Exploring the Terminal Pathway of Sex Pheromone Biosynthesis and Metabolism in the Silkworm
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Insect Rearing
2.2. Sample Collection
2.3. cDNA Library Preparation and Illumina Sequencing
2.4. Sequence Assembly and Annotation
2.5. Sequence Acquisition and Phylogenetic Analysis
2.6. Reverse Transcription-Polymerase Chain Reaction (RT-PCR) Analysis
2.7. Quantitative Real-Time PCR (qRT-PCR) Analysis
3. Results
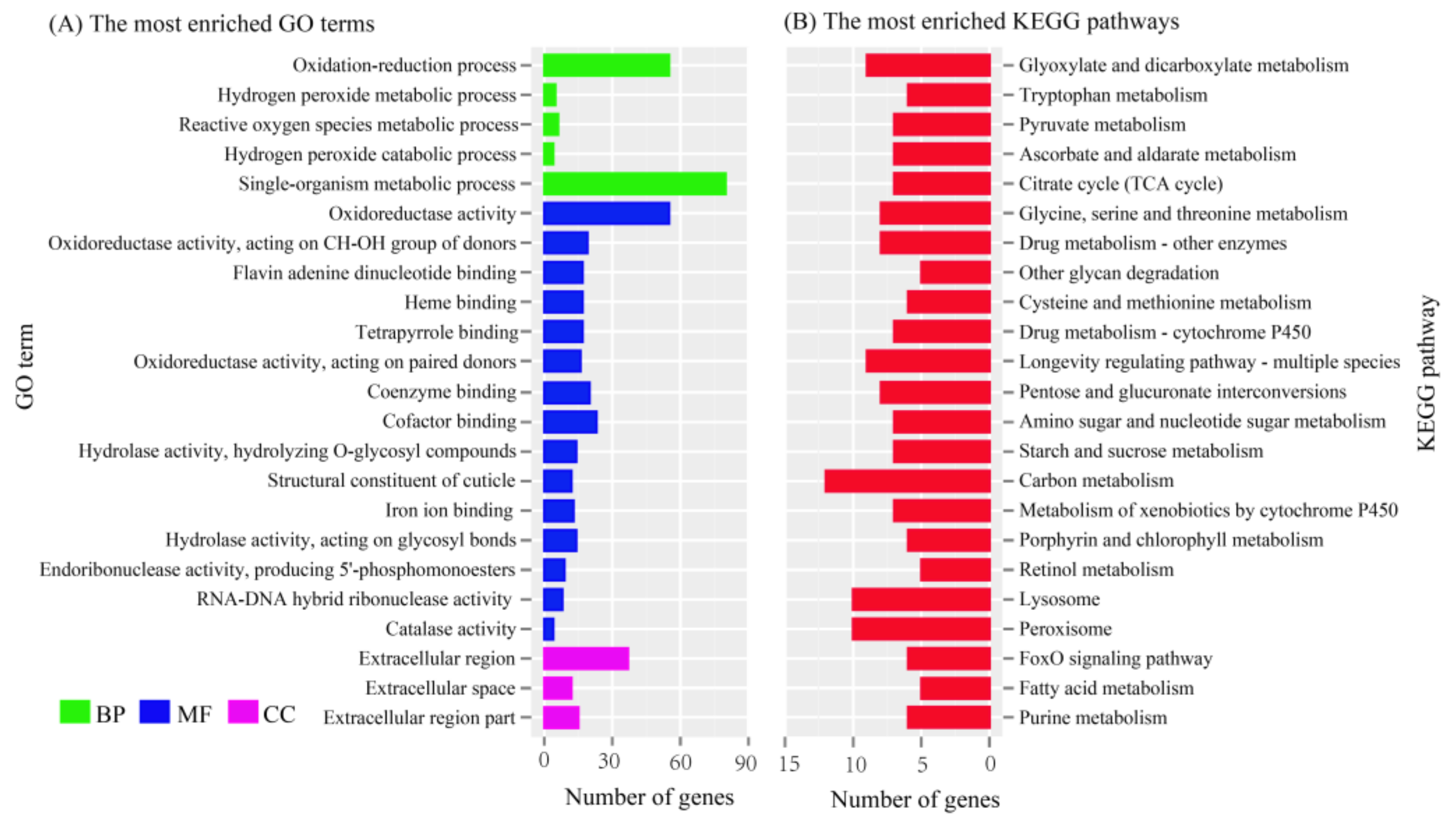
3.1. Transcriptome Sequencing, Assembly, and Functional Annotation
3.2. Differentially Expressed Genes (DEGs) between Bombyx mori and Bombyx mandarina
3.3. Identification of Candidate Genes involved in Pheromone Biosynthesis and Metabolism
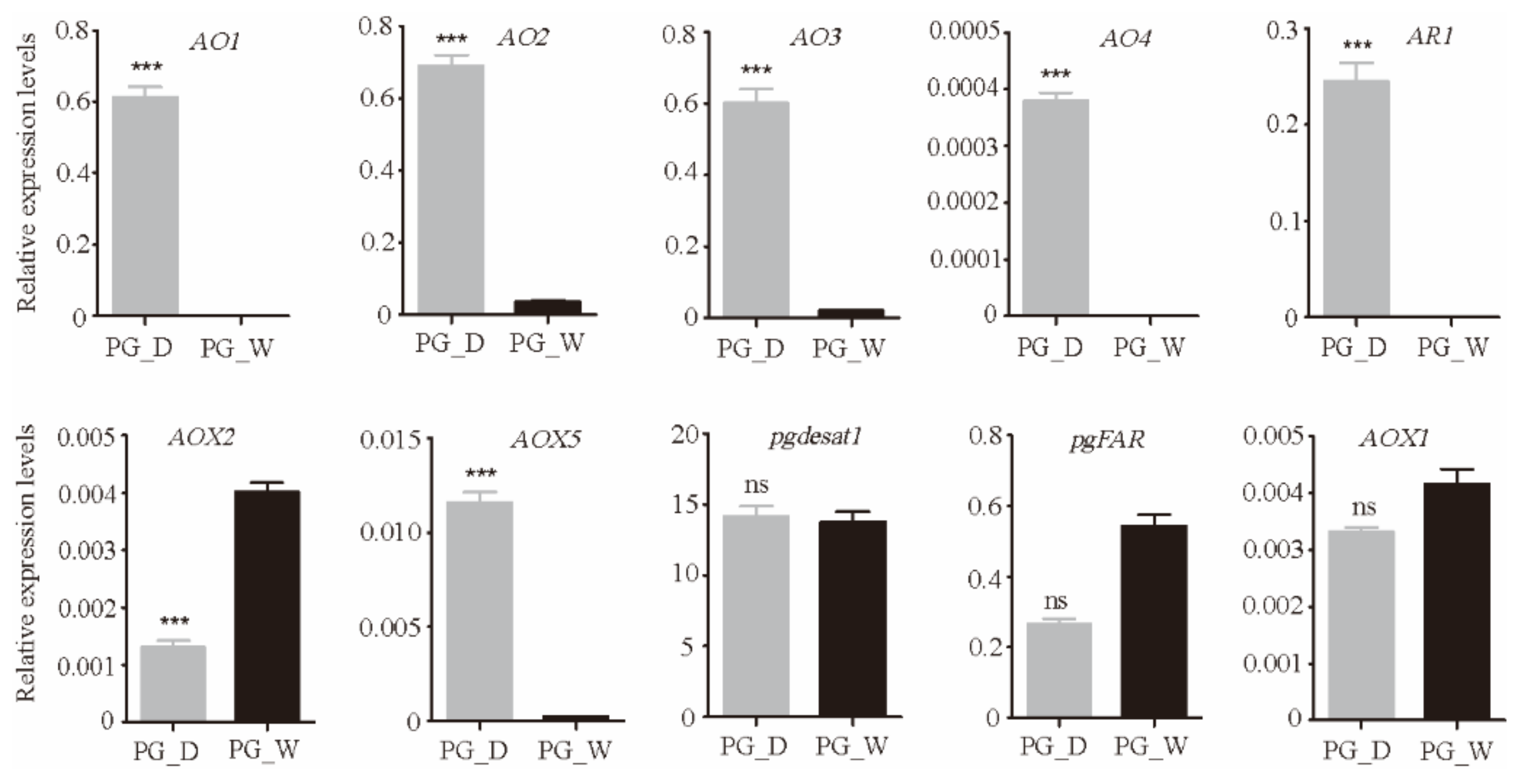
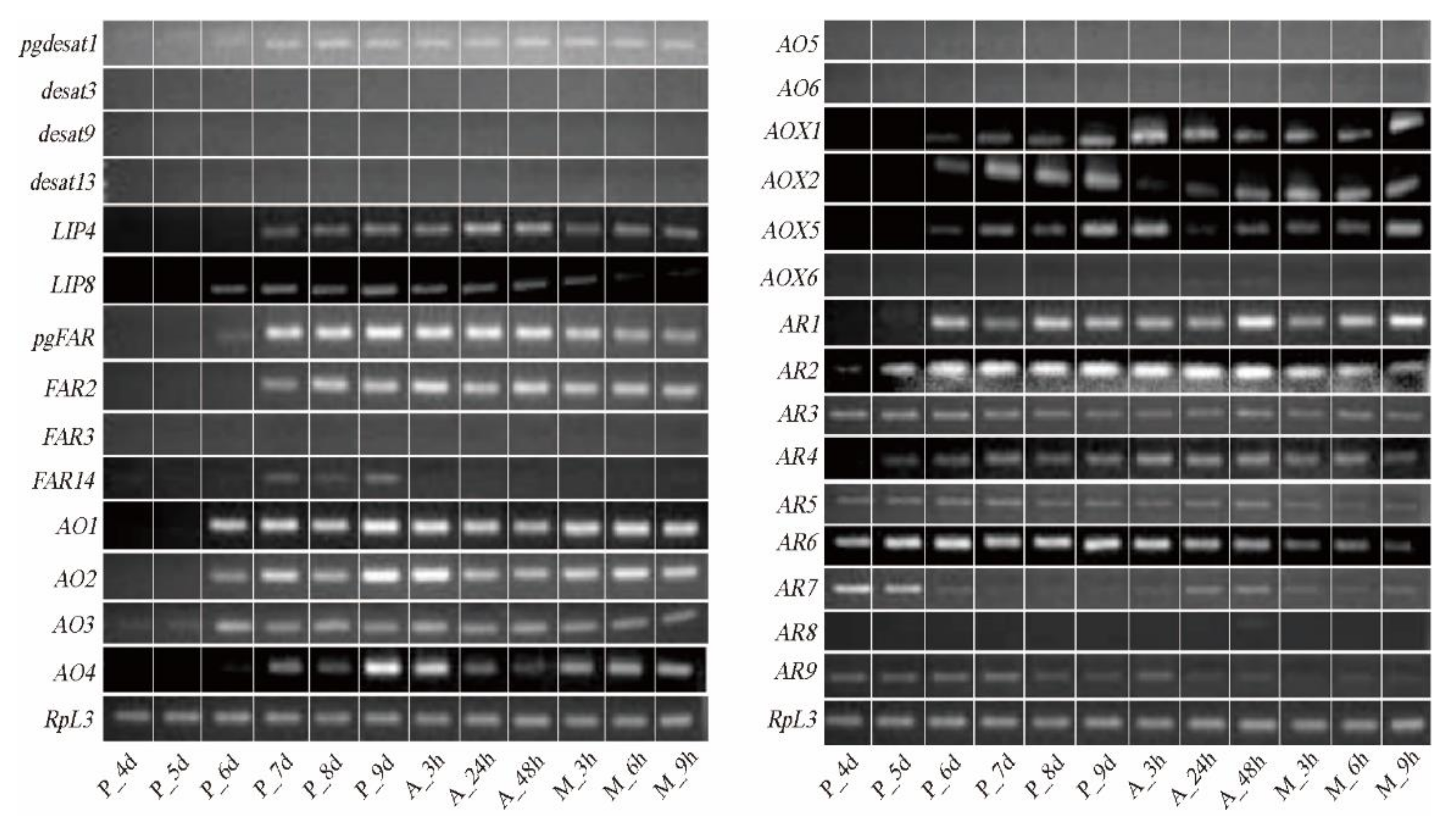
3.4. Genes involved in the Terminal Pathway of Bombykal Synthesis and Metabolism
3.4.1. Alcohol Oxidase (AO)
3.4.2. Aldehyde Reductase (AR)
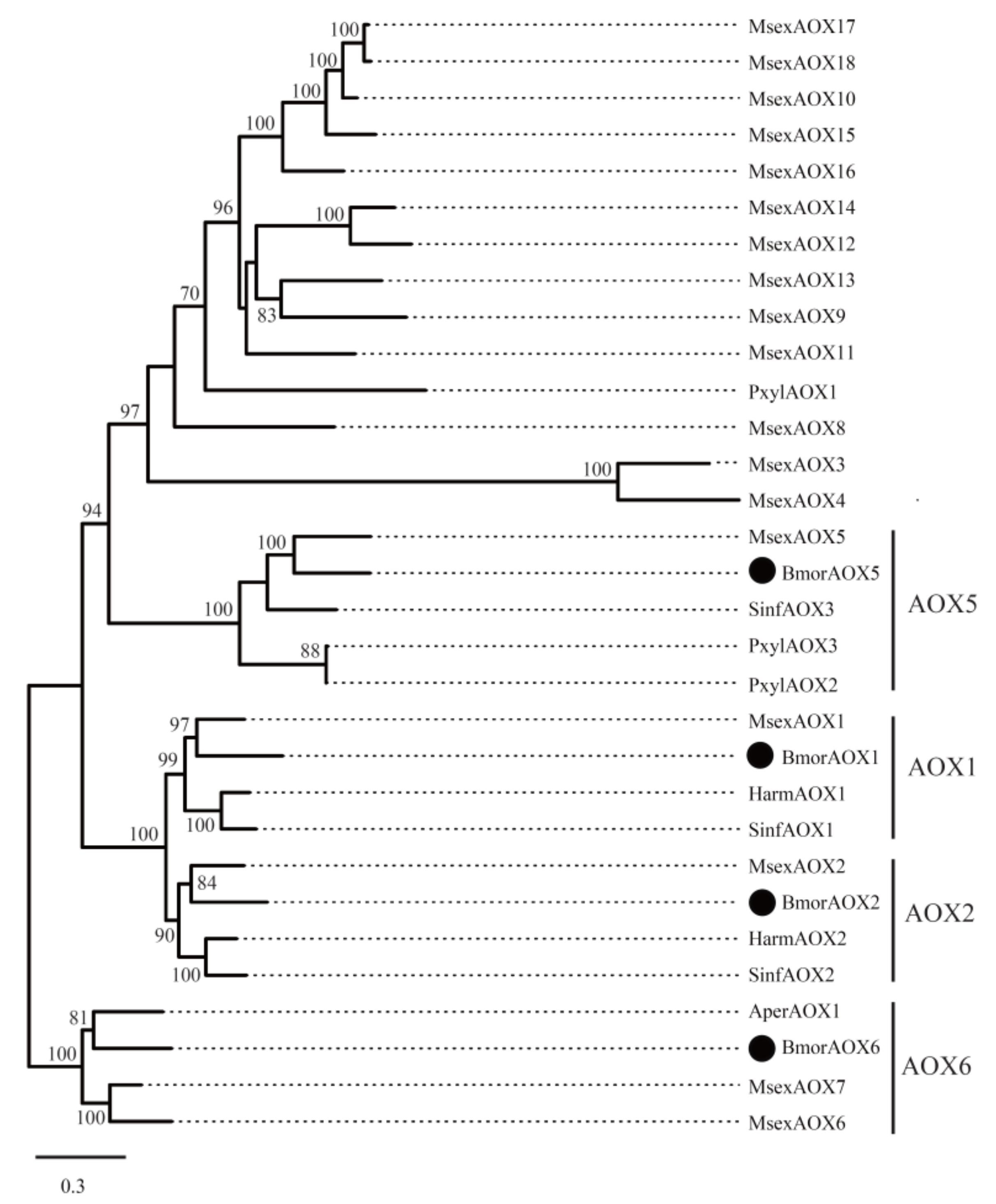
3.4.3. Aldehyde Oxidase (AOX)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Percy-Cunningham, J.E.; MacDonald, J.A. Biology and ultrastructure of sex pheromone-producing glands. In Pheromone Biochemistry; Prestwich, G.D., Blomquist, G.J., Eds.; Academic Press: Orlando, FL, USA, 1987; pp. 27–69. [Google Scholar]
- Cardé, R.T.; Cardé, A.M.; Hill, A.S.; Roelofs, W.L. Sex pheromone specificity as a reproductive isolating mechanism among the sibling species, Archips argyrospilus and A. mortuanus and other sympatric tortricine moths (Lepidoptera: Tortricidae). J. Chem. Ecol. 1977, 3, 71–84. [Google Scholar] [CrossRef]
- Millar, J. The devil is in the details. J. Chem. Ecol. 2014, 40, 517–518. [Google Scholar] [CrossRef]
- Jurenka, R. Insect pheromone biosynthesis. Top. Curr. Chem. 2004, 239, 97–132. [Google Scholar] [CrossRef] [PubMed]
- Roelofs, W.L.; Jurenka, R.A. Biosynthetic enzymes regulating ratios of sex pheromone components in female redbanded leafroller moths. Bioorg. Med. Chem. 1996, 4, 461–466. [Google Scholar] [CrossRef]
- Tillman, J.A.; Seybold, S.J.; Jurenka, R.A.; Blomquist, G.J. Insect pheromones--an overview of biosynthesis and endocrine regulation. Insect Biochem. Mol. Biol. 1999, 29, 481–514. [Google Scholar] [CrossRef]
- Jurenka, R. Regulation of pheromone biosynthesis in moths. Curr. Opin. Insect Sci. 2017, 24, 29–35. [Google Scholar] [CrossRef]
- Zhu, J.W.; Millar, J.; Löfstedt, C. Hormonal regulation of sex pheromone biosynthesis in the turnip moth, Agrotis segetum. Arch. Insect Biochem. Physiol. 1995, 30, 41–59. [Google Scholar] [CrossRef]
- Volpe, J.J.; Vagelos, P.R. Saturated fatty acid biosynthesis and its regulation. Annu. Rev. Biochem. 1973, 42, 21–60. [Google Scholar] [CrossRef]
- Wang, H.-L.; Liénard, M.A.; Zhao, C.-H.; Wang, C.-Z.; Löfstedt, C. Neofunctionalization in an ancestral insect desaturase lineage led to rare Δ6 pheromone signals in the Chinese tussah silkworm. Insect Biochem. Mol. Biol. 2010, 40, 742–751. [Google Scholar] [CrossRef]
- Liénard, M.A.; Lassance, J.-M.; Wang, H.-L.; Zhao, C.-H.; Piškur, J.; Johansson, T.; Löfstedt, C. Elucidation of the sex-pheromone biosynthesis producing 5,7-dodecadienes in Dendrolimus punctatus (Lepidoptera: Lasiocampidae) reveals Δ11- and Δ9-desaturases with unusual catalytic properties. Insect Biochem. Mol. Biol. 2010, 40, 440–452. [Google Scholar] [CrossRef]
- Hagström, A.K.; Albre, J.; Tooman, L.K.; Thirmawithana, A.H.; Corcoran, J.; Löfstedt, C.; Newcomb, R.D. A novel fatty acyl desaturase from the pheromone glands of Ctenopseustis obliquana and C. herana with specific Z5-desaturase activity on myristic acid. J. Chem. Ecol. 2014, 40, 63–70. [Google Scholar] [CrossRef] [Green Version]
- Xia, Y.-H.; Zhan, Y.-N.; Ding, B.-J.; Wang, H.-L.; Löfstedt, C. Multi-functional desaturases in two Spodoptera moths with Δ11 and Δ12 desaturation activities. J. Chem. Ecol. 2019, 45, 378–387. [Google Scholar] [CrossRef] [Green Version]
- Butenandt, A.; Beckman, R.; Stamm, D.; Hecker, E. Über den sexual-lockstoff des seidenspinner Bombyx mori, reidarstellung und constitution. Z. Naturf. B. 1959, 14, 283–284. [Google Scholar]
- Ramaswamy, S.B.; Randle, S.A.; Ma, W.K. Field evaluation of the sex pheromone components of Heliothis virescens (Lepidoptera, Noctuidae) in cone traps. Environ. Entomol. 1985, 14, 293–296. [Google Scholar] [CrossRef]
- Chisholm, M.D.; Steck, W.F.; Underhill, E.W.; Palaniswamy, P. Field trapping of diamondback moth Plutella xylostella using an improved four-component sex attractant blend. J. Chem. Ecol. 1983, 9, 113–1188. [Google Scholar] [CrossRef] [PubMed]
- Sofer, W.; Martin, P.F. Analysis of alcohol dehydrogenase gene expression in Drosophila. Ann. Rev. Genet. 1987, 21, 203–225. [Google Scholar] [CrossRef] [PubMed]
- Jurenka, R.A.; Roelofs, W.L. Characterization of the acetyltransferase used in pheromone biosynthesis in moths: Specificity for the Z isomer in tortricidae. Insect Biochem. 1989, 19, 639–644. [Google Scholar] [CrossRef]
- Li, R.T.; Ning, C.; Huang, L.Q.; Dong, J.F.; Li, X.; Wang, C.Z. Expressional divergences of two desaturase genes determine the opposite ratios of two sex pheromone components in Helicoverpa armigera and Helicoverpa assulta. Insect Biochem. Mol. Biol. 2017, 90, 90–100. [Google Scholar] [CrossRef]
- Rybczynski, R.; Vogt, R.G.; Lerner, M.R. Antennal-specific pheromone-degrading aldehyde oxidases from the moths Antheraea polyphemus and Bombyx mori. J. Biol. Chem. 1990, 265, 19712–19715. [Google Scholar] [CrossRef]
- Teal, P.E.A.; Tumlinson, J.H. The role of alcohols in pheromone biosynthesis by two noctuid moths that use acetate pheromone components. Arch. Insect Biochem. Physiol. 1987, 4, 261–269. [Google Scholar] [CrossRef]
- Matsumoto, S.; Hull, J.J.; Ohnishi, A.; Moto, K.; Fónagy, A. Molecular mechanisms underlying sex pheromone production in the silkmoth, Bombyx mori: Characterization of the molecular components involved in bombykol biosynthesis. J. Insect Physiol. 2007, 53, 752–759. [Google Scholar] [CrossRef]
- Matsumoto, S.; Ohnishi, A.; Lee, J.M.; Hull, J.J. Unraveling the pheromone biosynthesis activating neuropeptide (PBAN) signal transduction cascade that regulates sex pheromone production in moths. Vitam. Horm. 2010, 83, 425–445. [Google Scholar] [CrossRef]
- Groot, A.T.; Dekker, T.; Heckel, D.G. The genetic basis of pheromone evolution in moths. Annu. Rev. Entomol. 2016, 61, 99–117. [Google Scholar] [CrossRef]
- Moto, K.; Suzuki, M.G.; Hull, J.J.; Kurata, R.; Takahashi, S.; Yamamoto, M.; Okano, K.; Imai, K.; Ando, T.; Matsumoto, S. Involvement of a bifunctional fatty-acyl desaturase in the biosynthesis of the silkmoth, Bombyx mori, sex pheromone. Proc. Natl. Acad. Sci. USA 2004, 101, 8631–8636. [Google Scholar] [CrossRef] [Green Version]
- Ohnishi, A.; Hull, J.J.; Matsumoto, S. Targeted disruption of genes in the Bombyx mori sex pheromone biosynthetic pathway. Proc. Natl. Acad. Sci. USA 2006, 103, 4398–4403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moto, K.; Yoshiga, T.; Yamamoto, M.; Takahashi, S.; Okano, K.; Ando, T.; Nakata, T.; Matsumoto, S. Pheromone gland-specific fatty acyl reductase of the silkmoth, Bombyx mori. Proc. Natl. Acad. Sci. USA 2003, 100, 9156–9161. [Google Scholar] [CrossRef] [Green Version]
- Ohnishi, A.; Hashimoto, K.; Imai, K.; Matsumoto, S. Functional characterization of the Bombyx mori fatty acid transport protein (BmFATP) within the silkmoth pheromone gland. J. Biol. Chem. 2009, 284, 5128–5136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsumoto, S.; Yoshiga, T.; Yokoyama, N.; Iwanaga, M.; Koshiba, S.; Kigawa, T.; Hirota, H.; Yokoyama, S.; Okano, K.; Mita, K.; et al. Characterization of acyl-CoA-binding protein (ACBP) in the pheromone gland of the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 2001, 31, 603–609. [Google Scholar] [CrossRef]
- Du, M.; Yin, X.; Zhang, S.; Zhu, B.; Song, Q.; An, S. Identification of lipases involved in PBAN stimulated pheromone production in Bombyx mori using the DGE and RNAi approaches. PLoS ONE 2012, 7, e31045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roelofs, W.L.; Liu, W.; Hao, G.; Jiao, H.; Rooney, A.P.; Linn, C.E., Jr. Evolution of moth sex pheromones via ancestral genes. Proc. Natl. Acad. Sci. USA 2002, 99, 13621–13626. [Google Scholar] [CrossRef] [Green Version]
- Lassance, J.-M.; Liénard, M.A.; Antony, B.; Qian, S.; Fujii, T.; Tabata, J.; Ishikawa, Y.; Löfstedt, C. Functional consequences of sequence variation in the pheromone biosynthetic gene pgFAR for Ostrinia moths. Proc. Natl. Acad. Sci. USA 2013, 110, 3967–3972. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liénard, M.A.; Hagström, A.K.; Lassance, J.-M.; Löfstedt, C. Evolution of multicomponent pheromone signals in small ermine moths involves a single fatty-acyl reductase gene. Proc. Natl. Acad. Sci. USA 2010, 107, 10955–10960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antony, B.; Ding, B.-J.; Moto, K.; Aldosari, S.A.; Aldawood, A.S. Two fatty acyl reductases involved in moth pheromone biosynthesis. Sci. Rep. 2016, 6, 29927. [Google Scholar] [CrossRef] [Green Version]
- Qian, S.; Fujii, T.; Ito, K.; Nakano, R.; Ishikawa, Y. Cloning and functional characterization of a fatty acid transport protein (FATP) from the pheromone gland of a lichen moth, Eilema japonica, which secretes an alkenyl sex pheromone. Insect Biochem. Mol. Biol. 2011, 41, 22–28. [Google Scholar] [CrossRef]
- Luxova, A.; Svatos, A. Substrate specificity of membrae-bound alcohol oxidase from the tobacco hornworm moth (Manduca sexta) female pheromone glands. J. Mol. Cata. B Enzym. 2006, 38, 37–42. [Google Scholar] [CrossRef]
- Kiyota, R.; Arakawa, M.; Yamakawa, R.; Yasmin, A.; Ando, T. Biosynthetic pathways of the sex pheromone components and substrate selectivity of the oxidation enzymes working in pheromone glands of the fall webworm, Hyphantria cunea. Insect Biochem. Mol. Biol. 2011, 41, 362–369. [Google Scholar] [CrossRef]
- Teal, P.E.A.; Tumlinson, J.H. Terminal steps in pheromone biosynthesis by Heliothis virescens and H. zea. J. Chem. Ecol. 1986, 12, 353–366. [Google Scholar] [CrossRef]
- Vogel, H.; Heidel, A.J.; Heckel, D.G.; Groot, A.T. Transcriptome analysis of the sex pheromone gland of the noctuid moth Heliothis virescens. BMC Genom. 2010, 14, 11–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.-N.; Xia, Y.-H.; Zhu, J.-Y.; Li, S.-Y.; Dong, S.-L. Putative pathway of sex pheromone biosynthesis and degradation by expression patterns of genes identified from female pheromone gland and adult antenna of Sesamia inferens (Walker). J. Chem. Ecol. 2014, 40, 439–451. [Google Scholar] [CrossRef]
- Wang, Q.-H.; Gong, Q.; Fang, S.-M.; Liu, Y.-Q.; Zhang, Z.; Yu, Q.-Y. Identification of genes involved in sex pheromone biosynthesis and metabolic pathway in the Chinese oak silkworm, Antheraea pernyi. Int. J. Biol. Macromol. 2020, 163, 1487–1497. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K.; Higashiura, A.; Suzuki, M.; Shiotsuki, T.; Sugahara, R.; Fujii, T.; Nakagawa, A. Structural characterization of an aldo-keto reductase (AKR2E5) from the silkworm Bombyx mori. Biochem. Biophys. Res. Commun. 2016, 474, 104–110. [Google Scholar] [CrossRef] [Green Version]
- Choo, Y.-M.; Pelletier, J.; Atungulu, E.; Leal, W.S. Identification and characterization of an antennae-specific aldehyde oxidase from the navel orangeworm. PLoS ONE 2013, 8, e67794. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.-M.; He, M.; Wang, H.; Ma, Y.-F.; Dewer, Y.; Zhang, F.; He, P. A candidate aldehyde oxidase in the antennae of the diamondback moth, Plutella xylostella (L.), is potentially involved in the degradation of pheromones, plant-derived volatiles and the detoxification of xenobiotics. Pestic. Biochem. Physiol. 2021, 171, 104726. [Google Scholar] [CrossRef] [PubMed]
- Rybczynski, R.; Reagan, J.; Lerner, M.R. A pheromone-degrading aldehyde oxidase in the antennae of the moth Manduca sexta. J. Neurosci. 1989, 9, 1341–1353. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Yu, H.; Shen, Y.; Banno, Y.; Xiang, Z.; Zhang, Z. Phylogeny and evolutionary history of the silkworm. Sci. China Life Sci. 2012, 55, 483–496. [Google Scholar] [CrossRef] [Green Version]
- Daimon, T.; Fujii, T.; Fujii, T.; Yokoyama, T.; Katsuma, S.; Shinoda, T.; Shimada, T.; Ishikawa, Y. Reinvestigation of the sex pheromone of the wild silkmoth Bombyx mandarina: The effects of bombykal and bombykyl acetate. J. Chem. Ecol. 2012, 38, 1031–1035. [Google Scholar] [CrossRef]
- Kasang, G.; Kaissling, K.E.; Vostrowsky, O.; Bestmann, H.J. Bombykal, a second pheromone component of the silkworm moth Bombyx mori L. Angew. Chem. Lnt. Ed. Engl. 1978, 17, 60. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods. 2015, 12, 357–360. [Google Scholar] [CrossRef] [Green Version]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [Green Version]
- Darriba, D.; Posada, D.; Kozlov, A.M.; Stamatakis, A.; Morel, B.; Flouri, T. ModelTest-NG: A new and scalable tool for the selection of DNA and protein evolutionary models. Mol. Biol. Evol. 2020, 37, 291–294. [Google Scholar] [CrossRef] [Green Version]
- Kozlov, A.M.; Darriba, D.; Flouri, T.; Morel, B.; Stamatakis, A. RAxML-NG: A fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 2019, 35, 4453–4455. [Google Scholar] [CrossRef] [Green Version]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Ando, T.; Kasuga, K.; Yayma, Y.; Katoaka, H.; Suzuki, A. Termination of sex pheromone production in mated females of the silkworm moth. Arch. Insect Biochem. 1996, 31, 207–218. [Google Scholar] [CrossRef]
- Ahn, S.J.; Choi, M.Y.; Boo, K.S. Mating effect on sex pheromone production of the oriental tobacco budworm, Helicoverpa assulta. J. Asia-Pacif. Entomol. 2002, 5, 43–48. [Google Scholar] [CrossRef]
- Zhang, S.; Liu, X.; Zhu, B.; Yin, X.; Du, M.; Song, Q.; An, S. Identification of differentially expressed genes in the pheromone glands of mated and virgin Bombyx mori by digital gene expression profiling. PLoS ONE 2014, 9, e111003. [Google Scholar] [CrossRef]
- Teal, P.E.A.; Tumlinson, J.H. Properties of cuticular oxidases used for sex pheromone biosynthesis by Heliothis zea. J. Chem. Ecol. 1988, 14, 2131–2145. [Google Scholar] [CrossRef] [PubMed]
- He, P.; Zhang, Y.-F.; Hong, D.-Y.; Wang, J.; Wang, X.-L.; Zuo, L.-H.; Tang, X.-F.; Xu, W.-M.; He, M. A reference gene set for sex pheromone biosynthesis and degradation genes from the diamondback moth, Plutella xylostella, based on genome and transcriptome digital gene expression analyses. BMC Genom. 2017, 18, 219. [Google Scholar] [CrossRef] [Green Version]
- Garattini, E.; Terao, M. Aldehyde oxidase and its importance in novel drug discovery: Present and future challenges. Expert Opin. Drug Discov. 2013, 8, 641–654. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, Y.; Shen, G.; Mao, X.; Jiao, M.; Lin, Y. Identification and characterization of aldehyde oxidase 5 in the pheromone gland of the silkworm (Lepidoptera: Bombycidae). J. Insect Sci. 2020, 20, 31. [Google Scholar] [CrossRef] [PubMed]
- Pelletier, J.; Bozzolan, F.; Solvar, M.; François, M.-C.; Jacquin-Joly, E.; Maïbèche-Coisne, M. Identification of candidate aldehyde oxidases from the silkworm Bombyx mori potentially involved in antennal pheromone degradation. Gene 2007, 404, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Lin, Y.; Yang, C.W.; Wang, Y.X.; Xia, Q.Y. Identification and expression profiling of aldehyde oxidase genes in the silkworm, Bombyx mori. Acta Entomol. Sin. 2010, 53, 1–8. (In Chinese) [Google Scholar]



| Gene Name (Displayed/Total) | RPKM_D | RPKM_W | DEG | Functional Annotation |
|---|---|---|---|---|
| ACC1 (1/1) | 21.40 | 21.94 | n | acetyl-CoA carboxylase |
| FAS1 (1/2) | 284.73 | 175.52 | n | fatty acid synthase |
| pgdesat1 (1/8) | 7984.24 | 7534.97 | n | acyl CoA desaturase |
| BmFATP (1/6) | 125.52 | 289.58 | n | fatty acid transport protein |
| ACBP1 (2/2) | 31.77 | 19.69 | n | acyl-CoA-binding domain-containing protein |
| ACBP2 | 43.40 | 53.56 | n | acyl-CoA-binding domain-containing protein |
| LIP1 (4/16) | 234.39 | 131.44 | n | lipase 3 |
| LIP2 | 211.95 | 243.04 | n | lipase 3-like |
| LIP3 | 81.45 | 94.26 | n | lipase member H-A |
| LIP4 | 44.16 | 6.02 | y | lipase member H |
| pgFAR(2/18) | 215.88 | 734.50 | n | fatty-acyl reductase |
| FAR2 | 255.69 | 248.61 | n | putative fatty acyl-CoA reductase |
| AO1 (3/25) | 891.26 | 2.23 | y | 15-hydroxyprostaglandin dehydrogenase |
| AO2 | 925.77 | 93.66 | y | 15-hydroxyprostaglandin dehydrogenase |
| AO3 | 1036.26 | 124.93 | y | carbonyl reductase [NADPH] 3 |
| AR1 (2/11) | 582.61 | 66.52 | y | aldo-keto reductase AKR2E4-like |
| AR2 | 337.63 | 718.81 | n | aldo-keto reductase AKR2E4-like |
| AOX1 (3/4) | 357.31 | 318.79 | n | aldehyde oxidase 1 |
| AOX2 | 48.97 | 7.67 | y | xanthine dehydrogenase-like |
| AOX5 | 33.06 | 5.21 | y | xanthine dehydrogenase/oxidase |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Q.-H.; Gao, X.; Yu, H.-S.; Zhang, Z.; Yu, Q.-Y. Exploring the Terminal Pathway of Sex Pheromone Biosynthesis and Metabolism in the Silkworm. Insects 2021, 12, 1062. https://doi.org/10.3390/insects12121062
Wang Q-H, Gao X, Yu H-S, Zhang Z, Yu Q-Y. Exploring the Terminal Pathway of Sex Pheromone Biosynthesis and Metabolism in the Silkworm. Insects. 2021; 12(12):1062. https://doi.org/10.3390/insects12121062
Chicago/Turabian StyleWang, Qing-Hai, Xing Gao, Hong-Song Yu, Ze Zhang, and Quan-You Yu. 2021. "Exploring the Terminal Pathway of Sex Pheromone Biosynthesis and Metabolism in the Silkworm" Insects 12, no. 12: 1062. https://doi.org/10.3390/insects12121062
APA StyleWang, Q.-H., Gao, X., Yu, H.-S., Zhang, Z., & Yu, Q.-Y. (2021). Exploring the Terminal Pathway of Sex Pheromone Biosynthesis and Metabolism in the Silkworm. Insects, 12(12), 1062. https://doi.org/10.3390/insects12121062






