Evidence for the Involvement of the Chemosensory Protein AgosCSP5 in Resistance to Insecticides in the Cotton Aphid, Aphis gossypii
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Insect
2.2. Insecticides
2.3. Identification and Bioinformatics Analysis of AgosCSP Genes
2.4. Generation of Transgenic Drosophila Flies Expressing AgosCSP5
2.5. UAS/GAL4 Expression of AgosCSP5
2.6. Bioassays of A. Gossypii
2.7. Bioassays of Drosophila Flies
2.8. RNA Extraction, cDNA Synthesis, RT-PCR and RT-qPCR
2.9. Protein Structure Prediction and Refinement
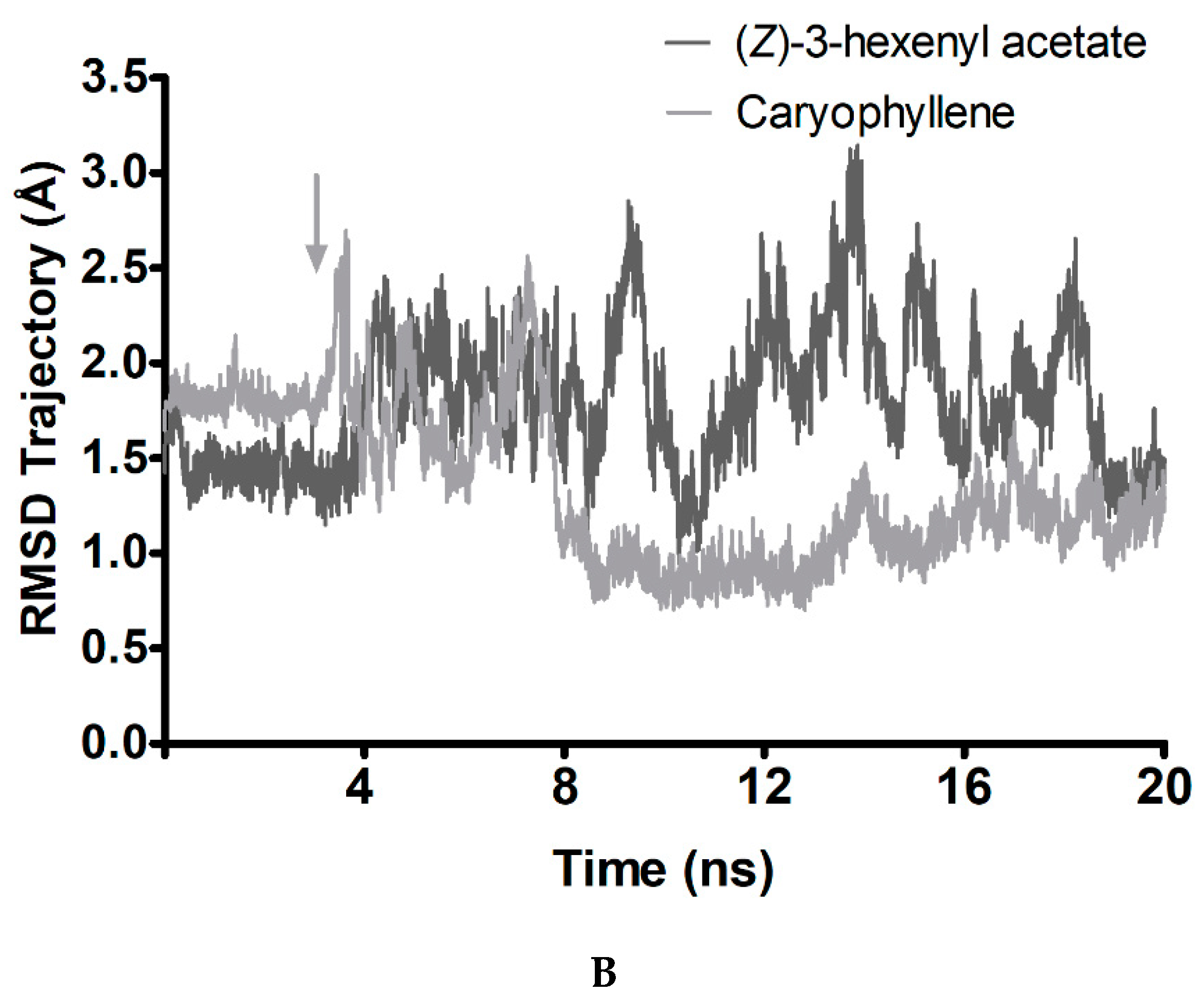
2.10. Molecular Docking and Complex Molecular Dynamics
3. Results
3.1. Sequence Annotation and Phylogenetic Analysis of A. Gossypii CSPs
3.2. Upregulation of A. Gossypii CSP Gene Expression by Insecticide Omethoate
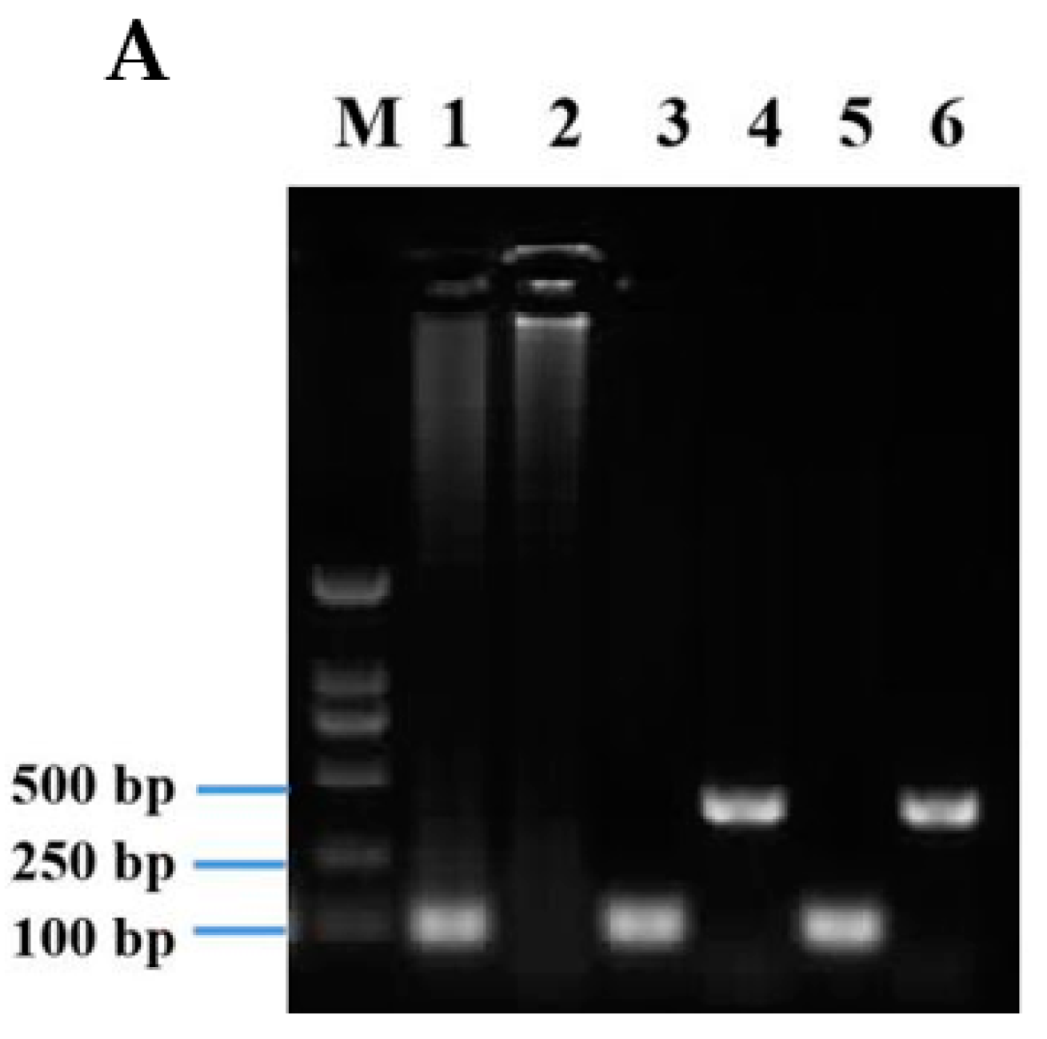
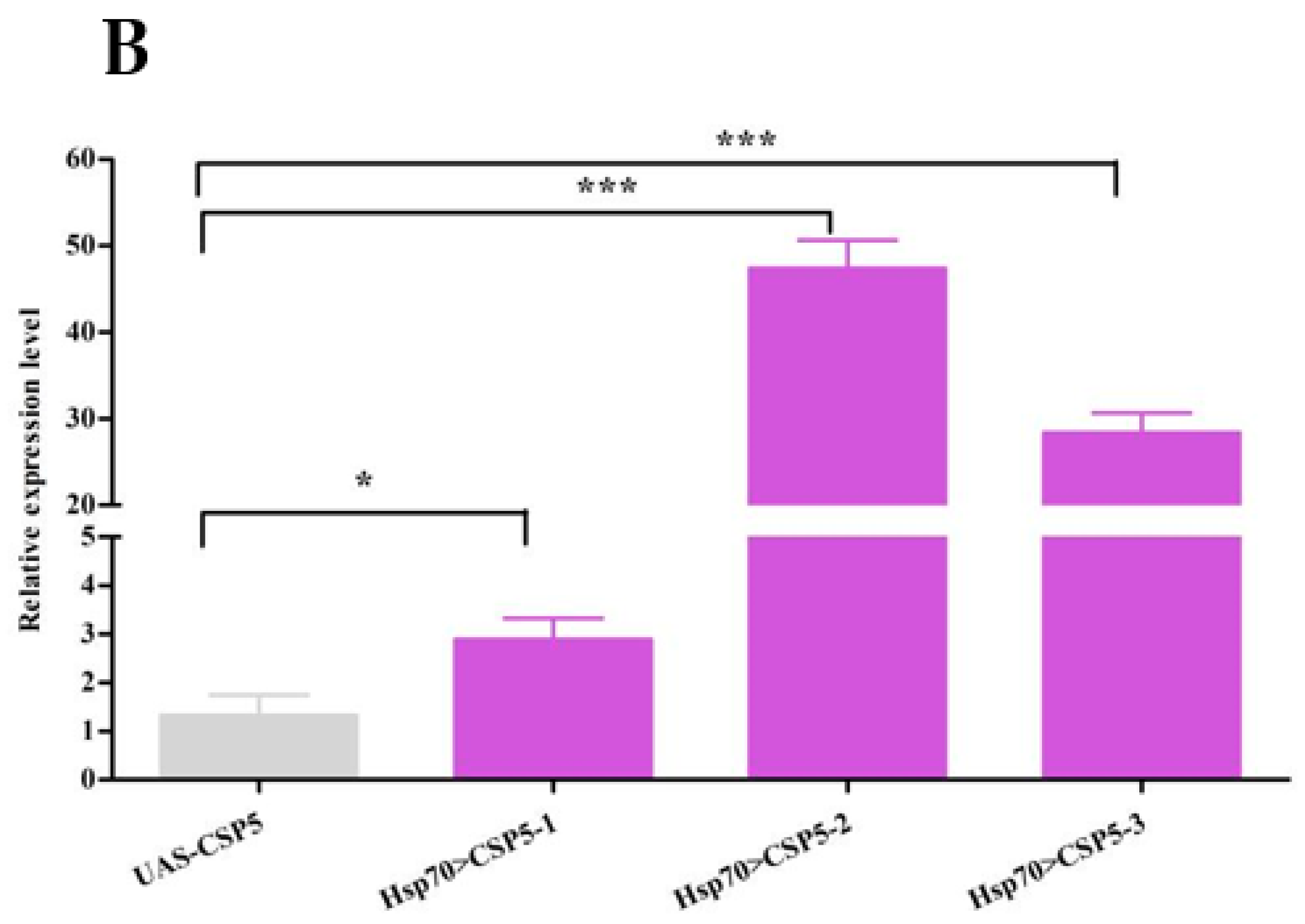
3.3. Expression of AgosCSP5 in Transgenic D. Melanogaster
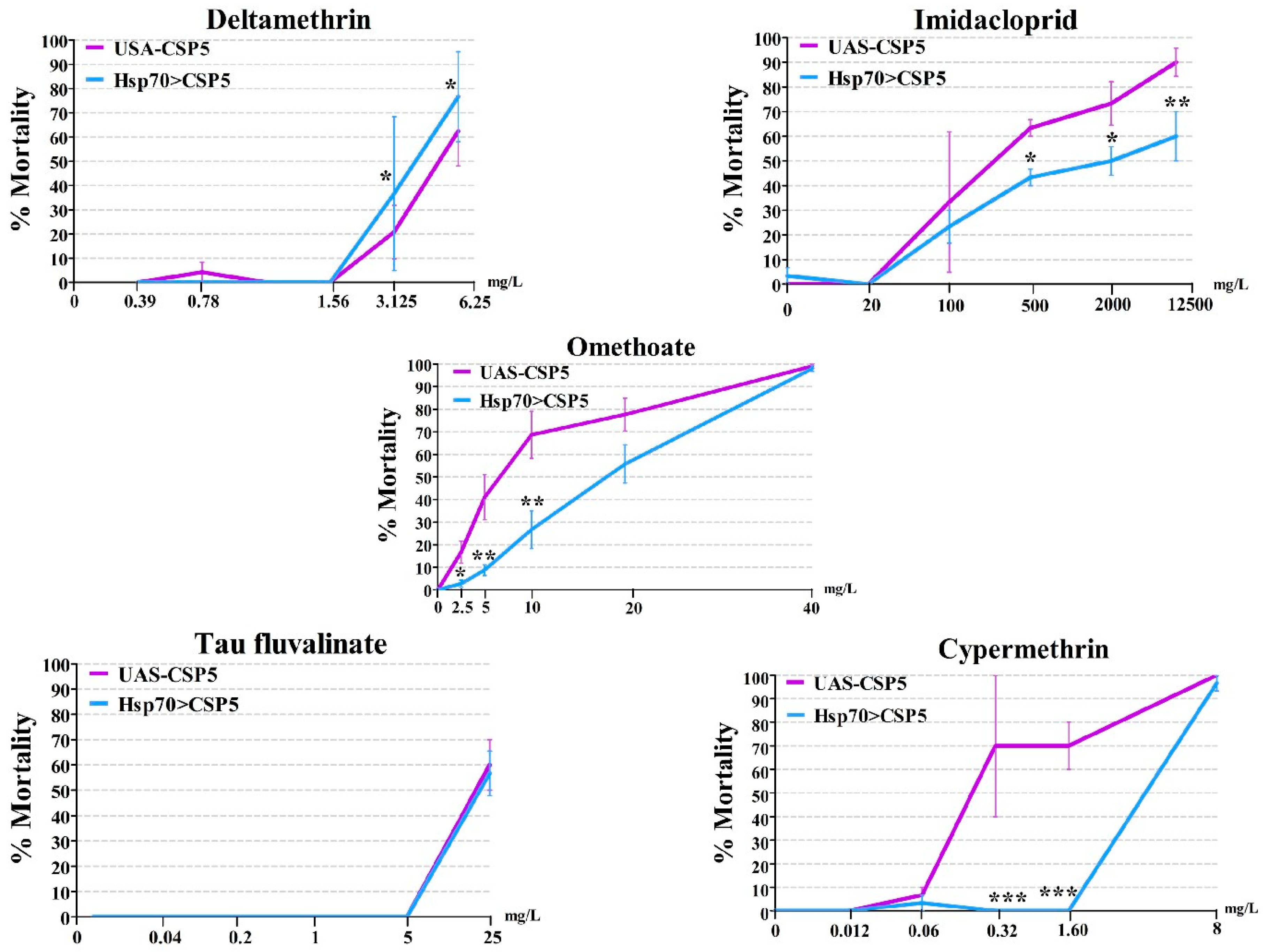
3.4. AgosCSP5 Confers the Resistance to Insecticides in Transgenic Drosophila Flies
3.5. AgosCSP5 Can Bind to Insecticides
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Venthur, H.; Zhou, J.J. Odorant receptors and odorant-binding proteins as insect pest control targets: A comparative analysis. Front. Physiol. 2018, 9, 1163. [Google Scholar] [CrossRef]
- Casida, J.E.; Durkin, K.A. Neuroactive insecticides: Targets, selectivity, resistance, and secondary effects. Annu. Rev. Entomol. 2013, 58, 99–117. [Google Scholar] [CrossRef]
- Zalucki, M.P.; Furlong, M. Behavior as a mechanism of insecticide resistance: Evaluation of the evidence. Curr. Opin. Insect Sci. 2017, 21, 19–25. [Google Scholar] [CrossRef] [Green Version]
- Balabanidou, V.; Grigoraki, L.; Vontas, J. Insect cuticle: A critical determinant of insecticide resistance. Curr. Opin. Insect Sci. 2018, 27, 68–74. [Google Scholar] [CrossRef]
- Ingham, V.A.; Wagstaff, S.; Ranson, H. Transcriptomic meta-signatures identified in Anopheles gambiae populations reveal previously undetected insecticide resistance mechanisms. Nat. Commun. 2018, 9, 5282. [Google Scholar] [CrossRef] [Green Version]
- Ingham, V.A.; Anthousi, A.; Douris, V.; Harding, N.J.; Lycett, G.; Morris, M.; Vontas, J.; Ranson, H. A sensory appendage protein protects malaria vectors from pyrethroids. Nature 2020, 577, 376–380. [Google Scholar] [CrossRef] [PubMed]
- Kaissling, K.E. Kinetics of olfactory responses might largely depend on the odorant-receptor interaction and the odorant deactivation postulated for flux detectors. J. Comp. Physiol. A Neuroethol. Sens. Neural. Behav. Physiol. 2013, 199, 879–896. [Google Scholar] [CrossRef] [PubMed]
- Leal, W.S. Odorant reception in insects: Roles of receptors, binding proteins, and degrading enzymes. Annu. Rev. Entomol. 2013, 58, 373–391. [Google Scholar] [CrossRef]
- Pelosi, P.; Zhou, J.J.; Ban, L.P.; Calvello, M. Soluble proteins in insect chemical communication. Cell Mol. Life Sci. 2006, 63, 1658–1676. [Google Scholar] [CrossRef]
- Vieira, F.G.; Rozas, J. Comparative genomics of the odorant-binding and chemosensory protein gene families across the Arthropoda: Origin and evolutionary history of the chemosensory system. Genome Biol. Evol. 2011, 3, 476–490. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.J. Odorant-binding proteins in insects. Vitam. Horm. 2010, 83, 241–272. [Google Scholar]
- Gu, S.H.; Wu, K.M.; Guo, Y.Y.; Field, M.L.; Pickett, J.A.; Zhang, Y.J.; Zhou, J.J. Identification and expression profiling of odorant binding proteins and chemosensory proteins between two wingless morphs and a winged morph of the cotton aphid Aphis gossypii Glover. PLoS ONE 2013, 8, e73524. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.L.; He, P.; Zhang, L.; Fang, S.Q.; Dong, S.L.; Zhang, Y.J.; Li, F. Large-scale identification of odorant-binding proteins and chemosensory proteins from expressed sequence tags in insects. BMC Genom. 2009, 10, 632. [Google Scholar] [CrossRef] [Green Version]
- Xuan, N.; Guo, X.; Xie, H.Y.; Lou, Q.N.; Lu, X.B.; Liu, G.X.; Picimbon, J.F. Increased expression of CSP and CYP genes in adult silkworm females exposed to avermectins. Insect Sci. 2015, 22, 203–219. [Google Scholar] [CrossRef]
- Bautista, M.A.M.; Bhandary, B.; Wijeratne, A.J.; Andrew, P.M.; Casey, W.H.; Omprakash, M. Evidence for trade-offs in detoxification and chemosensation gene signatures in Plutella xylostella. Pest Manag. Sci. 2015, 71, 423–432. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.X.; Xuan, N.; Chu, D.; Xie, H.Y.; Fan, Z.X.; Bi, Y.P.; Picimbon, J.F.; Qin, Y.C.; Zhong, S.T.; Li, Y.F.; et al. Biotype expression and insecticide response of Bemisia tabaci chemosensory protein-1. Arch. Insect Biochem. Physiol. 2014, 85, 137–151. [Google Scholar] [CrossRef]
- Xiong, W.; Gao, S.S.; Lu, Y.Y.; Wei, L.T.; Mao, J.J.; Xie, J.; Cao, Q.Q.; Liu, J.J.; Bi, J.X.; Song, X.W.; et al. Latrophilin participates in insecticide susceptibility through positively regulating CSP10 and partially compensated by OBPC01 in Tribolium castaneum. Pestic. Biochem. Physiol. 2019, 159, 107–117. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.D.; Mao, Y.W.; Zhang, L. Binding properties of four antennae-expressed chemosensory proteins (CSPs) with insecticides indicates the adaption of Spodoptera litura to environment. Pestic. Biochem. Physiol. 2018, 146, 43–51. [Google Scholar] [CrossRef]
- Lin, X.D.; Jiang, Y.; Zhang, L.; Cai, Y. Effects of insecticides chlorpyrifos, emamectin benzoate and fipronil on Spodoptera litura might be mediated by OBPs and CSPs. Bull. Entomol. Res. 2018, 108, 658–666. [Google Scholar] [CrossRef] [PubMed]
- Li, H.L.; Tan, J.; Song, X.M.; Wu, F.; Tang, M.Z.; Hua, Q.Y.; Zheng, H.Q.; Hu, F.L. Sublethal doses of neonicotinoid imidacloprid can interact with honeybee chemosensory protein 1 (CSP1) and inhibit its function. Biochem. Biophys. Res. Commun. 2017, 486, 391–397. [Google Scholar] [CrossRef]
- Cao, C.W.; Zhang, J.; Gao, X.W.; Liang, P.; Guo, H.L. Overexpression of carboxylesterase gene associated with organophosphorous insecticide resistance in cotton aphids, Aphis gossypii (Glover). Pesti. Biochem. Phys. 2008, 90, 175–180. [Google Scholar] [CrossRef]
- Chen, X.W.; Li, F.; Chen, A.Q.; Ma, K.S.; Liang, P.Z.; Liu, Y.; Song, D.L.; Gao, X.W. Both point mutations and low expression levels of the nicotinic acetylcholine receptor β1 subunit are associated with imidacloprid resistance in an Aphis gossypii (Glover) population from a Bt cotton field in China. Pestic. Biochem. Phys. 2017, 141, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.W.; Tang, C.Y.; Ma, K.S.; Xia, J.; Song, D.L.; Gao, X.W. Overexpression of UDP-glycosyltransferase potentially involved in insecticide resistance in Aphis gossypii Glover collected from Bt cotton fields in China. Pest Manag. Sci. 2019, 76, 1371–1377. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Shang, Q.L.; Fang, K.; Zhang, J.; Xi, J.H. Down-regulated transcriptional level of Ace1 combined with mutations in Ace1 and Ace2 of Aphis gossypii are related with omethoate resistance. Chem. Biol. Interact. 2010, 188, 553–557. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.J.; Vieira, F.G.; He, X.L.; Smadja, C.; Liu, R.; Rozas, J.; Field, L.M. Genome annotation and comparative analyses of the odorant-binding proteins and chemosensory proteins in the pea aphid Acyrthosiphon pisum. Insect Mol. Biol. 2010, 19, 113–122. [Google Scholar] [CrossRef] [Green Version]
- Li, F.; Ma, K.S.; Liang, P.Z.; Chen, X.W.; Liu, Y.; Gao, X.W. Transcriptional responses of detoxification genes to four plant allelochemicals in Aphis gossypii. J. Econ. Entomol. 2017, 110, 624–631. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [Green Version]
- Quan, Q.M.; Hu, X.; Pan, B.H.; Zeng, B.S.; Wu, N.N.; Fang, G.Q.; Cao, Y.H.; Chen, X.Y.; Li, X.; Huang, Y.P.; et al. Draft genome of the cotton aphid Aphis gossypii. Insect Biochem. Mol. Biol. 2019, 105, 25–32. [Google Scholar] [CrossRef]
- Miller, D.F.B.; Holtzman, S.L.; Kaufman, T.C. Customized microinjection glass capillary needles for P-element transformations in Drosophila melanogaster. Biotechniques 2002, 33, 366–372. [Google Scholar] [CrossRef] [Green Version]
- Pfafflfl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative pcr and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, Y. I-TASSER server: New development for protein structure and function predictions. Nucleic Acids Res. 2015, 43, W174–W181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kalé, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef] [Green Version]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Venthur, H.; Machuca, J.; Godoy, R.; Palma-Millanao, R.; Zhou, J.J.; Larama, G.; Bardehle, L.; Quiroz, A.; Ceballos, R.; Mutis, A. Structural investigation of selective binding dynamics for the pheromone-binding protein 1 of the grapevine moth, Lobesia botrana. Arch. Insect Biochem. Physiol. 2019, 101, e21557. [Google Scholar] [CrossRef]
- Li, H.L.; Ni, C.X.; Tan, J.; Zhang, L.Y.; Hu, F.L. Chemosensory proteins of the eastern honeybee, Apis cerana: Identification, tissue distribution and olfactory related functional characterization. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2016, 194–195, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Zhang, Q.; Xu, Q.X.; Xue, W.X.; Han, Z.L.; Sun, J.R.; Chen, J.L. Differential expression analysis of olfactory genes based on a combination of sequencing platforms and behavioral investigations in Aphidius gifuensis. Front Physiol. 2018, 9, 1679. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Wang, Z.W.; Wen, P.; Wei, W.; Chen, Y.; Ai, H.; Sun, W.B. Hydrocarbons mediate seed dispersal: A new mechanism of vespicochory. New Phytol. 2018, 220, 714–725. [Google Scholar] [CrossRef] [PubMed]
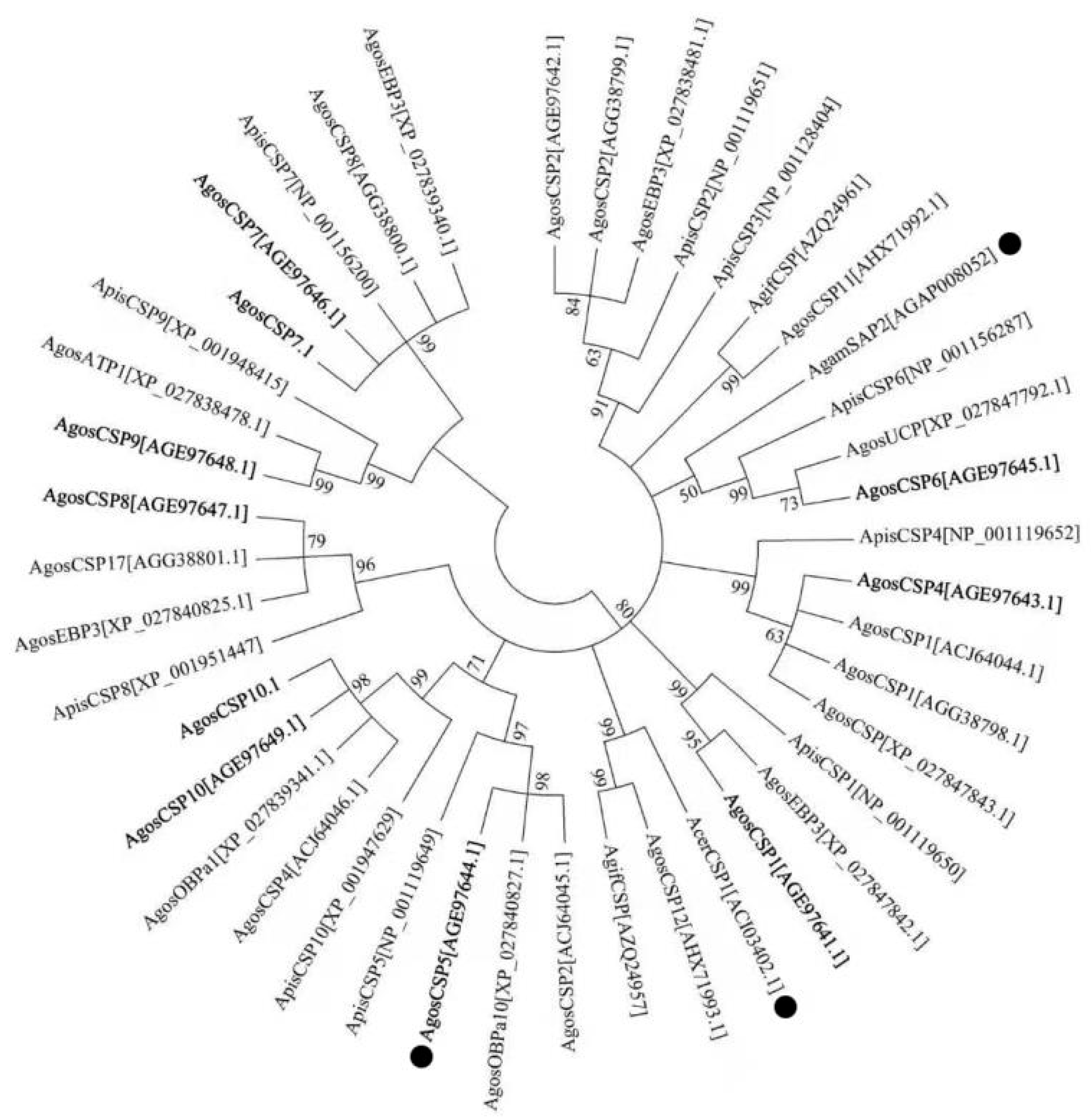


| Name Used in This Study | NCBI ID and Name of Identical Sequence Identified in Different Populations | Genome ID (Extron Position) | Strand | Scaffold ID A | Intron Size (bp) | Distance (bp) between CSPs on Same Scaffold C | |||
|---|---|---|---|---|---|---|---|---|---|
| Gu et al., 2013 [12] | Xu et al., 2009 [13] | Li et al., 2013 B | Quan et al., 2019 [29] | ||||||
| AgosCSP1 | AGE97641.1 CSP1 | n.d. | n.d. | XP_027847842.1 EBP3 | NW_021009645.1 (539802...539531, 533989...533748) | Plus/Minus | AgSCF3568 | 5542 | CSP4-CSP1-CSP6 16157, 16382 |
| AgosCSP2 | AGE97642.1 CSP2 | n.d. | AGG38799.1 CSP2 | XP_027838481.1 EBP3 | NW_021007053.1 (273051...273247, 273952...274160) | Plus/Plus | AgSCF0976 | 705 | CSP2-CSP9 248 |
| AgosCSP3 | n.d. | n.d. | n.d. n.d. | XP_027838481.1 EBP3 n. | NW_021007053.1 (273051...273247, 273952...274160) | Plus/Plus | AgSCF0976 | 705 | CSP2-CSP9 248 |
| AgosCSP4 | AGE97643.1 CSP4 | ACJ64044.1 CSP1 | AGG38798.1 CSP1 | XP_027847843.1 CSP | NW_021009645.1 (524668...524440, 523853...523645) | Plus/Minus | AgSCF3568 | 587 | CSP4-CSP1-CSP6 16157, 16382 |
| AgosCSP5 | AGE97644.1 CSP5 | ACJ64045.1 CSP2 | n.d. | XP_027840827.1 OBPa10 | NW_021007494.1 (888814...889061, 890414...890586) | Plus/Plus | AgSCF1417 | 1353 | CSP8-CSP 13830 |
| AgosCSP6 | AGE97645.1 CSP6 | n.d. | n.d. | XP_027847792.1 UCP | NW_021009645.1 (550130...549931, 548515...548313) | Plus/Minus | AgSCF3568 | 1416 | CSP4-CSP1-CSP6 16157, 16382 |
| AgosCSP7 | AGE97646.1 CSP7 | n.d. | AGG38800.1 CSP8 | XP_027839340.1 EBP3 | NW_021007172.1 (309846...309812, 303813...303527, 302266...302119) | Plus/Minus | AgSCF1095 | 5999, 1261 | CSP10-CSP7 136286 |
| AgosCSP8 | AGE97647.1 CSP8 | n.d. | AGG38801.1 CSP17 | XP_027840825.1 EBP3 | NW_021007494.1 (870401...870707, 874801...874984) | Plus/Plus | AgSCF1417 | 4094 | CSP8-CSP5 13830 |
| AgosCSP9 | AGE97648.1 CSP9 | n.d. | n.d. | XP_027838478.1 ATP1 | NW_021007053.1 (276758...276386, 274555...274408) | Plus/Minus | AgSCF0976 | 1831 | CSP2-CSP9 248 |
| AgosCSP10 | AGE97649.1 CSP10 | ACJ64046.1 CSP4 | n.d. | XP_027839341.1 OBPa10 | NW_021007172.1 (162107...162153, 162322...162542, 165646...165833) | Plus/Plus | AgSCF1095 | 169, 3104 | CSP10-CSP7 136286 |
| Na et al., 2014 B | Li et al., 2016 [37] | Fan et al., 2018 [38] | |||||||
| AgosCSP11 | AHX71992.1 CSP3 | n.d. | AZQ24961 AgifCSP | n.d. | n.d. | ||||
| AgosCSP12 | AHX71993.1 CSP4 | ACI03402.1 AcerCSP1 | AZQ24957 AgifCSP | n.d. | n.d. | ||||
| Strains | Number a | Slope ± SEM | LC50 (mg(a.i.)/L) (95% CL b) | Chi-Square (Df) c | Resistance Ratio d |
|---|---|---|---|---|---|
| Hsp70–GAL4 > UAS | 960 | 2.18 ± 0.15 | 5.76 (4.24–7.42) | 264.63 (50) | 1 |
| Hsp70–GAL4 > UAS–CSP5 | 1005 | 2.83 ± 0.17 | 15.02 (12.74–18.06) | 164.30 (50) | 2.61 |
| Ligands | Free Binding Energy (kJ mol−1) |
|---|---|
| Omethoate | −20.1 |
| Fluvalinate | −13.0 |
| Imidacloprid | −22.6 |
| Cypermethrin | −17.2 |
| Deltamethrin | −18.8 |
| Caryophyllene | −16.3 |
| (E)-β-farnesene | −21.3 |
| (Z)-3-hexenyl acetate | −18.4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, F.; Venthur, H.; Wang, S.; Homem, R.A.; Zhou, J.-J. Evidence for the Involvement of the Chemosensory Protein AgosCSP5 in Resistance to Insecticides in the Cotton Aphid, Aphis gossypii. Insects 2021, 12, 335. https://doi.org/10.3390/insects12040335
Li F, Venthur H, Wang S, Homem RA, Zhou J-J. Evidence for the Involvement of the Chemosensory Protein AgosCSP5 in Resistance to Insecticides in the Cotton Aphid, Aphis gossypii. Insects. 2021; 12(4):335. https://doi.org/10.3390/insects12040335
Chicago/Turabian StyleLi, Fen, Herbert Venthur, Shang Wang, Rafael A. Homem, and Jing-Jiang Zhou. 2021. "Evidence for the Involvement of the Chemosensory Protein AgosCSP5 in Resistance to Insecticides in the Cotton Aphid, Aphis gossypii" Insects 12, no. 4: 335. https://doi.org/10.3390/insects12040335







