Genetic Code Expansion System for Tight Control of Gene Expression in Bombyx mori Cell Lines
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Plasmids
2.2. Cell Transfection
2.3. Cell Imaging
2.4. Western Blotting
3. Results
3.1. Strategy for a Novel Genetic Code Expansion System to Control Gene Expression in B. mori Cell Lines
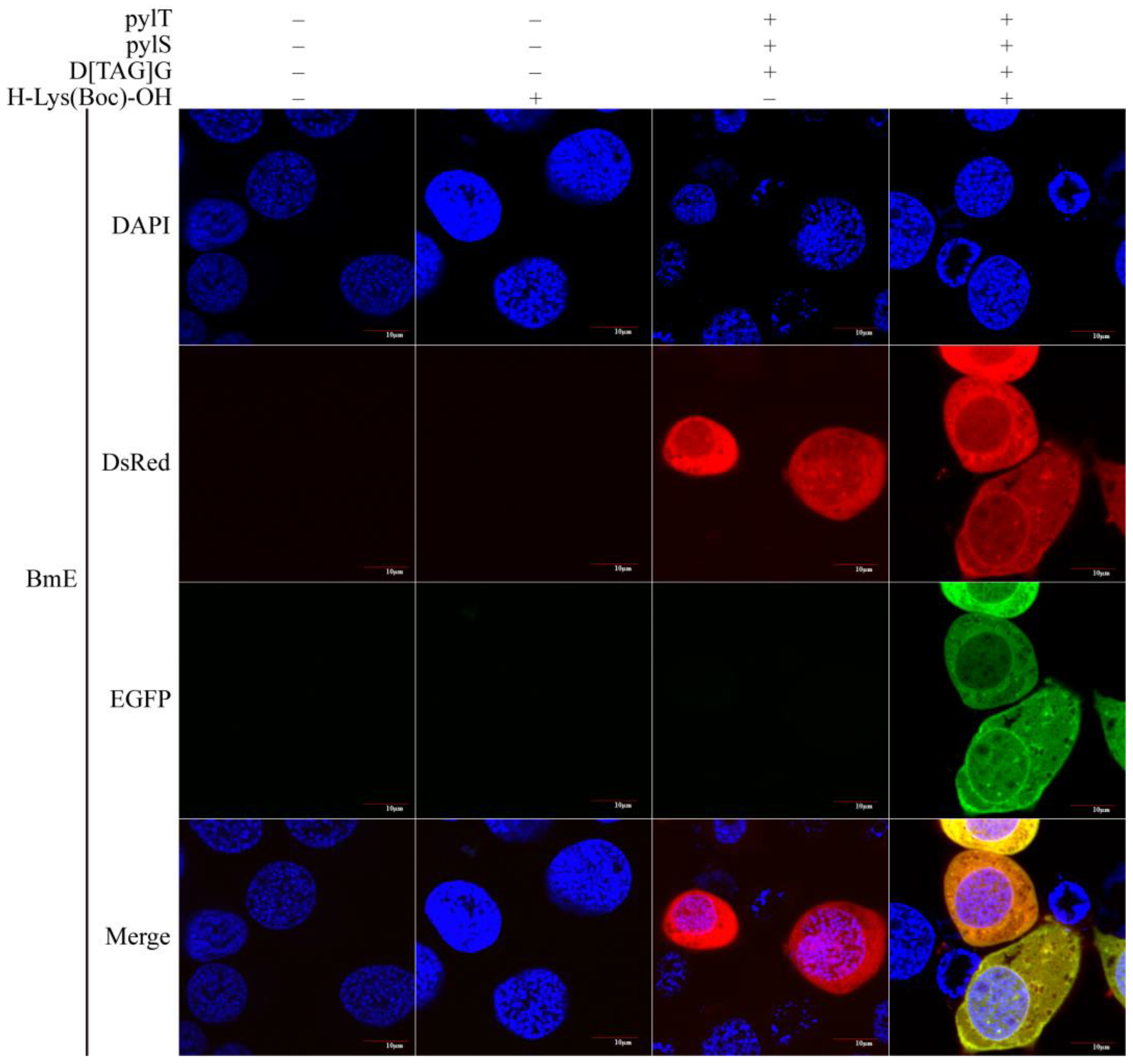
3.2. Testing of the Novel Genetic Code Expansion System in BmE Cell Line
3.3. Application of the Novel Genetic Code Expansion System in BmNs Cell Line
3.4. Efficient and Orthogonal Bombyx mori Genetic Code Extension System
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cambridge, S.B.; Geissler, D.; Calegari, F.; Anastassiadis, K.; Hasan, M.T.; Stewart, A.F.; Huttner, W.B.; Hagen, V.; Bonhoeffer, T. Doxycycline-dependent photoactivated gene expression in eukaryotic systems. Nat. Methods 2009, 6, 527–531. [Google Scholar] [CrossRef]
- González, F.; Zhu, Z.; Shi, Z.; Lelli, K.; Verma, N.; Li, Q.V.; Huangfu, D. An iCRISPR platform for rapid, multiplexable, and inducible genome editing in human pluripotent stem cells. Cell Stem Cell 2014, 15, 215–226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zetsche, B.; Volz, S.E.; Zhang, F. A split-Cas9 architecture for inducible genome editing and transcription modulation. Nat. Biotechnol. 2015, 33, 139–142. [Google Scholar] [CrossRef] [Green Version]
- Dow, L.E.; Fisher, J.; O’Rourke, K.P.; Muley, A.; Kastenhuber, E.R.; Livshits, G.; Tschaharganeh, D.F.; Socci, N.D.; Lowe, S.W. Inducible in vivo genome editing with CRISPR-Cas9. Nat. Biotechnol. 2015, 33, 390–394. [Google Scholar] [CrossRef] [Green Version]
- Ni, C.; Fox, K.J.; Prather, K.L.J. Substrate-activated expression of a biosynthetic pathway in Escherichia coli. Biotechnol. J. 2021. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Kobayashi, Y.; Huang, C.; Liu, B.; Qian, W.; Wan, F.; Schetelig, M.F. Highly Efficient Temperature Inducible CRISPR-Cas9 Gene Targeting in Drosophila suzukii. Int. J. Mol. Sci. 2021, 22, 6724. [Google Scholar] [CrossRef]
- Wang, X.; Han, J.; Zhang, X.; Ma, Y.; Lin, Y.; Wang, H.; Li, D.; Zheng, T.; Wu, F.; Ye, J.; et al. Reversible thermal regulation for bifunctional dynamic control of gene expression in Escherichia coli. Nat. Commun. 2021, 12, 1411. [Google Scholar] [CrossRef]
- Aratboni, H.A.; Rafiei, N.; Khorashad, L.K.; Lerma-Escalera, A.I.; Balderas-Cisneros, F.D.J.; Liu, Z.; Alemzadeh, A.; Shaji, S.; Morones-Ramírez, J.R. LED control of gene expression in a nanobiosystem composed of metallic nanoparticles and a genetically modified E. coli strain. J. Nanobiotechnol. 2021, 19, 190. [Google Scholar] [CrossRef] [PubMed]
- Yamada, M.; Suzuki, Y.; Nagasaki, S.C.; Okuno, H.; Imayoshi, I. Light Control of the Tet Gene Expression System in Mammalian Cells. Cell Rep. 2018, 25, 487–500.e6. [Google Scholar] [CrossRef] [Green Version]
- Li, F.; Lu, Z.; Wu, W.; Qian, N.; Wang, F.; Chen, T. Optogenetic gene editing in regional skin. Cell Res. 2019, 29, 862–865. [Google Scholar] [CrossRef]
- Wang, X.; Chen, X.; Yang, Y. Spatiotemporal control of gene expression by a light-switchable transgene system. Nat. Methods 2012, 9, 266–269. [Google Scholar] [CrossRef]
- Zhang, L.; Ward, J.D.; Cheng, Z.; Dernburg, A.F. The auxin-inducible degradation (AID) system enables versatile conditional protein depletion in C. elegans. Development 2015, 142, 4374–4384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, S.; Zhang, J.; Lu, W.; Liu, Y.; Xia, Q. SAA-Cas9: A tunable genome editing system with increased bio-safety and reduced off-target effects. J. Genet. Genom. 2019, 46, 145–148. [Google Scholar] [CrossRef] [PubMed]
- Ling, J.; O’Donoghue, P.; Söll, D. Genetic code flexibility in microorganisms: Novel mechanisms and impact on physiology. Nat. Rev. Microbiol. 2015, 13, 707–721. [Google Scholar] [CrossRef] [Green Version]
- Buckingham, R.H.; Kurland, C.G. Codon specificity of UGA suppressor tRNATrp from Escherichia coli. Proc. Natl. Acad. Sci. USA 1977, 74, 5496–5498. [Google Scholar] [CrossRef] [Green Version]
- Rogers, M.J.; Adachi, T.; Inokuchi, H.; Söll, D. Switching tRNAGln identity from glutamine to tryptophan. Proc. Natl. Acad. Sci. USA 1992, 89, 3463–3467. [Google Scholar] [CrossRef] [Green Version]
- Yamao, F.; Muto, A.; Kawauchi, Y.; Iwami, M.; Iwagami, S.; Azumi, Y.; Osawa, S. UGA is read as tryptophan in Mycoplasma capricolum. Proc. Natl. Acad. Sci. USA 1985, 82, 2306–2309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macino, G.; Coruzzi, G.; Nobrega, F.G.; Li, M.; Tzagoloff, A. Use of the UGA terminator as a tryptophan codon in yeast mitochondria. Proc. Natl. Acad. Sci. USA 1979, 76, 3784–3785. [Google Scholar] [CrossRef] [Green Version]
- Martin, N.C.; Pham, H.D.; Underbrink-Lyon, K.; Miller, D.l.; Donelson, J.E. Yeast mitochondrial tRNATrp can recognize the nonsense codon UGA. Nature 1980, 285, 579–581. [Google Scholar] [CrossRef]
- Köhrer, C.; Sullivan, E.L.; RajBhandary, U.L. Complete set of orthogonal 21st aminoacyl-tRNA synthetase-amber, ochre and opal suppressor tRNA pairs: Concomitant suppression of three different termination codons in an mRNA in mammalian cells. Nucleic Acids Res. 2004, 32, 6200–6211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Atkins, J.F.; Gesteland, R. Biochemistry. The 22nd amino acid. Science 2002, 296, 1409–1410. [Google Scholar] [CrossRef] [PubMed]
- Böck, A.; Forchhammer, K.; Heider, J.; Leinfelder, W.; Sawers, G.; Veprek, B.; Zinoni, F. Selenocysteine: The 21st amino acid. Mol. Microbiol. 1991, 5, 515–520. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Brock, A.; Chen, S.; Chen, S.; Schultz, P.G. Genetic incorporation of unnatural amino acids into proteins in mammalian cells. Nat. Methods 2007, 4, 239–244. [Google Scholar] [CrossRef] [PubMed]
- Chin, J.W.; Cropp, T.A.; Anderson, J.C.; Mukherji, M.; Zhang, Z.; Schultz, P.G. An expanded eukaryotic genetic code. Science 2003, 301, 964–967. [Google Scholar] [CrossRef]
- Zhang, Z.; Alfonta, L.; Tian, F.; Bursulaya, B.; Uryu, S.; King, D.S.; Schultz, P.G. Selective incorporation of 5-hydroxytryptophan into proteins in mammalian cells. Proc. Natl. Acad. Sci. USA 2004, 101, 8882–8887. [Google Scholar] [CrossRef] [Green Version]
- Bartoschek, M.D.; Ugur, E.; Nguyen, T.-A.; Rodschinka, G.; Wierer, M.; Lang, K.; Bultmann, S. Identification of permissive amber suppression sites for efficient non-canonical amino acid incorporation in mammalian cells. Nucleic Acids Res. 2021, 49, e62. [Google Scholar] [CrossRef]
- Koehler, C.; Sauter, P.F.; Wawryszyn, M.; Girona, G.E.; Gupta, K.; Landry, J.J.M.; Fritz, M.H.-Y.; Radic, K.; Hoffmann, J.-E.; Chen, Z.A.; et al. Genetic code expansion for multiprotein complex engineering. Nat. Methods 2016, 13, 997–1000. [Google Scholar] [CrossRef] [PubMed]
- Bianco, A.; Townsley, F.M.; Greiss, S.; Lang, K.; Chin, J.W. Expanding the genetic code of Drosophila melanogaster. Nat. Chem. Biol. 2012, 8, 748–750. [Google Scholar] [CrossRef]
- Mukai, T.; Wakiyama, M.; Sakamoto, K.; Yokoyama, S. Genetic encoding of non-natural amino acids in Drosophila melanogaster Schneider 2 cells. Protein Sci. 2010, 19, 440–448. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.J.; Kang, D.; Park, H.-S. Site-Specific Labeling of Proteins Using Unnatural Amino Acids. Mol. Cells 2019, 42, 386–396. [Google Scholar] [CrossRef]
- Xiang, Z.; Ren, H.; Hu, Y.S.; Coin, I.; Wei, J.; Cang, H.; Wang, L. Adding an unnatural covalent bond to proteins through proximity-enhanced bioreactivity. Nat. Methods 2013, 10, 885–888. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, T.-A.; Cigler, M.; Lang, K. Expanding the Genetic Code to Study Protein-Protein Interactions. Angew. Chem. 2018, 57, 14350–14361. [Google Scholar] [CrossRef]
- Peng, T.; Hang, H.C. Site-Specific Bioorthogonal Labeling for Fluorescence Imaging of Intracellular Proteins in Living Cells. J. Am. Chem. Soc. 2016, 138, 14423–14433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lang, K.; Davis, L.; Chin, J.W. Genetic encoding of unnatural amino acids for labeling proteins. Methods Mol. Biol. 2015, 1266, 217–228. [Google Scholar] [CrossRef]
- Elliott, T.S.; Bianco, A.; Townsley, F.M.; Fried, S.D.; Chin, J.W. Tagging and Enriching Proteins Enables Cell-Specific Proteomics. Cell Chem. Biol. 2016, 23, 805–815. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elliott, T.S.; Townsley, F.M.; Bianco, A.; Ernst, R.J.; Sachdeva, A.; Elsässer, S.J.; Davis, L.; Lang, K.; Pisa, R.; Greiss, S.; et al. Proteome labeling and protein identification in specific tissues and at specific developmental stages in an animal. Nat. Biotechnol. 2014, 32, 465–472. [Google Scholar] [CrossRef] [Green Version]
- Chen, B.; Zhang, N.; Xie, S.; Zhang, X.; He, J.; Muhammad, A.; Sun, C.; Lu, X.; Shao, Y. Gut bacteria of the silkworm Bombyx mori facilitate host resistance against the toxic effects of organophosphate insecticides. Environ. Int. 2020, 143, 105886. [Google Scholar] [CrossRef] [PubMed]
- Dunkelmann, D.L.; Willis, J.C.W.; Beattie, A.T.; Chin, J.W. Engineered triply orthogonal pyrrolysyl-tRNA synthetase/tRNA pairs enable the genetic encoding of three distinct non-canonical amino acids. Nat. Chem. 2020, 12, 535–544. [Google Scholar] [CrossRef]
- Karasaki, N.; Mon, H.; Takahashi, M.; Lee, J.M.; Koga, K.; Kawaguchi, Y.; Kusakabe, T. Establishment of tetracycline-inducible gene expression systems in the silkworm, Bombyx mori. Biotechnol. Lett. 2009, 31, 495–500. [Google Scholar] [CrossRef]
- Liu, J.; Hemphill, J.; Samanta, S.; Tsang, M.; Deiters, A. Genetic Code Expansion in Zebrafish Embryos and Its Application to Optical Control of Cell Signaling. J. Am. Chem. Soc. 2017, 139, 9100–9103. [Google Scholar] [CrossRef]
- Johnson, D.B.F.; Wang, C.; Xu, J.; Schultz, M.D.; Schmitz, R.J.; Ecker, J.R.; Wang, L. Release factor one is nonessential in Escherichia coli. ACS Chem. Biol. 2012, 7, 1337–1344. [Google Scholar] [CrossRef] [PubMed]
- Lajoie, M.J.; Rovner, A.J.; Goodman, D.B.; Aerni, H.-R.; Haimovich, A.D.; Kuznetsov, G.; Mercer, J.A.; Wang, H.H.; Carr, P.A.; Mosberg, J.A.; et al. Genomically recoded organisms expand biological functions. Science 2013, 342, 357–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, F.; Wei, Z.; Luo, Y.; Guo, H.; Zhang, G.; Xia, Q.; Wang, Y. SilkDB 3.0: Visualizing and exploring multiple levels of data for silkworm. Nucleic Acids Res. 2020, 48, D749–D755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Li, Y.; Liang, Y.; Wang, T.; Yang, C.; Ma, S.; Xia, Q. Comparative analysis of genome editing systems, Cas9 and BE3, in silkworms. Int. J. Biol. Macromol. 2020, 158, 486–492. [Google Scholar] [CrossRef]
- Komor, A.C.; Kim, Y.B.; Packer, M.S.; Zuris, J.A.; Liu, D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 2016, 533, 420–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teramoto, H.; Kojima, K. Production of Bombyx mori silk fibroin incorporated with unnatural amino acids. Biomacromolecules 2014, 15, 2682–2690. [Google Scholar] [CrossRef]
- Teramoto, H.; Nakajima, K.-I.; Kojima, K. Azide-Incorporated Clickable Silk Fibroin Materials with the Ability to Photopattern. ACS Biomater. Sci. Eng. 2016, 2, 251–258. [Google Scholar] [CrossRef] [PubMed]



| Gene Name | Primer Name | Sequence (5′ to 3′) |
|---|---|---|
| DsRed | DsRed-F (Xba I+Spe I) | GCTCTAGAGGGACTAGTATGGTGCGCTCCTCCAAG |
| DsRed-R (Sph I) | ACATGCATGCCTACAGGAACAGGTGGTGG | |
| EGFP | EGFP-F (Sph I) | ACATGCATGCATGGTGAGCAAGGGCGAG |
| EGFP-R (Hind III+BsrG I) | CCCAAGCTTTTACTTGTACAGCTCGTCCATG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, W.; Wang, R.; Wang, P.; Ma, S.; Xia, Q. Genetic Code Expansion System for Tight Control of Gene Expression in Bombyx mori Cell Lines. Insects 2021, 12, 1081. https://doi.org/10.3390/insects12121081
Lu W, Wang R, Wang P, Ma S, Xia Q. Genetic Code Expansion System for Tight Control of Gene Expression in Bombyx mori Cell Lines. Insects. 2021; 12(12):1081. https://doi.org/10.3390/insects12121081
Chicago/Turabian StyleLu, Wei, Ruolin Wang, Pan Wang, Sanyuan Ma, and Qingyou Xia. 2021. "Genetic Code Expansion System for Tight Control of Gene Expression in Bombyx mori Cell Lines" Insects 12, no. 12: 1081. https://doi.org/10.3390/insects12121081
APA StyleLu, W., Wang, R., Wang, P., Ma, S., & Xia, Q. (2021). Genetic Code Expansion System for Tight Control of Gene Expression in Bombyx mori Cell Lines. Insects, 12(12), 1081. https://doi.org/10.3390/insects12121081





