; Cao, W.X.; Lipshitz, H.D. The maternal-to-zygotic transition revisited. Development 2019, 146. [Google Scholar] [CrossRef] [Green Version]
- Horner, V.L.; Wolfner, M.F. Mechanical stimulation by osmotic and hydrostatic pressure activates Drosophila oocytes in vitro in a calcium-dependent manner. Dev. Biol. 2008, 316, 100–109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tadros, W.; Lipshitz, H.D. The maternal-to-zygotic transition: A play in two acts. Development 2009, 136, 3033–3042. [Google Scholar] [CrossRef] [Green Version]
- Chang, H.; Yeo, J.; Kim, J.G.; Kim, H.; Lim, J.; Lee, M.; Kim, H.H.; Ohk, J.; Jeon, H.Y.; Lee, H.; et al. Terminal Uridylyltransferases Execute Programmed Clearance of Maternal Transcriptome in Vertebrate Embryos. Mol. Cell 2018, 70, 72–82.e77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, B.; Wu, X.; Zhang, W.; Shen, W.; Sun, Q.; Liu, K.; Zhang, Y.; Wang, Q.; Li, Y.; Meng, A.; et al. Widespread Enhancer Dememorization and Promoter Priming during Parental-to-Zygotic Transition. Mol. Cell 2018, 72, 673–686.e676. [Google Scholar] [CrossRef] [Green Version]
- Atallah, J.; Lott, S.E. Evolution of maternal and zygotic mRNA complements in the early Drosophila embryo. PLoS Genet. 2018, 14, e1007838. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.X.; Kabelitz, S.; Gupta, M.; Yeung, E.; Lin, S.C.; Rammelt, C.; Ihling, C.; Pekovic, F.; Low, T.C.H.; Siddiqui, N.U.; et al. Precise Temporal Regulation of Post-transcriptional Repressors Is Required for an Orderly Drosophila Maternal-to-Zygotic Transition. Cell Rep. 2020, 31, 107783. [Google Scholar] [CrossRef]
- Benoit, B.; He, C.H.; Zhang, F.; Votruba, S.M.; Tadros, W.; Westwood, J.T.; Smibert, C.A.; Lipshitz, H.D.; Theurkauf, W.E. An essential role for the RNA-binding protein Smaug during the Drosophila maternal-to-zygotic transition. Development 2009, 136, 923–932. [Google Scholar] [CrossRef] [Green Version]
- Gallo, C.M.; Munro, E.; Rasoloson, D.; Merritt, C.; Seydoux, G. Processing bodies and germ granules are distinct RNA granules that interact in C. elegans embryos. Dev. Biol. 2008, 323, 76–87. [Google Scholar] [CrossRef] [Green Version]
- Rabani, M.; Pieper, L.; Chew, G.L.; Schier, A.F. A Massively Parallel Reporter Assay of 3 ‘ UTR Sequences Identifies In Vivo Rules for mRNA Degradation. Mol. Cell 2017, 68, 1083–1094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giraldez, A.J.; Mishima, Y.; Rihel, J.; Grocock, R.J.; Van Dongen, S.; Inoue, K.; Enright, A.J.; Schier, A.F. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science 2006, 312, 75–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramos, S.B.; Stumpo, D.J.; Kennington, E.A.; Phillips, R.S.; Bock, C.B.; Ribeiro-Neto, F.; Blackshear, P.J. The CCCH tandem zinc-finger protein Zfp36l2 is crucial for female fertility and early embryonic development. Development 2004, 131, 4883–4893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, H.L.; Nien, C.Y.; Liu, H.Y.; Metzstein, M.M.; Kirov, N.; Rushlow, C. The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature 2008, 456, 400–403. [Google Scholar] [CrossRef] [Green Version]
- Lee, M.T.; Bonneau, A.R.; Takacs, C.M.; Bazzini, A.A.; DiVito, K.R.; Fleming, E.S.; Giraldez, A.J. Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during the maternal-to-zygotic transition. Nature 2013, 503, 360–364. [Google Scholar] [CrossRef] [Green Version]
- De Iaco, A.; Planet, E.; Coluccio, A.; Verp, S.; Duc, J.; Trono, D. DUX-family transcription factors regulate zygotic genome activation in placental mammals. Nat. Genet. 2017, 49, 941–945. [Google Scholar] [CrossRef]
- De Iaco, A.; Coudray, A.; Duc, J.; Trono, D. DPPA2 and DPPA4 are necessary to establish a 2C-like state in mouse embryonic stem cells. Embo Rep. 2019, 20, e47382. [Google Scholar] [CrossRef]
- Eckersley-Maslin, M.; Alda-Catalinas, C.; Blotenburg, M.; Kreibich, E.; Krueger, C.; Reik, W. Dppa2 and Dppa4 directly regulate the Dux-driven zygotic transcriptional program. Genes Dev. 2019, 33, 194–208. [Google Scholar] [CrossRef] [Green Version]
- Hendrickson, P.G.; Dorais, J.A.; Grow, E.J.; Whiddon, J.L.; Lim, J.W.; Wike, C.L.; Weaver, B.D.; Pflueger, C.; Emery, B.R.; Wilcox, A.L.; et al. Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat. Genet. 2017, 49, 925–934. [Google Scholar] [CrossRef]
- Gao, L.; Wu, K.L.; Liu, Z.B.; Yao, X.L.; Yuan, S.L.; Tao, W.R.; Yi, L.Z.; Yu, G.L.; Hou, Z.Z.; Fan, D.D.; et al. Chromatin Accessibility Landscape in Human Early Embryos and Its Association with Evolution. Cell 2018, 173, 248–259. [Google Scholar] [CrossRef] [Green Version]
- Goldsmith, M.R.; Shimada, T.; Abe, H. The genetics and genomics of the silkworm, Bombyx mori. Annu. Rev. Entomol. 2005, 50, 71–100. [Google Scholar] [CrossRef] [PubMed]
- Xia, Q.; Cheng, D.; Duan, J.; Wang, G.; Cheng, T.; Zha, X.; Liu, C.; Zhao, P.; Dai, F.; Zhang, Z.; et al. Microarray-based gene expression profiles in multiple tissues of the domesticated silkworm, Bombyx mori. Genome Biol. 2007, 8, R162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamauchi, H.; Yoshitake, N. Developmental stages of ovarian follicles of the silkworm, Bombyx mori L. J. Morphol. 1984, 179, 21–31. [Google Scholar] [CrossRef]
- Mizoguchi, A.; Ohashi, Y.; Hosoda, K.; Ishibashi, J.; Kataoka, H. Developmental profile of the changes in the prothoracicotropic hormone titer in hemolymph of the silkworm Bombyx mori: Correlation with ecdysteroid secretion. Insect Biochem. Mol. Biol. 2001, 31, 349–358. [Google Scholar] [CrossRef]
- Swevers, L.; Eystathioy, T.; Iatrou, K. The orphan nuclear receptors BmE75A and BmE75C of the silkmoth Bombyx mori: Hornmonal control and ovarian expression. Insect Biochem. Mol. Biol. 2002, 32, 1643–1652. [Google Scholar] [CrossRef]
- Swevers, L.; Iatrou, K. The ecdysone regulatory cascade and ovarian development in lepidopteran insects: Insights from the silkmoth paradigm. Insect Biochem. Mol. Biol. 2003, 33, 1285–1297. [Google Scholar] [CrossRef]
- Sonobe, H.; Yamada, R. Ecdysteroids during early embryonic development in silkworm Bombyx mori: Metabolism and functions. Zool. Sci. 2004, 21, 503–516. [Google Scholar] [CrossRef] [PubMed]
- Ito, Y.; Yasuda, A.; Sonobe, H. Synthesis and phosphorylation of ecdysteroids during ovarian development in the silkworm, Bombyx mori. Zool. Sci. 2008, 25, 721–727. [Google Scholar] [CrossRef]
- Ito, Y.; Sonobe, H. The role of ecdysteroid 22-kinase in the accumulation of ecdysteroids in ovary of silkworm Bombyx mori. Ann. NY Acad. Sci. 2009, 1163, 421–424. [Google Scholar] [CrossRef]
- Niwa, Y.S.; Niwa, R. Transcriptional regulation of insect steroid hormone biosynthesis and its role in controlling timing of molting and metamorphosis. Dev. Growth Differ. 2016, 58, 94–105. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Sun, W.; Sun, B.Y.; Xiao, Y.; Zhang, Z. The dynamic landscape of gene regulation during Bombyx mori oogenesis. BMC Genom. 2017, 18, 714. [Google Scholar] [CrossRef]
- Zhang, M.R.; Xu, P.Z.; Pang, H.L.; Chen, T.; Zhang, G.Z. Expression Analysis of mRNA Decay of Maternal Genes during Bombyx mori Maternal-to-Zygotic Transition. Int. J. Mol. Sci. 2019, 20, 5651. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.R.; Qin, S.; Xu, P.Z.; Zhang, G.Z. Identifying potential maternal genes of Bombyx mori using digital gene expression profiling. PLoS ONE 2018, 13, e0192745. [Google Scholar] [CrossRef]
- Guo, H.; Jiang, L.; Xia, Q. Selection of reference genes for analysis of stress-responsive genes after challenge with viruses and temperature changes in the silkworm Bombyx mori. Mol. Genet. Genom. 2016, 291, 999–1004. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Kawashima, M. KEGG mapping tools for uncovering hidden features in biological data. Protein Sci. 2021. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.Z.; Zhang, M.R.; Wang, X.Y.; Wu, Y.C. Precocious Metamorphosis of Silkworm Larvae Infected by BmNPV in the Latter Half of the Fifth Instar. Front. Physiol. 2021, 12, 650972. [Google Scholar] [CrossRef]
- Ohnishi, E.; Hiramoto, M.; Fujimoto, Y.; Kakinuma, K.; Ikekawa, N. Isolation and identification of major ecdysteroid conjugates from the ovaries of Bombyx mori. Insect Biochem. 1989, 19, 95–101. [Google Scholar] [CrossRef]
- Makka, T.; Seino, A.; Tomita, S.; Fujiwara, H.; Sonobe, H. A possible role of 20-hydroxyecdysone in embryonic development of the silkworm Bombyx mori. Arch. Insect Biochem. Physiol. 2002, 51, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Kozlova, T.; Thummel, C.S. Essential roles for ecdysone signaling during Drosophila mid-embryonic development. Science 2003, 301, 1911–1914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.F.; Zhang, Z.; Sun, W. Ecdysone oxidase and 3-dehydroecdysone-3beta-reductase contribute to the synthesis of ecdysone during early embryonic development of the silkworm. Int. J. Biol. Sci. 2018, 14, 1472–1482. [Google Scholar] [CrossRef] [Green Version]
- Hanaoka, K.; Onishi, E. Changes in ecdysone titre during pupal-adult development in the silkworm, Bombyx mori. J. Insect Physiol. 1974, 20, 2375–2384. [Google Scholar] [CrossRef]
- Carter, J.M.; Baker, S.C.; Pink, R.; Carter, D.R.; Collins, A.; Tomlin, J.; Gibbs, M.; Breuker, C.J. Unscrambling butterfly oogenesis. BMC Genom. 2013, 14, 283. [Google Scholar] [CrossRef] [Green Version]
- Blythe, S.A.; Wieschaus, E.F. Zygotic Genome Activation Triggers the DNA Replication Checkpoint at the Midblastula Transition. Cell 2015, 160, 1169–1181. [Google Scholar] [CrossRef] [Green Version]
- Briggs, J.A.; Weinreb, C.; Wagner, D.E.; Megason, S.; Peshkin, L.; Kirschner, M.W.; Klein, A.M. The dynamics of gene expression in vertebrate embryogenesis at single-cell resolution. Science 2018, 360, aar5780. [Google Scholar] [CrossRef] [Green Version]
- Sankar, A.; Lerdrup, M.; Manaf, A.; Johansen, J.V.; Gonzalez, J.M.; Borup, R.; Blanshard, R.; Klungland, A.; Hansen, K.; Andersen, C.Y.; et al. KDM4A regulates the maternal-to-zygotic transition by protecting broad H3K4me3 domains from H3K9me3 invasion in oocytes. Nat. Cell Biol. 2020, 22, 380–388. [Google Scholar] [CrossRef] [PubMed]
- Swevers, L.; Iatrou, K. The ecdysone agonist tebufenozide (RH-5992) blocks the progression into the ecdysteroid-induced regulatory cascade and arrests silkmoth oogenesis at mid-vitellogenesis. Insect Biochem. Molec. 1999, 29, 955–963. [Google Scholar] [CrossRef]
- Ballut, L.; Marchadier, B.; Baguet, A.; Tomasetto, C.; Seraphin, B.; Le Hir, H. The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nat. Struct. Mol. Biol. 2005, 12, 861–869. [Google Scholar] [CrossRef]
- Choe, J.; Ryu, I.; Park, O.H.; Park, J.; Cho, H.; Yoo, J.S.; Chi, S.W.; Kim, M.K.; Song, H.K.; Kim, Y.K. eIF4AIII enhances translation of nuclear cap-binding complex-bound mRNAs by promoting disruption of secondary structures in 5′UTR. Proc. Natl. Acad. Sci. USA 2014, 111, E4577–E4586. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, A.; Amikura, R.; Hanyu, K.; Kobayashi, S. Me31B silences translation of oocyte-localizing RNAs through the formation of cytoplasmic RNP complex during Drosophila oogenesis. Development 2001, 128, 3233–3242. [Google Scholar] [CrossRef] [PubMed]
- Gotze, M.; Dufourt, J.; Ihling, C.; Rammelt, C.; Pierson, S.; Sambrani, N.; Temme, C.; Sinz, A.; Simonelig, M.; Wahle, E. Translational repression of the Drosophila nanos mRNA involves the RNA helicase Belle and RNA coating by Me31B and Trailer hitch. RNA 2017, 23, 1552–1568. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| No. of Maternal Genes | Name of Maternal Genes | |
|---|---|---|
| Upward trend | 18 | me31B, lok, vri, CycB, aub, BAEE, PPAE, Sod2, Smg, eIF4AIII, rod, babo, h, Chc, wbl, ndl(osp), pip, dpp |
| Downward trend | 9 | Egfr, Su(var)205, proPPAE, PAH, esc, Nelf-E, Nelf-A, tld, sog |
| Flattening trend | 39 | Pabn2, Eif-4a, Bin1, bai, tud, gammaTub, Mat89Ba, Pc, Btk29A, Src64B, shot, spoon(AKAP1), Msp300(nesprin-1), TPH1, vfl, KCNQ, Sel(cnpy1), Hip14(ZDHHC17), Hp1b-l, spz, mamo, tkv, hb, snk, asp, proSP7, glo(hnRNPF), Csp(DnaJ-7), pie, SPE, sax, gro, SPE-like, Dif, Th, Src42A, Dst, Snap25, Moe |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, M.; Xu, P.; Chen, T. Hemolymph Ecdysteroid Titer Affects Maternal mRNAs during Bombyx mori Oogenesis. Insects 2021, 12, 969. https://doi.org/10.3390/insects12110969
Zhang M, Xu P, Chen T. Hemolymph Ecdysteroid Titer Affects Maternal mRNAs during Bombyx mori Oogenesis. Insects. 2021; 12(11):969. https://doi.org/10.3390/insects12110969
Chicago/Turabian StyleZhang, Meirong, Pingzhen Xu, and Tao Chen. 2021. "Hemolymph Ecdysteroid Titer Affects Maternal mRNAs during Bombyx mori Oogenesis" Insects 12, no. 11: 969. https://doi.org/10.3390/insects12110969
APA StyleZhang, M., Xu, P., & Chen, T. (2021). Hemolymph Ecdysteroid Titer Affects Maternal mRNAs during Bombyx mori Oogenesis. Insects, 12(11), 969. https://doi.org/10.3390/insects12110969





