Disentangling Ethiopian Honey Bee (Apis mellifera) Populations Based on Standard Morphometric and Genetic Analyses
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Morphometric Analysis
2.2. Molecular Analyses
2.3. Statistical Analyses
3. Results
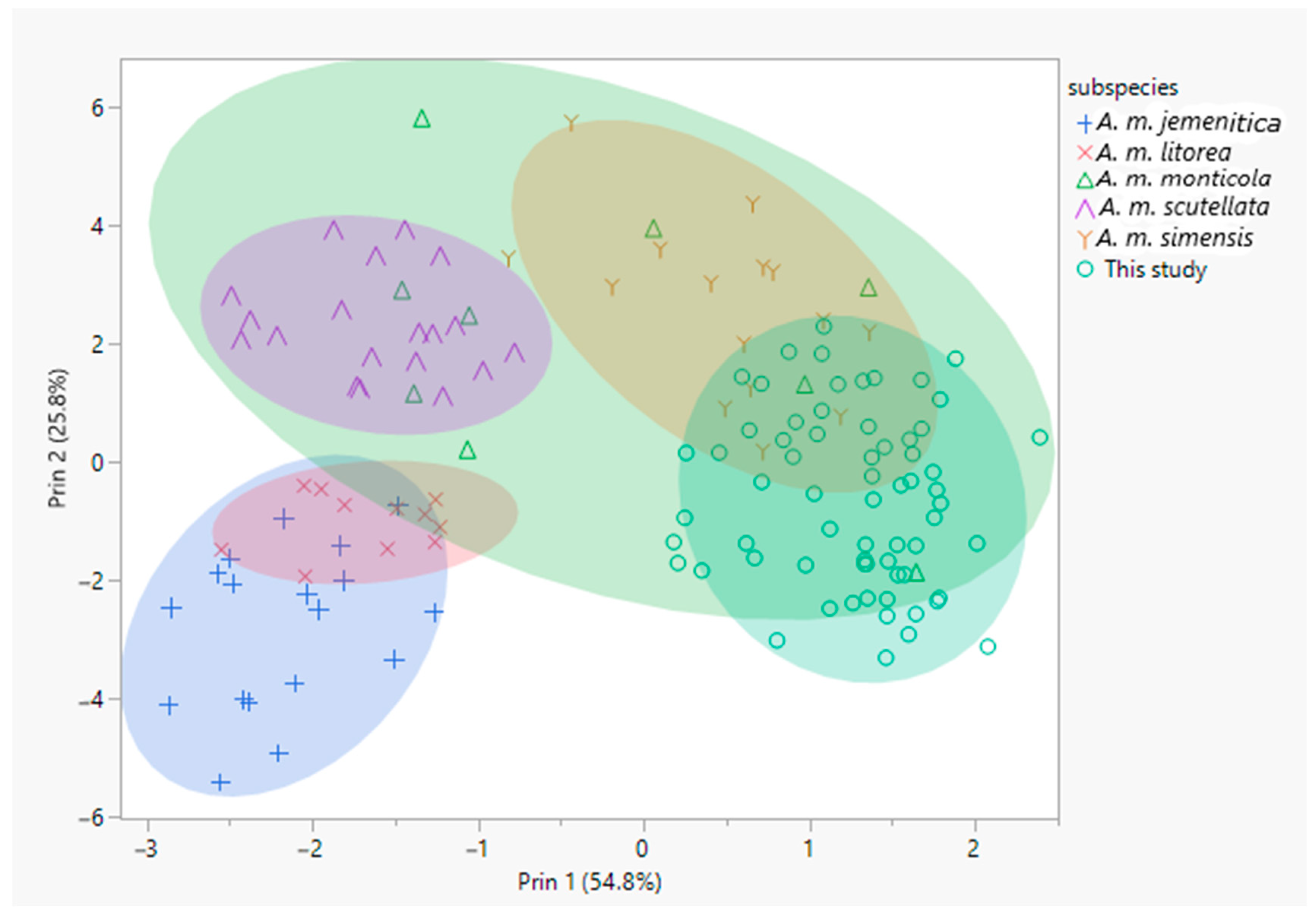
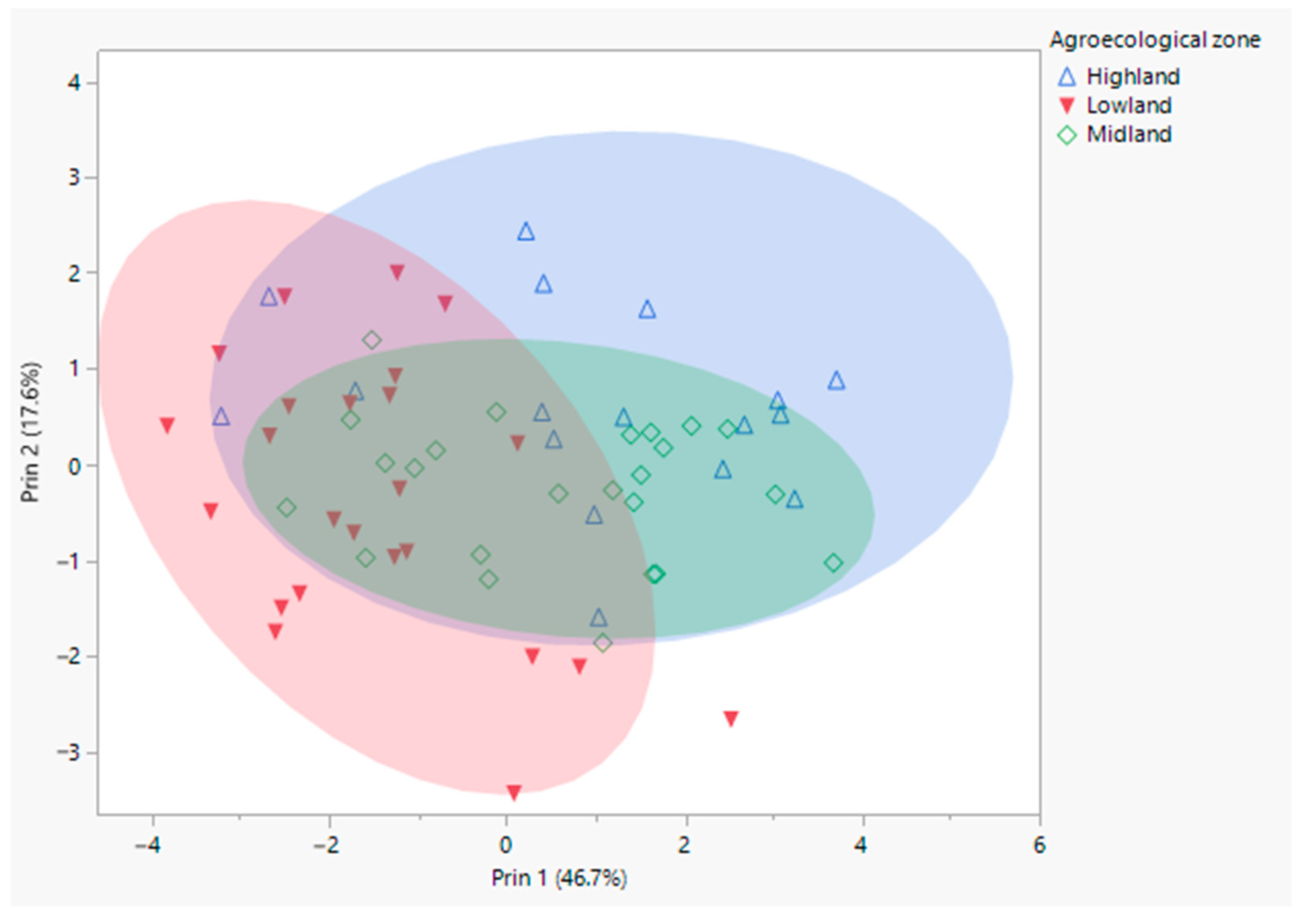
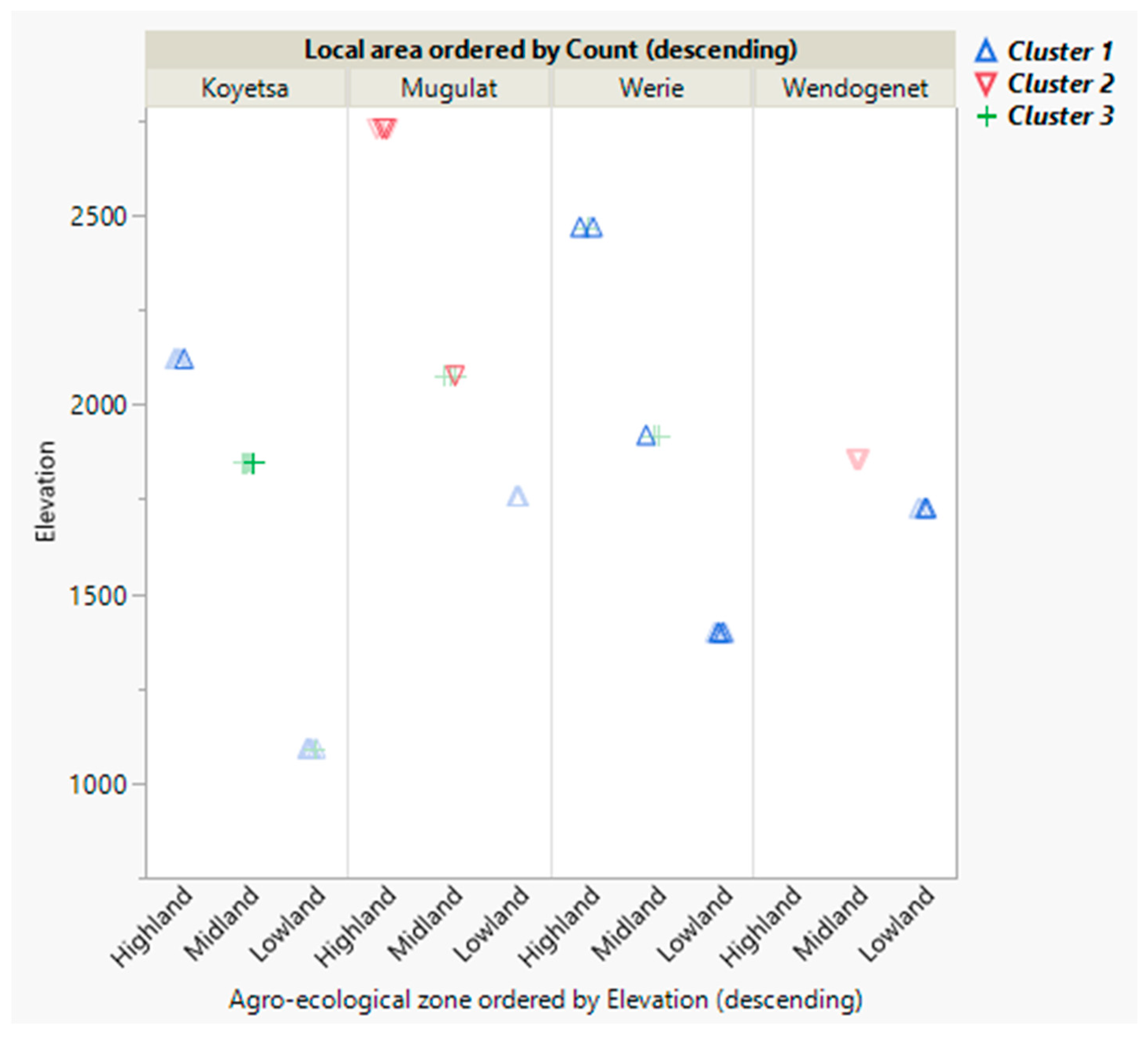
3.1. Morphometric Analyses
3.2. Genetic Analyses
3.3. Integrated Analysis Using Morphometrics and Molecular Data
4. Discussion
4.1. Morphometric Analyses
4.2. Genetic Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ruttner, F. Biogeography and Taxonomy of Honey Bees; Springer: Berlin, Germany, 1988. [Google Scholar]
- Garnery, L.; Cornuet, J.-M.; Solignac, M. Evolutionary History of the Honey Bee Apis Mellifera Inferred from Mitochondrial DNA Analysis. Mol. Ecol. 1992, 1, 145–154. [Google Scholar] [CrossRef] [PubMed]
- Franck, P.; Garnery, L.; Loiseau, A.; Oldroyd, B.P.; Hepburn, H.R.; Soligna, M.; Cornuet, J.M. Genetic Diversity of the Honeybee in Africa: Microsatellite and Mitochondrial Data. Heredity 2001, 86, 420–430. [Google Scholar] [CrossRef]
- Weinstock, G.M.; Robinson, G.E.; Gibbs, R.A.; Worley, K.C.; Evans, J.D.; Maleszka, R.; Robertson, H.M.; Weaver, D.B.; Beye, M.; Bork, P.; et al. Insights into Social Insects from the Genome of the Honeybee Apis Mellifera. Nature 2006, 443, 931–949. [Google Scholar] [CrossRef]
- Zayed, A.; Whitfield, C.W. A Genome-Wide Signature of Positive Selection in Ancient and Recent Invasive Expansions of the Honey Bee Apis Mellifera. Proc. Natl. Acad. Sci. USA 2008. [Google Scholar] [CrossRef]
- Wallberg, A.; Schöning, C.; Webster, M.T.; Hasselmann, M. Two Extended Haplotype Blocks Are Associated with Adaptation to High Altitude Habitats in East African Honey Bees. PloS Genet. 2017, 13, e1006792. [Google Scholar] [CrossRef] [PubMed]
- Cridland, J.M.; Tsutsui, N.D.; Ramírez, S.R. The Complex Demographic History and Evolutionary Origin of the Western Honey Bee, Apis Mellifera. Genome Biol. Evol. 2017. [Google Scholar] [CrossRef]
- Dogantzis, K.A.; Zayed, A. Recent Advances in Population and Quantitative Genomics of Honey Bees. Curr. Opin. Insect Sci. 2019, 31, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Keller, L.; Reeve, H.K. Genetic Variability, Queen Number, and Polyandry in Social Hymenoptera. Evolution 1994, 48, 694–704. [Google Scholar] [CrossRef]
- Tarpy, D.R.; Page, R.E. No Behavioral Control over Mating Frequency in Queen Honey Bees (Apis Mellifera L.): Implications for the Evolution of Extreme Polyandry. Am. Nat. 2000, 155, 820–827. [Google Scholar] [CrossRef] [PubMed]
- McNally, L.C.; Schneider, S.S. Seasonal Cycles of Growth, Development and Movement of the African Honey Bee, Apis Mellifera Scutellata, in Africa. Insectes Sociaux 1992, 39, 167–179. [Google Scholar] [CrossRef]
- Fuller, Z.L.; Niño, E.L.; Patch, H.M.; Bedoya-Reina, O.C.; Baumgarten, T.; Muli, E.; Mumoki, F.; Ratan, A.; McGraw, J.; Frazier, M.; et al. Genome-Wide Analysis of Signatures of Selection in Populations of African Honey Bees (Apis Mellifera) Using New Web-Based Tools. BMC Genom. 2015. [Google Scholar] [CrossRef]
- Arias, M.C.; Sheppard, W.S. Molecular Phylogenetics of Honey Bee Subspecies (Apis Mellifera, L.) Inferred from Mitochondrial DNA Sequence. Mol. Phylogenet. Evol. 1996, 5, 557–566. [Google Scholar] [CrossRef]
- Alburaki, M.; Bertrand, B.; Legout, H.; Moulin, S.; Alburaki, A.; Sheppard, W.S.; Garnery, L. A Fifth Major Genetic Group among Honeybees Revealed in Syria. BMC Genet. 2013, 14. [Google Scholar] [CrossRef] [PubMed]
- Meixner, M.D.; Leta, M.A.; Koeniger, N.; Fuchs, S. The Honey Bees of Ethiopia Represent a New Subspecies of Apis Mellifera-Apis Mellifera Simensis n. Ssp. Apidologie 2011, 42, 425–437. [Google Scholar] [CrossRef]
- Hailu, T.G.; D’ALvise, P.; Tofilski, A.; Fuchs, S.; Greiling, J.; Rosenkranz, P.; Hasselmann, M.H. Insights into Ethiopian Honey Bee Diversity Based on Wing Geomorphometric and Mitochondrial DNA Analyses. Apidologie 2020. [Google Scholar] [CrossRef]
- Hepburn, H.R.; Radloff, S.E. Biogeographical Correlates of Population Variance in the Honeybees (Apis Mellifera L.) of Africa. Apidologie 1997, 28, 243–258. [Google Scholar] [CrossRef][Green Version]
- Amssalu, N.; Radloff, H. Multivariate Morphometric Analysis of Honeybees (Apis Mellifera) in the Ethiopian Region. Apidologie 2004, 35, 71–81. [Google Scholar] [CrossRef]
- Radloff, S.E.; Hepburn, H.R. Multivariate Analysis of Honeybees, Apis Mellifera Linnaeus (Hymenoptera: Apidae), of the Horn of Africa. Afr. Entomol. 1997, 5, 57–64. [Google Scholar]
- Hepburn, H.R.; Radloff, S.E.; Oghiakhe, S. Mountain Honeybees of Africa. Apidologie 2000, 31, 205–221. [Google Scholar] [CrossRef][Green Version]
- Mulder, H.A.; Bijma, P.; Hill, W.G. Prediction of Breeding Values and Selection Responses with Genetic Heterogeneity of Environmental Variance. Genetics 2007, 175, 1895–1910. [Google Scholar] [CrossRef]
- Gruber, K.; Schöning, C.; Otte, M.; Kinuthia, W.; Hasselmann, M. Distinct Subspecies or Phenotypic Plasticity? Genetic and Morphological Differentiation of Mountain Honey Bees in East Africa. Ecol. Evol. 2013, 3, 3204–3218. [Google Scholar] [CrossRef]
- Meixner, M.D.M.; Riasc, M.C.A.; Heppardb, W.S.S. Mitochondrial DNA Polymorphisms in Honey Bee Subspecies from Kenya. Apidologie 2000, 31, 181–190. [Google Scholar] [CrossRef]
- Money, T.G.A.; Sproule, M.K.J.; Cross, K.P.; Robertson, X.R.M. Octopamine Stabilizes Conduction Reliability of an Unmyelinated Axon during Hypoxic Stress. J. Neurophysiol. 2016, 116, 949–959. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, G.A.B.; Robertson, R.M. A Role for Octopamine in Coordinating Thermoprotection of an Insect Nervous System. J. Therm. Biol. 2006, 31, 149–158. [Google Scholar] [CrossRef]
- Blenau, W.; Scheiner, R.; Plu, S. Beha v Ioural Pharmacology of Octopamine, Tyramine and Dopamine in Honey Bees. Behav. Brain Res. 2002, 136, 545–553. [Google Scholar]
- Hammer, M.; Menzel, R. Multiple Sites of Associative Odor Learning as Revealed by Local Brain Microinjections of Octopamine in Honeybees; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1998. [Google Scholar]
- Erber, J.; Kloppenburg, P. The Modulatory Effects of Serotonin and Octopamine in the Visual System of the Honey Bee (Apis Mellifera L.). J. Comp. Physiol. A 1995, 176, 119–129. [Google Scholar] [CrossRef]
- Behrends, A.; Scheiner, R. Octopamine Improves Learning in Newly Emerged Bees but Not in Old Foragers. J. Exp. Biol. 2012, 1076–1083. [Google Scholar] [CrossRef]
- Cook, C.N.; Brent, C.S.; Breed, M.D. Octopamine and Tyramine Modulate the Thermoregulatory Fanning Response in Honey Bees (Apis Mellifera). J. Exp. Biol. 2017, 220, 1925–1930. [Google Scholar] [CrossRef]
- Bastide, H.; Yassin, A.; Johanning, E.J.; Pool, J.E. Pigmentation in Drosophila Melanogaster Reaches Its Maximum in Ethiopia and Correlates Most Strongly with Ultra-Violet Radiation in Sub-Saharan Africa. BMC Evol. Biol. 2014, 14, 1–14. [Google Scholar] [CrossRef]
- Michener, C.D. The Brazilian Bee Problem. Annu. Rev. Entomol. 1975, 20, 399–416. [Google Scholar] [CrossRef]
- Meixner, M.D.; Costa, C.; Kryger, P.; Hatjina, F.; Bouga, M.; Ivanova, E.; Büchler, R. Conserving Diversity and Vitality for Honey Bee Breeding Conserving Diversity and Vitality for Honey Bee Breeding. J. Apic. Res. 2010, 49, 85–92. [Google Scholar] [CrossRef]
- Cánovas, F.; De La Rúa, P.; Serrano, J.; Galián, J. Microsatellite Variability Reveals Beekeeping Influences on Iberian Honeybee Populations. Apidologie 2011. [Google Scholar] [CrossRef]
- Espregueira Themudo, G.; Rey-Iglesia, A.; Robles Tascón, L.; Bruun Jensen, A.; da Fonseca, R.R.; Campos, P.F. Declining Genetic Diversity of European Honeybees along the Twentieth Century. Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Teweldemedhn, G.; Yayneshet, T. Honeybee Colony Marketing Practices in Werieleke District Of The Tigray Region, Ethiopia. Bee World 2014, 91, 30–35. [Google Scholar] [CrossRef]
- Central Statistical Agency (CSA). Agricultural Sample Survey 2016/17 [2009 e.c.]: Report on Livestock and Livestock Characteristics; Central Statistical Agency: Addis Ababa, Ethiopia, 2017; Volume II. [Google Scholar]
- Meixner, M.D.; Pinto, M.A.; Bouga, M.; Kryger, P.; Ivanova, E.; Fuchs, S. Standard Methods for Characterising Subspecies and Ecotypes of Apis Mellifera. J. Apic. Res. 2013, 52, 1–28. [Google Scholar] [CrossRef]
- Nawrocka, A.; Kandemir, İ.; Fuchs, S.; Tofilski, A. Computer Software for Identification of Honey Bee Subspecies and Evolutionary Lineages. Apidologie 2018, 49, 172–184. [Google Scholar] [CrossRef]
- Tofilski, A. DrawWing, a Program for Numerical Description of Insect Wings. J. Insect Sci. 2004, 4, 1–5. [Google Scholar] [CrossRef]
- Garnery, L.; Solignac, M.; Celebrano, G.; Cornuet, J. A Simple Test Using Restricted PCR-Amplifled Mitochondrial DNA to Study the Genetic Structure of Apis Mellifera L. Experientia 1993, 49, 1016–1021. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mat, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DNA Sequence Polymorphism Analysis of Large Datasets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Tajima, F. Evolutionary Relationship of DNA Sequences in Finite Populations. Genetics 1983, 437–460. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The Neighbor-Joining Method: A New Method for Reconstructing Phylogenetic Trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef] [PubMed]
- Watterson, G.A. On the Number of Segregating Sites in Genetical Models without Recombination. Theor. Popul. Biol. 1975, 276, 256–276. [Google Scholar] [CrossRef]
- Tajima, F. Statistical Method for Testing the Neutral Mutation Hypothesis by DNA Polymorphism. Genetics 1989, 123, 607–612. [Google Scholar] [CrossRef]
- Wright, S. The Genetic Structure of Populations. Eugenics 1951, 15, 323–354. [Google Scholar] [CrossRef] [PubMed]
- Hudson, R.R.; Boos, D.D.; Kaplan, N.L. A Statistical Test for Detecting Geographic Subdivision’. Mol. Biol. Evol. 1992, 9, 138–151. [Google Scholar]
- Hudson, R.R.; Slatkint, M.; Maddison, W.P. Estimation of Levels of Gene Flow from DNA Sequence Data. Genet. Soc. Am. 1992, 132, 583–589. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M. Estimation of the Number of Nucleotide Substitutions in the Control Region of Mitochondrial DNA in Humans and Chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef]
- Schreiber, J.B. Latent Class Analysis: An Example for Reporting Results. Res. Soc. Adm. Pharm. 2017, 13, 1196–1201. [Google Scholar] [CrossRef] [PubMed]
- Radloff, S.; Hepburn, R. Population Structure and Morphometric Variance of the Apis Mellifera Scutellata Group of Honeybees in Africa. Genet. Mol. Biol. 2000, 23, 305–316. [Google Scholar] [CrossRef]
- Abou-Shaara, H.F. Thermal Tolerance Characteristics of Two Honey Bee Races. J. Agric. Urban Entomol. 2015, 31, 1–8. [Google Scholar] [CrossRef]
- Nuru, A. Selling Honeybee Colonies as a Source of Income for Subsistence Beekeepers. Bees Dev. 2002, 64. Available online: http://www.beesfordevelopment.org/documents/s/selling-honeybee-colonies-as-a-source-of-income-for-subsistence-beekeepers/ (accessed on 23 February 2021).
- Teweldemedhn, G.; Yayneshet, T. Honeybee Colony Marketing and Its Implications for Queen Rearing and Beekeeping Development in Tigray, Ethiopia. Int. J. Livest. Prod. 2014, 5, 117–128. [Google Scholar] [CrossRef]
- Harpur, B.A.; Minaei, S.; Kent, C.F.; Zayed, A. Management Increases Genetic Diversity of Honey Bees via Admixture. Mol. Ecol. 2012, 21, 4414–4421. [Google Scholar] [CrossRef]
- Harpur, B.A.; Minaei, S. Admixture Increases Diversity in Managed Honey Bees: Reply to De La Rúa et al. (2013). Mol. Ecol. 2013, 22, 3211–3215. [Google Scholar] [CrossRef]
- De la Rúa, P.; Jaffé, R.; Muñoz, I.; Serrano, J.; Moritz, B.F. Conserving Genetic Diversity in the Honeybee: Comments on Harpur et Al. (2012). Mol. Ecol. Resour. 2013, 22, 3208–3210. [Google Scholar] [CrossRef]
- Wallberg, A.; Glémin, S.; Webster, M.T. Extreme Recombination Frequencies Shape Genome Variation and Evolution in the Honeybee, Apis Mellifera. PLoS Genet. 2015, 11, 1–27. [Google Scholar] [CrossRef] [PubMed]
| Subspecies | Femur | Tibia | Metatarsus Length | Metatarsus Width | Length of Hind Leg | Forewing Length | Forewing Width | Cubital Vein Distance a | Cubital Vein Distance b | Cubital Index |
|---|---|---|---|---|---|---|---|---|---|---|
| A. m. jemenitica | 2.36 | 2.92 | 1.82 | 1.04 | 7.11 | 8.13 | 2.80 | 0.42 | 0.19 | 2.26 |
| A. m. litorea | 2.42 | 2.99 | 1.85 | 1.05 | 7.26 | 8.37 | 2.90 | 0.44 | 0.19 | 2.33 |
| A. m. monticola | 2.46 | 3.07 | 1.95 | 1.09 | 7.49 | 8.83 | 3.01 | 0.49 | 0.22 | 2.23 |
| A. m. scutellata | 2.53 | 3.13 | 1.95 | 1.10 | 7.60 | 8.65 | 2.98 | 0.46 | 0.20 | 2.38 |
| A. m. simensis | 2.51 | 3.08 | 1.98 | 1.08 | 7.57 | 8.70 | 3.05 | 0.51 | 0.23 | 2.22 |
| This study | 2.40 | 2.93 | 1.90 | 1.02 | 7.27 | 8.29 | 3.02 | 0.51 | 0.23 | 2.24 |
| Highland | 2.41 ab | 2.96 a | 1.91 a | 1.05 a | 7.29 a | 8.43 a | 3.05 a | 0.51 a | 0.24 a | 2.20 a |
| Lowland | 2.38 b | 2.88 b | 1.88 a | 1.00 b | 7.26 a | 8.16 c | 2.98 b | 0.51 a | 0.23 a | 2.19 a |
| Midland | 2.41 a | 2.95 a | 1.91 a | 1.02 c | 7.28 a | 8.31 b | 3.03 a | 0.52 a | 0.23 a | 2.30 a |
| Significance (AEZ) | * | ** | * | ** | ** | ** |
| Country | Geographic Region | Local Area | AEZ | Number of Segregating Sites (S) | Nucleotide Diversity | Tajima’s D | ||
|---|---|---|---|---|---|---|---|---|
| Average Number of Differences (k) | Average Number of Pairwise Differences (π) | Watterson Estimator (θw) | ||||||
| Ethiopia | North | Mugulat | Highland | 66 | 24.75 | 0.0156 | 0.0172 | −0.179 |
| 0.0 | 0.0 | 0.0 | 0.001 | |||||
| Midland | 111 | 33.47 | 0.0211 | 0.0277 | −1.014 | |||
| 0.0 | 0.0 | 0.0 | 0.0 | |||||
| Lowland | 79 | 28.62 | 0.0180 | 0.0212 | −0.663 | |||
| 5 | 1.6 | 0.004 | 0.005 | |||||
| Total Mugulat | 173 | 29.98 | 0.0189 | 0.0296 | −1.543 | |||
| 5 | 0.56 | 0.001 | 0.002 | |||||
| Werie | Highland | 75 | 27.10 | 0.0171 | 0.0207 | −0.762 | ||
| 1 | 0.4 | 0.001 | 0.001 | |||||
| Midland | 73 | 29.40 | 0.0185 | 0.0223 | −0.697 | |||
| 1 | 0.6 | 0.001 | 0.001 | |||||
| Lowland | 95 | 29.20 | 0.0184 | 0.0220 | −0.486 | |||
| 0.0 | 0.0 | 0.0 | 0.0 | |||||
| Total Werie | 169 | 30.41 | 0.0192 | 0.0299 | −1.294 | |||
| 2 | 0.36 | 0.001 | 0.001 | |||||
| Koyetsa | Highland | 82 | 28.31 | 0.0178 | 0.0212 | −0.486 | ||
| 6 | 3.06 | 0.006 | 0.006 | |||||
| Midland | 106 | 31.03 | 0.0196 | 0.0252 | −0.861 | |||
| 5 | 2.67 | 0.006 | 0.005 | |||||
| Lowland | 92 | 29.44 | 0.0186 | 0.0232 | −0.585 | |||
| 4 | 2.13 | 0.005 | 0.004 | |||||
| Total Koyetsa | 181 | 30.21 | 0.0190 | 0.0313 | −1.392 | |||
| 7 | 2.4 | 0.005 | 0.004 | |||||
| Total North | 354 | 30.65 | 0.0193 | 0.0460 | −2.045 | |||
| 8 | 1.61 | 0.004 | 0.004 | |||||
| South | Wendogenet | Midland | 64 | 25.62 | 0.0161 | 0.0196 | −0.095 | |
| 2 | 0.8 | 0.002 | 0.002 | |||||
| Lowland | 92 | 29.31 | 0.0185 | 0.0225 | −0.368 | |||
| 0.0 | 0.0 | 0.0 | 0.0 | |||||
| Total South | 122 | 29.20 | 0.0184 | 0.0248 | −0.752 | |||
| 2 | 0.4 | 0.001 | 0.002 | |||||
| Total Ethiopia | 391 | 30.44 | 0.0192 | 0.0486 | −2.09 | |||
| 10 | 1.5 | 0.003 | 0.005 | |||||
| Mau | MF | 69 | 23.89 | 0.0151 | 0.0161 | −0.128 | ||
| MS | 124 | 28.81 | 0.0182 | 0.0243 | −0.841 | |||
| Kenya | Mount Kenya | MKS | 118 | 30.71 | 0.0194 | 0.0230 | −0.515 | |
| Savanna | 183 | 30.68 | 0.0196 | 0.0287 | −1.111 | |||
| Total Kenya | 207 | 29.91 | 0.0189 | 0.0302 | −1.349 | |||
| Country | AEZ | Number of Segregating Sites (S) | Nucleotide Diversity | Tajima’s D | ||
|---|---|---|---|---|---|---|
| Average Number of Nucleotide Differences (k) | Average Number of Pairwise Difference (π) | Watterson Estimator (θw) | ||||
| Ethiopia | Highland | 142 | 28.10 | 0.0177 | 0.0258 | −1.099 |
| 6 | 1.57 | 0.003 | 0.004 | |||
| Midland | 166 | 30.48 | 0.0192 | 0.0308 | −1.432 | |
| 7 | 1.43 | 0.003 | 0.004 | |||
| Lowland | 234 | 30.91 | 0.0195 | 0.0364 | −1.418 | |
| 5 | 1.5 | 0.003 | 0.003 | |||
| Kenya | Mountain forest | 69 | 23.89 | 0.0151 | 0.0161 | −0.128 |
| Savanna | 183 | 30.68 | 0.0196 | 0.0287 | −1.111 | |
| This Study | All Highlands | All Midlands | All Lowlands | MF | MS | MKS | All Savannah | |
|---|---|---|---|---|---|---|---|---|
| This study | 0.005 *** | 0.005 *** | 0.007 *** | 0.006 *** | ||||
| 0.006 ** | 0.009 *** | 0.006 ** | 0.007 *** | |||||
| All highlands | 0.004 * | 0.006 ** | 0.018 *** | 0.013 *** | 0.015 *** | 0.009 *** | ||
| 0.005 * | 0.012 *** | 0.023 *** | 0.013 ** | 0.015 ** | 0.008 * | |||
| All midlands | 0.0149 | 0.005 ** | 0.011 ** | 0.010 ** | 0.012 *** | 0.008 *** | ||
| 0.0215 | 0.007 *** | 0.012 ** | 0.013 *** | 0.011 ** | 0.009 *** | |||
| All lowlands | 0.0236 | 0.0153 | 0.012 *** | 0.010 *** | 0.012 *** | 0.008 *** | ||
| 0.0797 | 0.0297 | 0.017 *** | 0.025 *** | 0.015 *** | 0.017 *** | |||
| MF | 0.0542 | 0.0775 | 0.0463 | 0.062 | 0.025 *** | 0.026 *** | 0.015 *** | |
| 0.0697 | 0.1093 | 0.0468 | 0.101 | 0.031 ** | 0.029 ** | 0.017 ** | ||
| MS | 0.0332 | 0.0356 | 0.0415 | 0.039 | 0.0766 | 0.015 *** | ||
| 0.0411 | 0.0487 | 0.04 | 0.0732 | 0.1014 | 0.021** | |||
| MKS | 0.0496 | 0.057 | 0.0545 | 0.0547 | 0.0947 | 0.055 | ||
| 0.0874 | 0.0413 | 0.0671 | 0.1647 | 0.1205 | 0.0923 | |||
| 0.02767 | 0.03223 | 0.03196 | 0.03324 | 0.0798 | ||||
| All savannah | 0.03957 | 0.02033 | 0.03206 | 0.09598 | 0.08487 |
| (a) Analysis based on a nuclear marker on chromosome seven, denoted as r7-frag, using Ethiopian and Kenyan honey bee samples. | |||||||||||||||
| Local Area | AEZ | Mugulat | Werie | Koyetsa | Wendogenet | Mau | Mount Kenya | ||||||||
| Highland | Midland | Lowland | Highland | Midland | Lowland | Highland | Midland | Lowland | Midland | Lowland | MF | MS | MKS | ||
| Mugulat | Highland | 0.007 ns | 0.022 * | 0.044 *** | 0.012 ns | 0.031 ** | 0.031 ** | 0.015 * | 0.027 *** | 0.042 * | 0.024 ** | 0.024 * | 0.019 ** | 0.022 ** | |
| 0.005 ns | 0.026 * | 0.052 * | 0.012 ns | 0.033 * | 0.020 * | 0.012 ns | 0.035 ** | 0.041 * | 0.029 ** | 0.035 * | 0.018 * | 0.019 ** | |||
| Midland | 0.02 | 0.009 ns | 0.029 ** | −0.006 ns | 0.021 * | 0.006 ns | −0.001 ns | 0.011 ** | 0.031 * | 0.009 ns | 0.016 * | 0.012 * | 0.009 * | ||
| −0.01 | 0.008 ns | 0.041 * | 0.008 ns | 0.020 * | 0.004 ns | 0.002 ns | 0.019 *** | 0.027 * | 0.013 * | 0.022 * | 0.012 * | 0.006 ns | |||
| Lowland | 0.07 | 0.04 | 0.039 * | 0.011 ns | 0.033 ** | 0.018 ** | 0.012 * | 0.018 * | 0.047 * | 0.018 * | 0.026 * | 0.013 * | 0.014 * | ||
| 0.081 | 0.013 | 0.062 ** | 0.010 ns | 0.033 ** | 0.015 ns | 0.013 ns | 0.026 ** | 0.043 * | 0.022 * | 0.033 * | 0.023 ** | 0.013 * | |||
| Werie | Highland | 0.094 | 0.081 | 0.076 | 0.011 ns | 0.035 ** | 0.018 * | 0.023 ** | 0.021 * | 0.053 * | 0.029 ** | 0.054 *** | 0.025 *** | 0.023 ** | |
| 0.164 | 0.180 | 0.240 | 0.028 ns | 0.057 ** | 0.034 * | 0.041 ** | 0.062 ** | 0.062 * | 0.064 ** | 0.074 ** | 0.030 ** | 0.039 *** | |||
| Midland | 0.013 | −0.013 | 0.025 | 0.024 | 0.020 * | 0.010 ns | 0.0004 ns | 0.008 ns | 0.035 ns | 0.006 ns | 0.023 * | 0.012 * | 0.012 * | ||
| 0.008 | −0.037 | 0.013 | 0.133 | 0.016 ns | 0.003 ns | 0.002 ns | 0.010 ns | 0.025 ns | 0.006 ns | 0.022 ns | 0.014 * | 0.011 ns | |||
| Lowland | 0.091 | 0.069 | 0.118 | 0.120 | 0.078 | 0.020 * | 0.019 ** | 0.019 * | 0.035 * | 0.021 ** | 0.038 *** | 0.026 *** | 0.028 *** | ||
| 0.109 | 0.055 | 0.110 | 0.219 | 0.028 | 0.020 * | 0.021 * | 0.022 * | 0.037 ** | 0.026 ** | 0.043 *** | 0.033 ** | 0.029 ** | |||
| Koyetsa | Highland | 0.070 | 0.012 | 0.061 | 0.041 | 0.060 | 0.10 | 0.001 ns | 0.006 ns | 0.034 * | 0.008 ns | 0.031 *** | 0.012 ** | 0.009 ns | |
| 0.066 | −0.001 | 0.042 | 0.095 | 0.001 | 0.05 | 0.0001 ns | 0.011 ns | 0.021 ns | 0.015 ns | 0.029 * | 0.015 * | 0.011 ns | |||
| Midland | 0.037 | −0.022 | 0.042 | 0.076 | −0.005 | 0.07 | 0.01 | 0.011 * | 0.027 * | 0.009 ns | 0.018 * | 0.010 * | 0.013 * | ||
| 0.025 | −0.031 | 0.039 | 0.173 | −0.022 | 0.06 | −0.01 | 0.017 ** | 0.024 * | 0.013 * | 0.017 * | 0.011 * | 0.011 * | |||
| Lowland | 0.108 | 0.039 | 0.064 | 0.039 | 0.026 | 0.07 | 0.03 | 0.04 | 0.031 * | 0.010 ns | 0.029 ** | 0.017 *** | 0.015 ** | ||
| 0.190 | 0.107 | 0.108 | 0.272 | 0.021 | 0.08 | 0.06 | 0.10 | 0.034 * | 0.009 ns | 0.037 ** | 0.036 *** | 0.023 ** | |||
| Wendogenet | Midland | 0.016 | 0.059 | 0.113 | 0.091 | 0.040 | 0.06 | 0.09 | 0.05 | 0.07 | 0.034 * | 0.054 ** | 0.029 ** | 0.030 ** | |
| 0.103 | 0.088 | 0.118 | 0.175 | 0.033 | 0.07 | 0.04 | 0.05 | 0.09 | 0.036 ** | 0.048 ** | 0.028 ** | 0.031 ** | |||
| Lowland | 0.074 | 0.023 | 0.060 | 0.083 | 0.005 | 0.08 | 0.03 | 0.02 | 0.04 | 0.09 | 0.032 ** | 0.016 ** | 0.015 ** | ||
| 0.142 | 0.057 | 0.060 | 0.285 | −0.002 | 0.07 | 0.05 | 0.06 | 0.01 | 0.09 | 0.036 ** | 0.035 *** | 0.020 ** | |||
| Mau | MF | 0.061 | 0.032 | 0.068 | 0.142 | 0.063 | 0.10 | 0.10 | 0.03 | 0.09 | 0.11 | 0.10 | 0.025 *** | 0.026 *** | |
| 0.097 | 0.051 | 0.080 | 0.277 | 0.039 | 0.13 | 0.07 | 0.02 | 0.16 | 0.11 | 0.14 | 0.031 ** | 0.029 *** | |||
| MS | 0.057 | 0.037 | 0.057 | 0.087 | 0.056 | 0.088 | 0.035 | 0.023 | 0.067 | 0.092 | 0.050 | 0.077 | 0.015 ** | ||
| 0.064 | 0.066 | 0.137 | 0.121 | 0.098 | 0.148 | 0.053 | 0.035 | 0.243 | 0.109 | 0.212 | 0.121 | 0.021 ** | |||
| Mount Kenya | MKS | 0.103 | 0.021 | 0.054 | 0.110 | 0.070 | 0.123 | 0.030 | 0.046 | 0.076 | 0.124 | 0.055 | 0.095 | 0.055 | |
| 0.077 | −0.003 | 0.039 | 0.187 | 0.047 | 0.106 | 0.006 | 0.028 | 0.139 | 0.124 | 0.094 | 0.101 | 0.092 | |||
| (b) Analysis based on the mitochondrial COI-COII region using Ethiopian honey bee samples. | |||||||||||||||
| Local Area | AEZ | Mugulat | Werie | Koyetsa | Wendogenet | ||||||||||
| Highland | Midland | Lowland | Highland | Midland | Lowland | Highland | Midland | Lowland | Midland | Lowland | |||||
| Mugulat | Highland | nc | 0.025 ns | 0.057 ns | 0.214 ns | nc | 0.233 ns | 0.298 ns | 0.450 ns | 0.014 ns | nc | ||||
| 0.103 * | 0.004 ns | 0.066 ns | 0.080 | 0.138 * | 0.096 | 0.101 ns | 0.178 ns | 0.023 ns | 0.031 ns | ||||||
| Midland | 0.0000 | 0.025 ns | 0.057 ns | 0.214 ns | nc | 0.233 ns | 0.298 ns | 0.450 ns | 0.087 ns | nc | |||||
| 0.1857 | 0.085 ns | 0.075 ns | 0.243 ** | 0.106 ns | 0.194 * | 0.190 * | 0.296 * | 0.200 ** | 0.126 * | ||||||
| Lowland | 0.0000 | 0.0000 | −0.033 ns | −0.008 ns | 0.025 ns | 0.020 ns | 0.054 ns | 0.145 ns | −0.028 ns | −0.024 ns | |||||
| nc | 0.063 | 0.086 * | 0.096 * | 0.147 * | 0.031 ns | 0.042 ns | 0.094 ns | 0.033 ns | 0.006 ns | ||||||
| Werie | Highland | 0.0000 | 0.0000 | 0.0000 | 0.069 ns | 0.057 ns | 0.116 ns | 0.191 ns | 0.329 ns | −0.012 ns | 0.000 ns | ||||
| 0.1858 | 0.0615 | 0.0970 | 0.115 * | 0.161 * | 0.099 ns | 0.104 ns | 0.203 * | 0.131 ** | 0.042 ns | ||||||
| Midland | 0.2500 | 0.2500 | nc | 0.1667 | 0.214 ns | 0.096 ns | 0.124 ns | 0.268 ns | 0.043 ns | 0.156 ns | |||||
| 0.1901 | 0.3232 | 0.0719 | 0.1667 | 0.066 ns | 0.102 ns | 0.097 ns | 0.192 * | 0.042 ns | 0.047 ns | ||||||
| Lowland | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.2500 | 0.233 ns | 0.298 ns | 0.450 ns | 0.087 ns | nc | |||||
| 0.3333 | 0.4316 | 0.2111 | 0.2521 | 0.1122 | 0.155 ** | 0.148 * | 0.245 * | 0.065 ns | 0.105 ns | ||||||
| Koyetsa | Highland | 0.3429 | 0.3429 | 0.0533 | 0.2973 | 0.2143 | 0.3429 | −0.068 ns | −0.049 ns | 0.105 ns | 0.180 ns | ||||
| 0.2686 | 0.3558 | 0.1110 | 0.2553 | 0.2552 | 0.3119 | −0.060 ns | −0.061 ns | 0.106 ns | 0.057 ns | ||||||
| Midland | 0.4286 | 0.4286 | 0.1136 | 0.3947 | 0.2576 | 0.4286 | nc | −0.071 ns | 0.155 ns | 0.245 ns | |||||
| 0.2704 | 0.3503 | 0.1211 | 0.2685 | 0.2533 | 0.3082 | nc | −0.048 ns | 0.113 ns | 0.071 ns | ||||||
| Lowland | 0.6000 | 0.6000 | 0.2723 | 0.5581 | 0.4605 | 0.6000 | nc | nc | 0.278 ns | 0.401 ns | |||||
| 0.4276 | 0.5225 | 0.2606 | 0.4399 | 0.4223 | 0.4743 | nc | nc | 0.201 ns | 0.153 ns | ||||||
| Wendogenet | Midland | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.1250 | 0.0000 | 0.2927 | 0.3659 | 0.5217 | 0.030 ns | ||||
| 0.0698 | 0.3099 | 0 | 0.2473 | 0.0741 | 0.2376 | 0.3219 | 0.3268 | 0.4842 | |||||||
| Lowland | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.2500 | 0.0000 | 0.3429 | 0.4286 | 0.6000 | 0.0000 | |||||
| 0.0614 | 0.0843 | nc | 0.0160 | 0.0250 | 0.1680 | 0.2160 | 0.2361 | 0.3959 | nc | ||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hailu, T.G.; D'Alvise, P.; Hasselmann, M. Disentangling Ethiopian Honey Bee (Apis mellifera) Populations Based on Standard Morphometric and Genetic Analyses. Insects 2021, 12, 193. https://doi.org/10.3390/insects12030193
Hailu TG, D'Alvise P, Hasselmann M. Disentangling Ethiopian Honey Bee (Apis mellifera) Populations Based on Standard Morphometric and Genetic Analyses. Insects. 2021; 12(3):193. https://doi.org/10.3390/insects12030193
Chicago/Turabian StyleHailu, Teweldemedhn Gebretinsae, Paul D'Alvise, and Martin Hasselmann. 2021. "Disentangling Ethiopian Honey Bee (Apis mellifera) Populations Based on Standard Morphometric and Genetic Analyses" Insects 12, no. 3: 193. https://doi.org/10.3390/insects12030193
APA StyleHailu, T. G., D'Alvise, P., & Hasselmann, M. (2021). Disentangling Ethiopian Honey Bee (Apis mellifera) Populations Based on Standard Morphometric and Genetic Analyses. Insects, 12(3), 193. https://doi.org/10.3390/insects12030193








