Simple Summary
Atkinsoniella is a large genus of almost 99 species across the world within the subfamily Cicadellinae, which is a large subfamily, comprising more than 2400 species of approximately 330 genera. Some of the Cicadellinae distributed worldwide are known as important agricultural pests. To better understand the mitogenomic characteristics of the genus Atkinsoniella and reveal phylogenetic relationships, the complete mitochondrial genomes of Atkinsoniella grahami and Atkinsoniella xanthonota were sequenced and comparatively analyzed in this study. The mitogenomes of these two Atkinsoniella species were found to be highly conserved, similarly to other Cicadellidae, except for the secondary structure of trnaS1, which formed a loop with the dihydrouridine (DHC) arm. This phenomenon has also been observed in other insect mitogenomes. Phylogenetic analyses, based on mitogenomes using both the maximum likelihood (ML) and Bayesian inference (BI) methods of three datasets, supported the monophyly of Cicadellinae, as well as the other subfamilies, and produced a well-resolved framework of Cicadellidae and valuable data for the phylogenetic study of Cicadellinae.
Abstract
The complete mitochondrial genomes of Atkinsoniella grahami and Atkinsoniella xanthonota were sequenced. The results showed that the mitogenomes of these two species are 15,621 and 15,895 bp in length, with A+T contents of 78.6% and 78.4%, respectively. Both mitogenomes contain 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), 2 ribosomal RNA genes (rRNAs), and a control region (CR). For all PCGs, a standard start ATN codon (ATT, ATG, or ATA) was found at the initiation site, except for ATP8, for which translation is initiated with a TTG codon. All PCGs terminate with a complete TAA or TAG stop codon, except for COX2, which terminates with an incomplete stop codon T. All tRNAs have the typical cloverleaf secondary structure, except for trnS, which has a reduced dihydrouridine arm. Furthermore, these phylogenetic analyses were reconstructed based on 13 PCGs and two rRNA genes of 73 mitochondrial genome sequences, with both the maximum likelihood (ML) and Bayesian inference (BI) methods. The obtained mitogenome sequences in this study will promote research into the classification, population genetics, and evolution of Cicadellinae insects in the future.
1. Introduction
The leafhopper subfamily Cicadellinae is distributed worldwide and contains around 2400 species, represented by approximately 330 genera [1]. In China, 26 genera and 315 species of Cicadellinae have been recorded [2]. Some Cicadellinae insects are of considerable economic importance as they feed on sap in the xylem of woody and herbaceous plants and, via this process, are able to transmit phytopathogenic bacterium and plant viruses to crops, ornamental plants, and weeds [3,4,5,6]. Accurate identification of insects is extremely important for pest control. However, the taxonomic status of some species, based on morphology, remains controversial. Thus, molecular data have been considered as a useful adjunct to the identification and phylogenetic analysis of insects.
The mitochondrial genomes of insects are typically 14.5–17 kb circular double-stranded molecules that contain 37 genes, including 13 protein-coding genes (PCGs), 2 ribosomal RNA genes (rRNAs), 22 transfer RNA genes (tRNAs), and a control region that contains the initial sites for replication and transcription [7,8,9]. With the rapid development of next-generation sequencing technology, the cost of sequencing has been drastically reduced. Mitochondrial genomes are widely used for studying the evolutionary genomics and phylogenetic relationships of different taxonomic levels in insects due to their high genome copy numbers, simple genetic structure [7], maternal inheritance [10], conserved gene components [11], and relatively high evolutionary rate [12]. To date, more than 110 complete or near-complete mitogenomes of Cicadellidae species have been published, most of which have been widely used for evolutionary studies. However, the mitogenomes of only six species of the subfamily Cicadellinae (Bothrogonia ferruginea, B. qiongana, Cicadella viridis, Cofana yasumatsui, Cuerna sp., and Homalodisca vitripennis) (Table 1) have been sequenced and annotated, while none of them belong to the genus Atkinsoniella.

Table 1.
Mitochondrial genomes used for the phylogenetic analyses in this study.
Atkinsoniella is a large genus within the subfamily Cicadellinae, established with A. decisa Distant 1908 as its type species, and comprising almost 99 valid species worldwide, with 88 species occurring in China [1,2]. Atkinsoniella is mainly distributed in the Oriental realm, with a few in the Palearctic realm. Most of them are polyphagous and often feed on weeds and trees, and they typically live in the damp environment of mountain forests. Their bodies are mostly black with light spots, or light with black, red, yellow, or orange markings. Additionally, they have sexual dimorphism and polymorphism, which makes morphological identification difficult. A. grahami Young, 1986 (syn. A. nigroscuta Zhang and Kuoh, 1993 [47] and A. furcula Yang and Li, 2002 [48]) are widely distributed species in China that have been recorded in several Chinese provinces, including Yunnan, Sichuan, Henan, Shaanxi, Gansu, Hubei, Hunan, Guangdong, Hainan, Chongqing, and Guizhou. Meanwhile, to date, A. xanthonota has only been reported in Yunnan province in China [2].
To date, no mitogenome of Atkinsoniella has been sequenced. This lack of mitogenomic data has limited the understanding of the evolution of Cicadellinae at the genomic level. Therefore, the mitogenomes of A. grahami and A. xanthonota were sequenced and analyzed to help us understand the mitogenomic structures and phylogenetic relationships within this group. Hopefully, this study will be valuable for the taxonomy and phylogeny of Cicadellidae insects in the future.
2. Materials and Methods
2.1. Sample Collection and Mitogenome Sequencing
The specimens of A. grahami used in this study were collected from the Tangjiahe National Natural Reserve (E: 104°45′48″, N: 32°35′11.8428″, H: 1726 m), Sichuan Province, China on 26 July 2017 (collector: Hong-Li He). The A. xanthonota specimens were collected from Jinping County (E: 103°13′48″, N: 22°46′55″, H: 1809 m), Yunnan Province, China on 18 July 2018 (collector: Jia-Jia Wang). All of the fresh specimens were immediately preserved in 100% ethanol and stored at −20 °C in the laboratory before DNA extraction. The identification of specimens was based on morphological characteristics, especially the male genitalia, described by Yang et al. [2]. Total genomic DNA was extracted from tissues of the head and thorax muscle of a single adult specimen using a DNeasy® Tissue Kit (Qiagen, Hilden, Germany), according to the manufacturer’s protocol. Voucher DNA (stored at −20 °C) and external genitalia (preserved in glycerol) were deposited at the Institute of Entomology, Guizhou University, Guiyang, China (GUGC). The total genomic DNA of A. grahami and A. xanthonota were used for library preparation and next-generation sequencing (Illumina NovaSeq6000 platform, Berry Genomic, Beijing, China) with a paired-end 150 sequencing strategy. The clean, next-generation sequencing results were assembled using NOVOPlasty 2.7.2 [49] based on the mitochondrial COX1 gene fragment (MG397188, submitted by Dewaard) downloaded from GenBank as a starting reference.
2.2. Sequence Annotation and Analysis
The initial annotation of the mitogenomes was carried out using Mitoz 2.4-alpha [50], with the invertebrate mitochondrial genetic codes. The locations and secondary structures of the tRNA genes were reconfirmed and predicted using the MITOS web server (http://mitos.bioinf.uni-leipzig.de/index.py, accessed on 6 October 2020) [51] and the tRNAscan-SE search server (http://lowelab.ucsc.edu/tRNAscan-SE/, accessed on 6 October 2020) [52,53]. The start codon, stop codon, and length of the 13 PCGs were manually checked and adjusted by comparison with the published Cicadellinae mitogenome sequences B. ferruginea (KU167550) and H. vitripennis (NC_006899). Open reading frames (ORFs) were also confirmed based on the invertebrate mitochondrial genetic code. The two rRNA genes were assumed to extend to the boundaries of the locations of adjacent tRNA genes (trnL1 and trnV), and then compared with the homologous rRNA genes of other published Cicadellidae species to define the right boundary of s-rRNA abuts to the control region. The circular mitogenome maps were visualized using OGDRAW (https://chlorobox.mpimp-golm.mpg.de/OGDraw.html, accessed on 9 October 2020) [54]. Strand asymmetry was calculated according to the formulas: AT skew = [A − T] / [A + T] and GC skew = [G − C] / [G + C] [55]. The nucleotide composition statistics and relative synonymous codon usage (RSCU) values of each PCG were computed with MEGA 6.0 [56]. Tandem repeats in the control region were identified using the Tandem Repeats Finder program (http://tandem.bu.edu/trf/trf.html, accessed on 23 October 2020) [57]. The newly sequenced mitogenome sequences of A. grahami and A. xanthonota were submitted to GenBank with the accession numbers MW533712 and MW533713, respectively.
2.3. Phylogenetic Analyses
In the phylogenetic analyses, 65 mitogenomes of 64 available leafhopper species (including two newly sequenced species), representing 11 subfamilies of the family Cicadellidae (Deltocephalinae, Iassinae, Coelidiinae, Macropsinae, Megophthalminae, Idiocerinae, Evacanthinae, Cicadellinae, Typhlocybinae, Ledrinae, and Mileewinae) and six treehopper species of Aetalioninae, Centrotinae, and Smiliinae were selected as the ingroup. In addition, Callitettix braconoides (NC_025497) and Magicicada tredecim (NC_041652) from the respective families Cercopidae and Cicadidae were used as the outgroup (Table 1). Three datasets were concatenated for phylogenetic analysis: (1) AA: amino acid sequences of the PCGs; (2) PCG12: first and second codon positions of the PCGs; (3) PCG12RNA: the first and the second codon positions of the PCGs and two rRNA genes. Since some rRNA genes of the mitogenome sequences were unavailable, the number of species used for the PCG12RNA dataset was different to that of the PCG12 and AA datasets.
The alignments of 13 PCGs (without stop codons) were aligned using the MASCE [58] algorithm in PhyloSuite 1.2.1 [59], with the invertebrate mitochondrial genetic code. The rRNA genes were aligned with MAFFT 7.313 [60] using the Q-INS-I strategy; gaps and ambiguous sites were removed using the Gblocks 0.91b [61,62] algorithm in PhyloSuite 1.2.1 and default settings, with the exception of AA gap positions, which were toggled to “none”. Then, the alignments of each individual gene were concatenated as different datasets using Geneious prime 2020.2.4.
The best schemes for the partition and substitution models were determined in PartitionFinder v.2.1.1 with the corrected Akaike information criterion (AICc) and greedy search algorithm [63]. The starting partitions used to initiate the PartitionFinder analysis are listed in Table S1. Maximum likelihood (ML) analysis was conducted using IQ-TREE v.1.6.8 [64]. Branch support was estimated with 10,000 replicates of ultrafast bootstrap. Bayesian inference (BI) analysis was performed on MrBayes 3: Bayesian phylogenetic inference under mixed [65] with the default settings, by simulating four independent runs for 100 million generations with sampling every 1000 generations, the initial 25% of samples were discarded as burn-in.
3. Results and Discussion
3.1. Mitogenome Organization and Nucleotide Composition
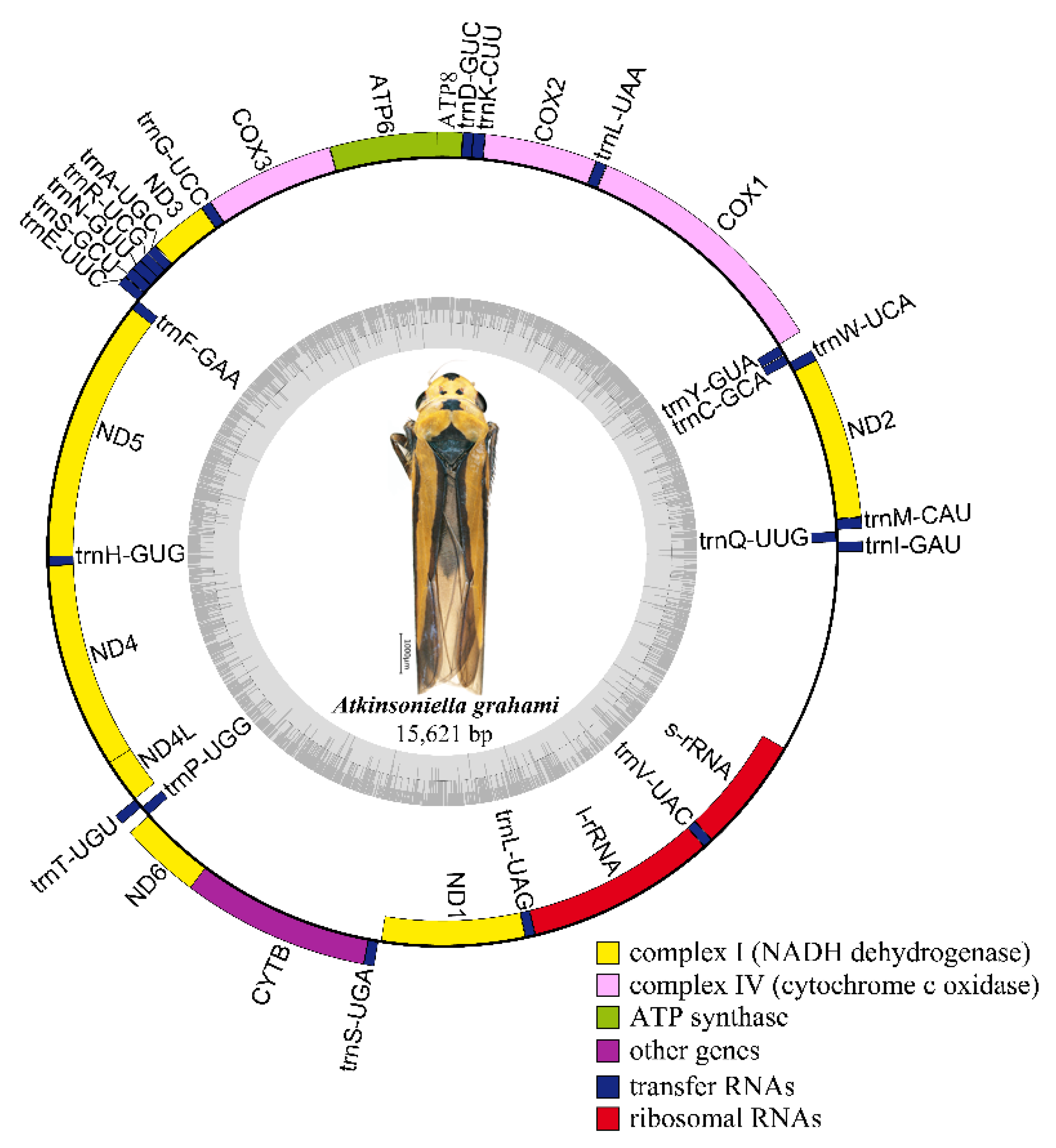
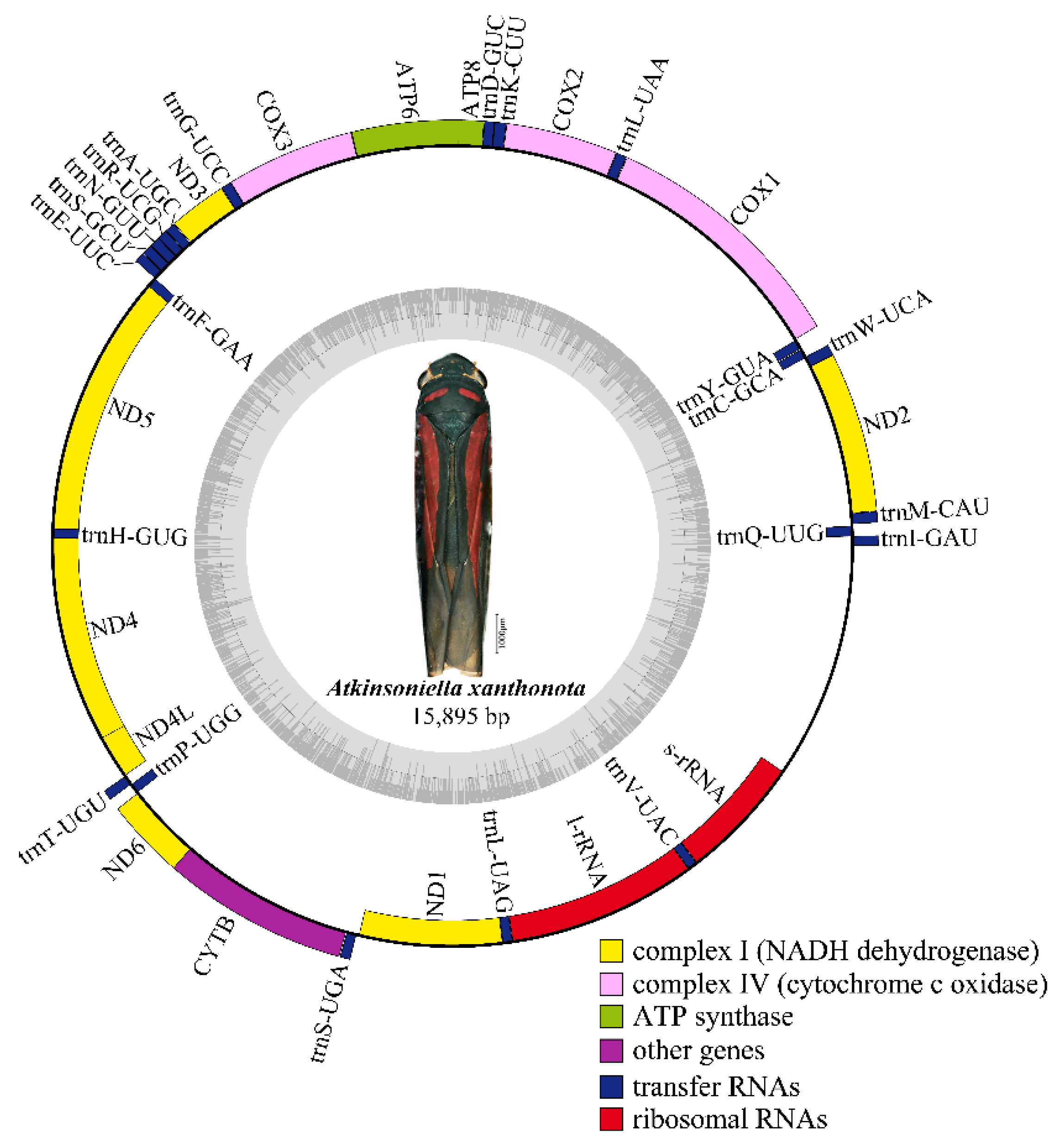
Nine complete or partial mitogenomes were analyzed: the two mitogenomes newly sequenced in this study and another seven mitogenomes downloaded from GenBank without any revision of annotations. The length of the entire mitogenome sequences ranged from 12,696 to 15,895 bp, and contained 37 genes (13 PCGs, 22 tRNA genes, and 2 rRNA genes) and a control region. The Cuerna sp. (KX437741) and Cicadella viridis (KY752061) mitogenomes were incomplete. The A. grahami and A. xanthonota mitogenomes are closed circular molecules 15,621 and 15,895 bp in length, respectively (Figure 1 and Figure 2). The variation in mitogenome size among the different Cicadellinae insects is mainly due to the variable number of repeats in the control region. These nine Cicadellinae mitogenome sequences showed identical gene orders, with the typical gene arrangement of insects. A total of 14 genes: four PCGs (ND5, ND4, ND4L, and ND1), eight tRNAs (trnQ, trnC, trnY, trnF, trnH, trnT, trnP, and trnL), and two rRNAs (l-rRNA and s–rRNA), were encoded on the minority strand (N-strand), while the other 23 genes (nine PCGs and 14 tRNAs) were encoded on the majority strand (J-strand) (Table 2 and Table 3).
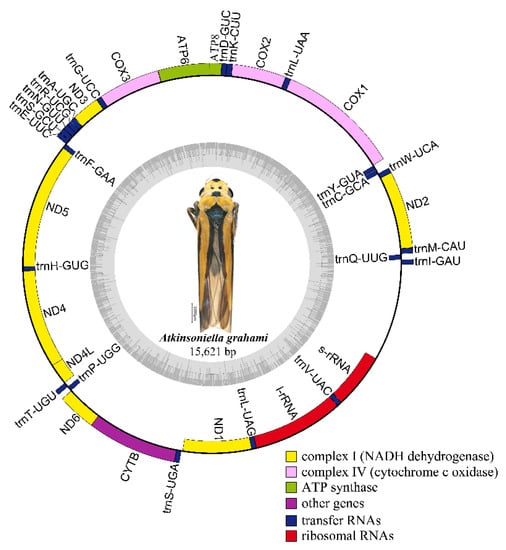
Figure 1.
Circular map of the mitochondrial genome of Atkinsoniella grahami. Genes are represented by different color blocks. Color blocks outside the circle indicate that the genes are on the majority strand (J-strand); those within the circle indicate the genes are located on the minority strand (N-strand).
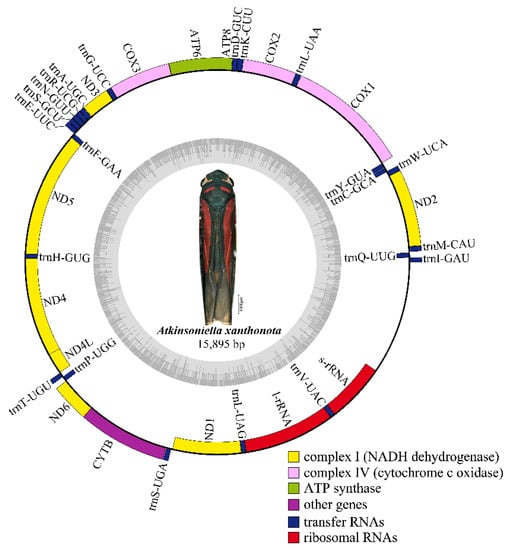
Figure 2.
Circular map of the mitochondrial genome of Atkinsoniella xanthonota. Genes are represented with different color blocks. Color blocks outside the circle indicate the genes are on the majority strand (J-strand); those within the circle indicate the genes are located on the minority strand (N-strand).

Table 2.
Annotations for the Atkinsoniella grahami mitochondrial genomes.

Table 3.
Annotations for the Atkinsoniella xanthonota mitochondrial genomes.
The overall nucleotide composition of A. grahami was determined as A: 41.9%, T: 36.6%, C: 11.6%, and G: 9.9%, while it was A: 41.8%, T: 36.7%, C: 11.7%, and G: 9.9% in A. xanthonota. Similar to the other Cicadellinae mitogenomes, these two mitogenomes were both consistently AT nucleotide biased, with 78.6% in A. grahami and 78.4% in A. xanthonota. The A+T content of the rRNAs was the highest (82.1% in A. grahami and 81.9% in A. xanthonota), while the A+T content of the PCGs was the lowest (77.5% in A. grahami and 77.4% in A. xanthonota) (Table 4). The AT skew was 0.068 for A. grahami and 0.065 for A. xanthonota, indicating a slightly higher occurrence of A compared to T nucleotides. Similar results were also observed for the entire mitogenomes of the seven other Cicadellinae, where all of the AT skews were positive and all of the GC skews were negative (Table 5).

Table 4.
Nucleotide composition of the Atkinsoniella grahami and Atkinsoniella xanthonota mitogenomes.

Table 5.
Nucleotide composition of the Cicadellinae mitochondrial genomes of Atkinsoniella grahami, Atkinsoniella xanthonota, Bothrogonia ferruginea, Bothrogonia qiongana, Cicadella viridis, Cofana yasumatsui, Cuerna sp., and Homalodisca vitripennis.
3.2. Overlapping and Intergenic Spacer Regions
The mitogenomes of A. grahami and A. xanthonota were found to have a total of 66 bp (sum of 15 locations) and 64 bp (sum of 14 locations) overlaps between genes, respectively. The longest observed overlaps were both 20 bp, located between the trnF and ND5 genes. The total length of the intergenic spacers was 38 bp for A. grahami and 41 bp for A. xanthonota. All of the intergenic spacers of the two mitogenomes range from 1 to 15 bp. The longest intergenic spacer situated between trnS and ND1 was found in both the A. grahami and A. xanthonota mitogenomes. There is a 2 bp overlap in A. grahami and a 3 bp intergenic spacer region in A. xanthonota between CYTB and trnS. Aside from this difference, all of the overlapping and intergenic spacers were identical.
3.3. Protein-Coding Genes and Codon Usage
The 13 PCGs of A. grahami encode 3645 amino acids with a total length of 10,935 bp (excluding the stop codons), and those of A. xanthonota encode 3643 amino acids with a total length of 10,929 bp (excluding the stop codons). The A+T contents of the 13 PCGs were 77.5% and 77.4% in the in A. grahami and A. xanthonota mitogenomes, respectively. Moreover, the A+T content of the third codon position was higher than that of the first and second codon positions in these newly sequenced mitogenomes. The 13 PCGs in A. grahami and A. xanthonota both showed a negative AT skew (−0.150 and −0.152, respectively) and a positive GC skew (0.012 and 0.013, respectively). Nine PCGs were encoded on the J-strand and four PCGs (ND5, ND4, ND4L, and ND1) were encoded on the N-strand.
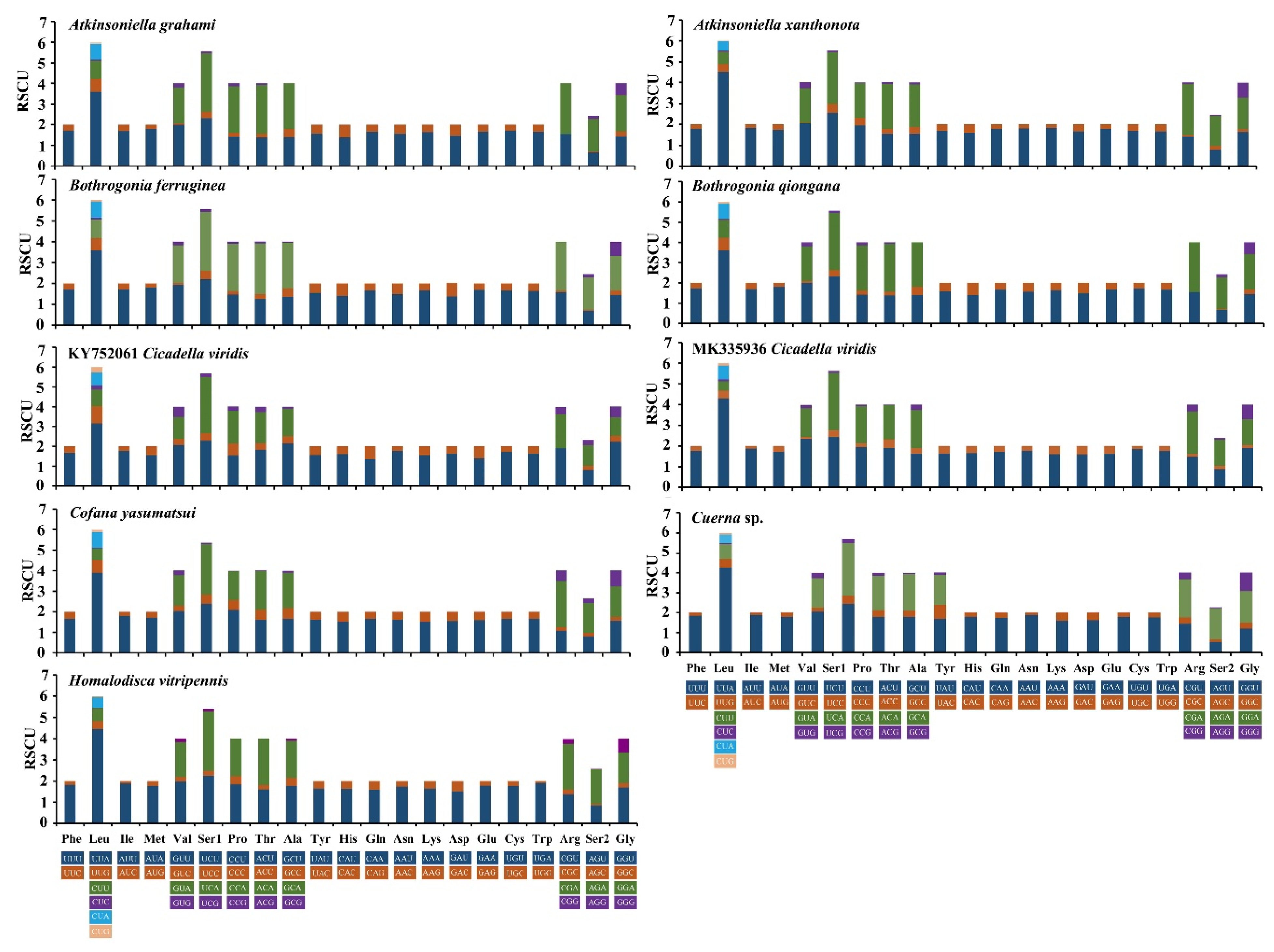
These two newly sequenced Atkinsoniella mitogenomes exhibited similar start and stop codons. Translation of all PCGs is initiated with typical ATN codons (ATT, ATG, or ATA), except for ATP8, which is initiated with a TTG codon (Table 2 and Table 3). While 12 PCGs terminated with the complete stop codon TAA/TAG, the truncated stop codon T was also detected in the COX2 gene in the two Atkinsoniella mitogenomes. Such an incomplete stop codon is commonly found in insect mitogenomes and is presumed to be generated by the polyadenylation process [9]. Meanwhile, the stop codons of ATP8 and CYTB differed between these two species. A. grahami used TAA and TAG as the stop codons for ATP8 and CYTB, respectively, while A. xanthonota used TAG and TAA as the stop codons of ATP8 and CYTB, respectively. In the seven other sequenced Cicadellinae mitogenomes that were examined, similar start and stop codons were found, except for the incomplete stop codon observed in COX1 of Homalodisca vitripennis, ND3 and CYTB of B. ferruginea, and COX3 of Cicadella viridis (Table S2). In these nine mitogenomes, the termination codon TAA occurred more frequently than TAG. Meanwhile, all COX2 ended with incomplete T or TA. The relative synonymous codon usage (RSCU) of nine sequenced Cicadellinae mitogenomes (of eight species) was calculated and drawn (Figure 3). The results show that the nine mitogenomes share the same codon families and similar RSCU features. The four most frequently used codons observed for these nine mitogenomes are UUU-Phe, UUA-Leu, AUU-Ile, and AUA-Met, which were composed of A and U.
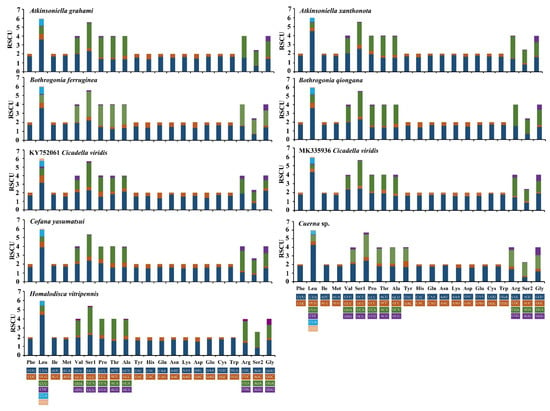
Figure 3.
Relative synonymous codon usage (RSCU) in protein-coding genes (PCGs) of the mitogenomes of Atkinsoniella grahami, Atkinsoniella xanthonota, and other six Cicadellinae species. Codon families are indicated in boxes below the x-axis; the colors correspond to the stacked columns, and frequency of RSCU is provided on the y-axis.
3.4. Transfer and Ribosomal RNA Genes
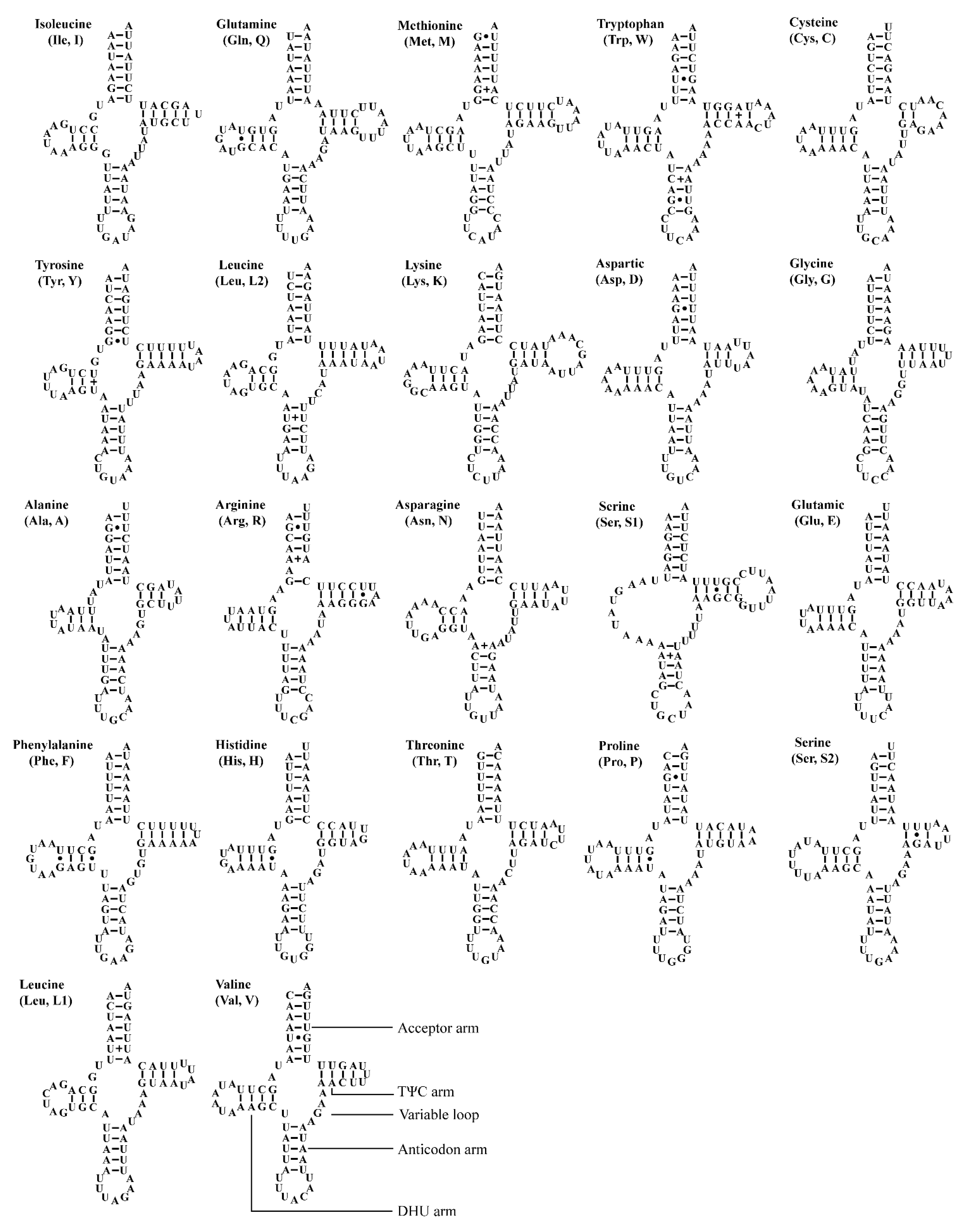
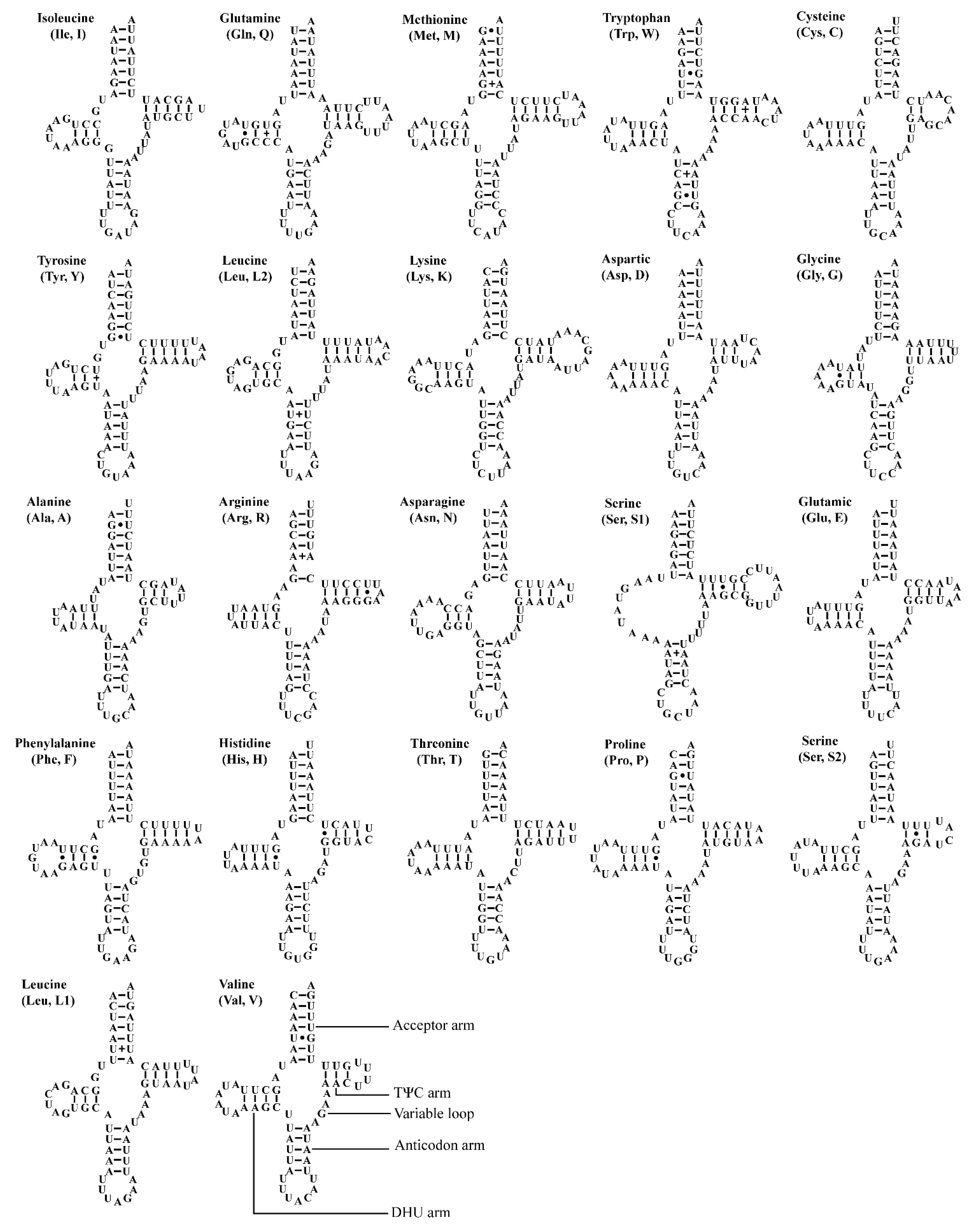
The two Atkinsoniella mitogenomes both contain the typical number of 22 tRNAs, eight of which were encoded by N-strand and 14 encoded by J-strand, ranging from 61 bp (trnA, trnH) to 71 bp (trnK) in length (Table 2 and Table 3). The total length of tRNA sequence was 1426 and 1423 bp, accounting for 9.13% and 8.95% of the whole mitogenome in A. grahami and A. xanthonota, respectively. All tRNAs of these two mitogenomes indicated a positive AT skew (0.022 and 0.018, respectively) and a positive GC skew (0.152 and 0.137, respectively). Most tRNAs exhibited typical cloverleaf secondary structures, except for trnS1 (GCU), which lacks a dihydrouridine (DHU) arm, and instead being replaced by a simple loop. Moreover, there were some kinds of unmatched base pairs (Figure 4 and Figure 5). These two structural characteristics are also present in other leafhoppers [40,48,49,66,67,68,69]. In the predicted secondary structure, the length of the anticodon loop of all tRNAs was highly conserved for 7 bp, compared to the variable sizes of the DHU and TΨC loops.
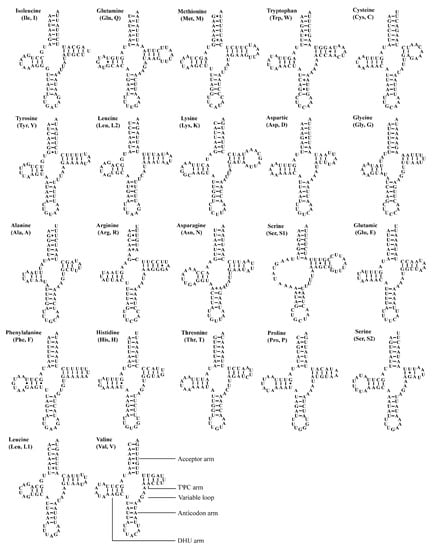
Figure 4.
Predicted secondary cloverleaf structure for the tRNAs of Atkinsoniella grahami. The tRNA arms are illustrated as for trnV. Dashes (–), solid dots (•), and pluses (+) indicate the Watson–Crick base pairings, G–U bonds, and mismatches, respectively.
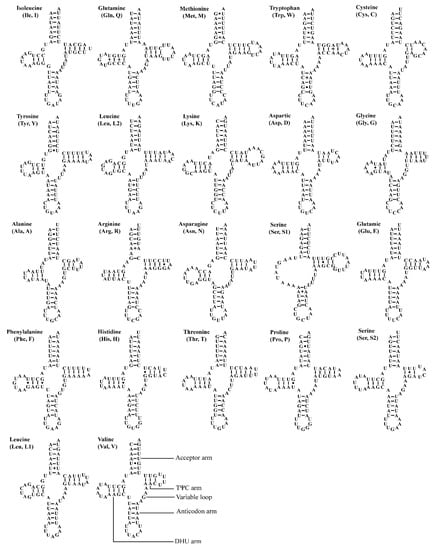
Figure 5.
Predicted secondary cloverleaf structure for the tRNAs of Atkinsoniella xanthonota. The tRNA arms are illustrated as for trnV. Dashes (–), solid dots (•), and pluses (+) indicate the Watson–Crick base pairings, G–U bonds, and mismatches, respectively.
Two rRNA genes (l-rRNA and s-rRNA) were recognized in the two newly sequenced mitogenomes. l-rRNA was located between the trnL1 and trnV. The s-rRNA was located between trnV and the control region. The length of two rRNA genes were 1215 bp (l-rRNA) and 740 bp (s-rRNA) in A. grahami, and 1217 bp (l-rRNA) and 798 bp (s-rRNA) in A. xanthonota. The A+T content region of the rRNAs in Cicadellinae mitogenomes ranged from 78.3% to 82.1% (Table 5). The A+T content reached 82.1% in A. grahami and 81.9% in A. xanthonota. Additionally, the two rRNAs in these two mitogenomes showed a negative AT skew (−0.119 in A. grahami and −0.117 in A. xanthonota) and a positive GC skew (0.234 in A. grahami and 0.231 in A. xanthonota).
3.5. Control Region
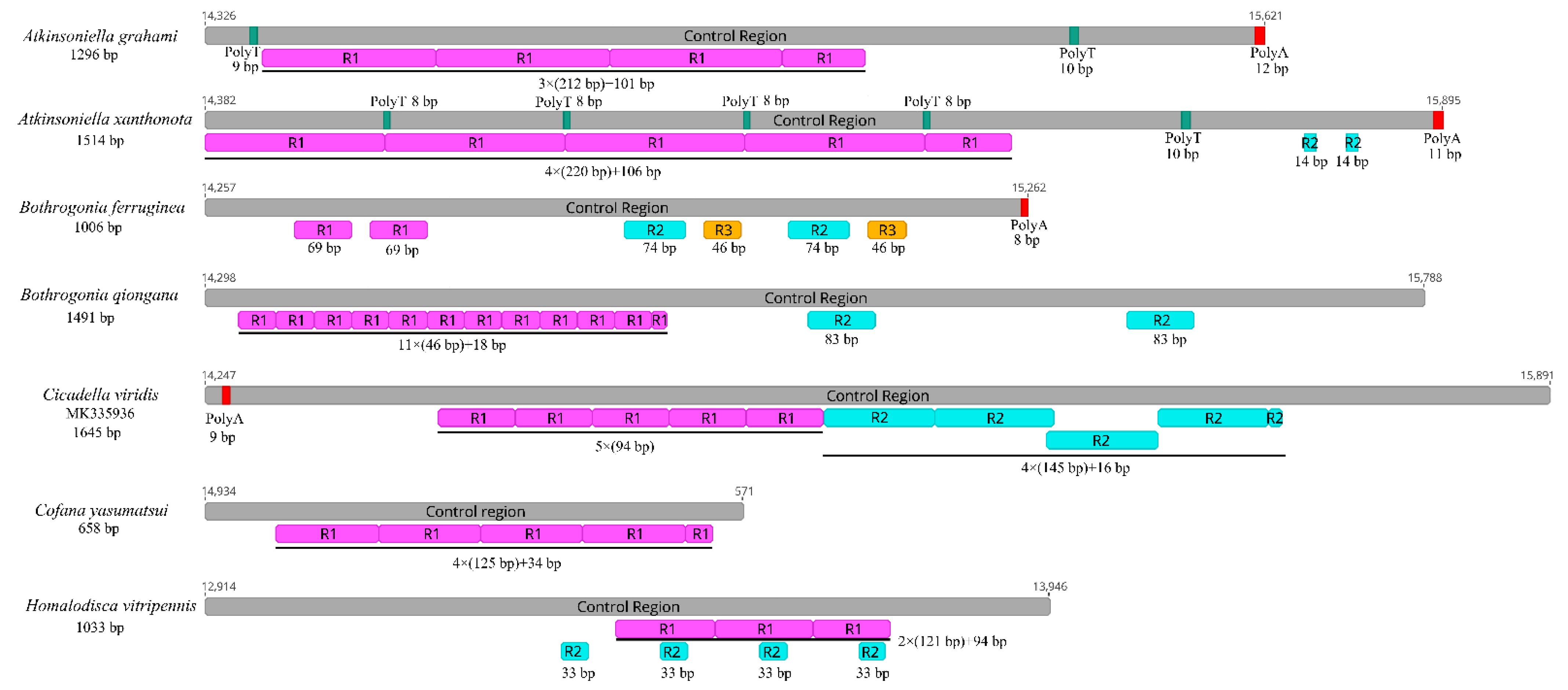
The control region is the longest non-coding region, plays an indispensable role in the study of molecular evolution, and contains regulatory functions for replication and transcription [7,8,70]. The control regions in the Cicadellinae mitogenomes were not highly conserved and were located between the s-rRNA and trnI genes, and ranged from 658 to 1645 bp in size (Figure 6). The length of the control regions in the two Atkinsoniella mitogenomes was 1296 bp in A. grahami and 1514 bp in A. xanthonota (Figure 1, Table 2 and Table 3). The A+T content was 80.4% and 79.9%, respectively. The AT and GC skews were both positive, with values of 0.052 and 0.071 in A. grahami, and 0.029 and 0.118 in A. xanthonota, indicating that A and G were more abundant than T and C. The Cicadellinae mitogenomes had one to three types of tandem repeat units, ranging from 14 to 220 bp. One tandem repeat (212 bp) was found in the control region of the A. grahami mitogenome, and two tandem repeats (220 and 14 bp) were found in the A. xanthonota mitogenome (Figure 6).
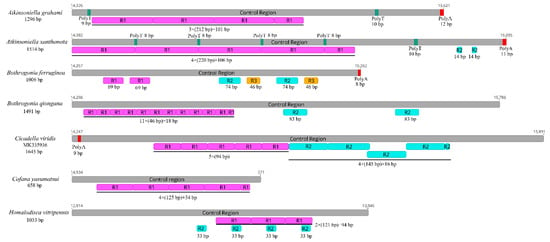
Figure 6.
Organization of the control regions in the mitochondrial genomes of Atkinsoniella graham, Atkinsoniella xanthonota, and the five other Cicadellinae species. The gray blocks refer to the control region. The numbers located at the start and end of gray blocks refer to the location of control region in each mitogenome. R refers to repeat unit, with the number indicating the number of repeats. The red and green blocks refer to the structures of poly (A) and poly (T), respectively.
Furthermore, poly-A regions were found at the end of the control region in A. grahami, A. xanthonota, and B. ferruginea. This has also been observed in other insect mitogenomes [71,72,73,74,75]. Two and five poly-T regions were also found in A. grahami and A. xanthonota, respectively. The results indicate that the length, nucleotide sequences, and copy numbers of the tandem repeat units in the control region were variable among known Cicadellinae mitogenomes.
3.6. Phylogenetic Analyses
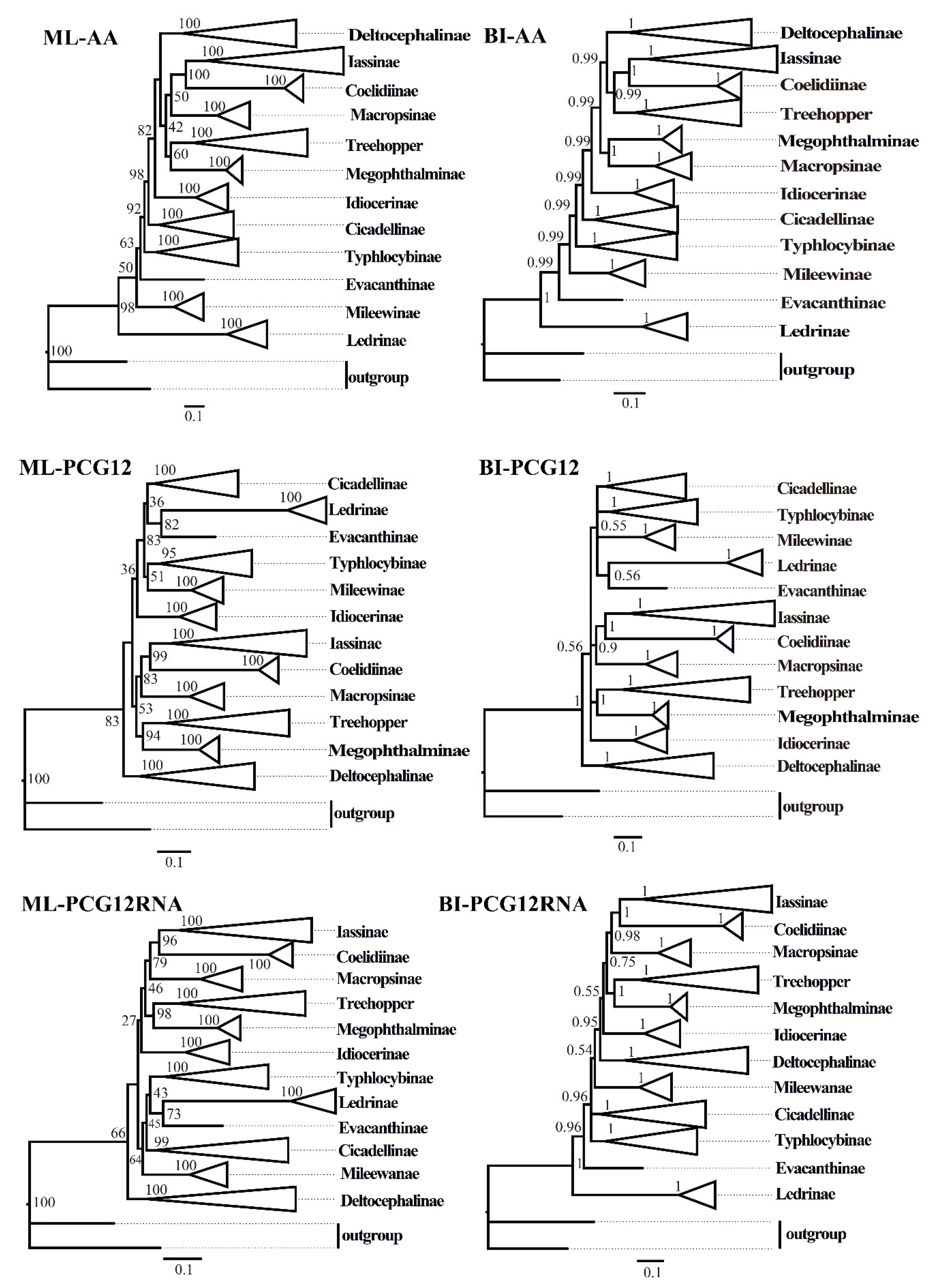
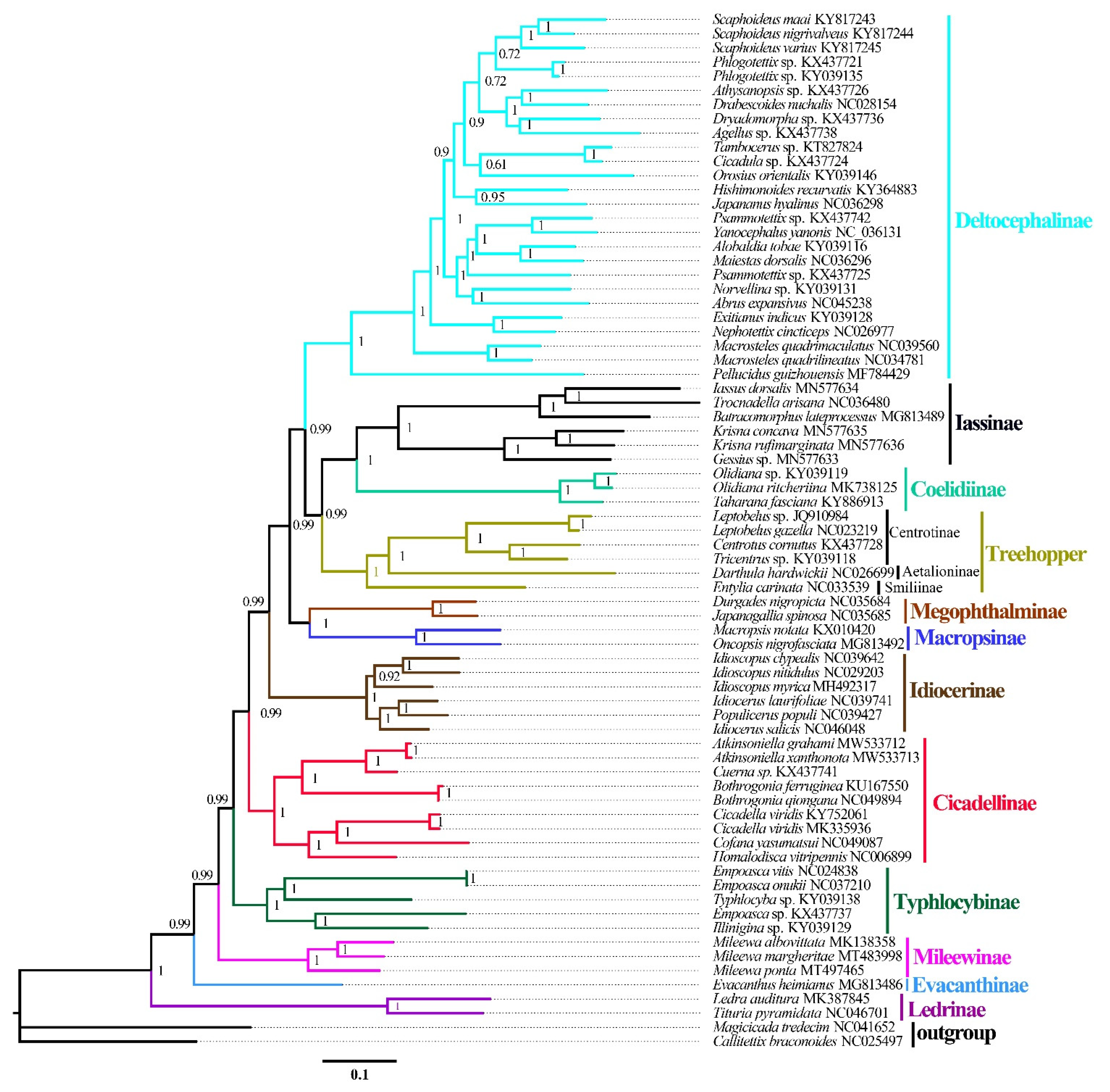
ML and BI analyses were used to reconstruct the phylogenetic relationships among the 65 species of the 11 subfamilies of leafhoppers, 6 species of treehoppers, and 2 outgroups, under the best partitioning scheme and models selected by PartitionFinder (Table S3). Six phylogenetic trees (ML-AA, BI-AA, ML-PCG12, BI-PCG12, ML-PCG12RNA, and BI-PCG12RNA) were established, with most nodes receiving high nodal support values and a few nodes receiving only moderate or low support in the ML and BI trees (Figure 7 and Figure 8 and Figures S1–S5). In the obtained topology, each subfamily was consistently recovered as monophyletic in different analyses, while the relationships among subfamilies were discrepant across analyses. The comparative analysis of these six phylogenetic trees showed that treehoppers share a common ancestor with the subfamilies Deltocephalinae, Iassinae, Coelidiinae, Macropsinae, Megophthalminae, Idiocerinae, Evacanthinae, Cicadellinae, Typhlocybinae, Ledrinae, and Mileewinae. The results of this study support the point that treehoppers are derived from paraphyletic Cicadellidae, which has been reported in former studies [30,37,38,76,77,78].
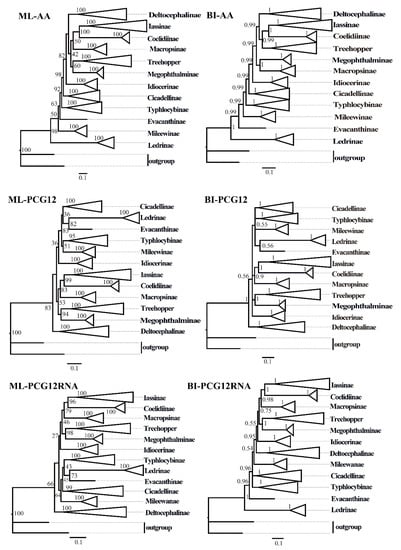
Figure 7.
The phylogenetic trees of leafhoppers were inferred from different mitochondrial genome datasets using maximum likelihood (ML) and Bayesian inference (BI) methods. AA: amino acid sequences of the protein-coding genes (PCGs) from 73 species; PCG12: first and second codon positions of PCGs from 73 species; PCG12RNA: the first and the second codon positions of the PCGs and two rRNA genes from 67 species. Numbers on each node correspond to the bootstrap support values (BS) and Bayesian posterior probability (PP).
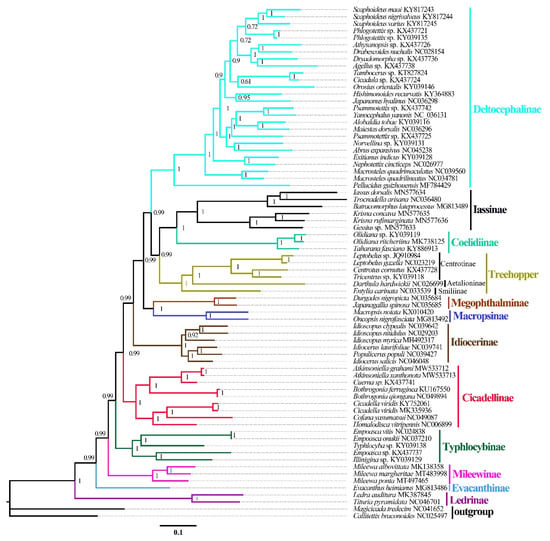
Figure 8.
Phylogenetic trees inferred by Bayesian inference (BI) based on the amino acids (AA). Numbers on each node correspond to the Bayesian posterior probability (PP) for 100 million generations.
Moreover, within the family Cicadellidae, some relationships were highly supported and constant. Iassinae and Coelidiinae were sister groups with high support values (bootstrap support values (BS) ≥ 96, Bayesian posterior probability (PP) = 1) among the six phylogenetic trees, which is consistent with the results of previous studies based on mitogenomes [26,31,32,33]. And Macropsinae emerged as a sister group with Iassinae and Coelidiinae in all phylogenetic trees except for the BI-AA tree. Meanwhile, most nodes within each subfamily received high support and the relationships within Coelidiinae, Typhlocybinae, and Mileewinae were consistent in all analyses.
Within the subfamily Cicadellinae, the phylogenetic relationships indicated that Cicadellinae was consistently a monophyletic group. This is different from the previous studies based on mitogenomes reported by Wang [30,33], which suggested Cicadellinae was not a monophyletic group. All of the ML and BI analyses suggested that the relationships within Cicadellinae were ((H. vitripennis + (Co. yasumatsui + Ci. viridis)) + ((B. ferruginea + B. qiongana) + (Cuerna sp. + (A. grahami + A. xanthonota)))) based on AA and PCG12, and ((H. vitripennis + (Co. yasumatsui + Ci. viridis)) + ((B. ferruginea + B. qiongana) + (A. grahami + A. xanthonota))) based on PCG12RNA with high support values (Figure 8 and Figures S1–S5). The newly sequenced A. grahami grouped with A. xanthonota. The inferred relationships among the genus of Cicadellinae were highly stable; Homalodisca, Cofana, and Cicadella clustered to a branch, forming a sister group with the clade that Atkinsoniella and Bothrogonia formed. Additionally, Mileewinae and Cicadellinae were divided into two separate clades in this study within all phylogenetic trees, except for the BI-PCG12 tree, which formed polytomies, potentially due to inadequate data for ascertaining how these lineages are related (Figure 7and Figures S2). These results support the point of Young [79], and Linnavuori and Delong [80] that Mileewinae is a separate group from Cicadellinae.
4. Conclusions
In this study, the complete mitochondrial genome sequences of A. grahami and A. xanthonota were provided, with a comparative analysis within the available mitogenome sequences of Cicadellinae and a phylogenetic analysis of Cicadellidae. This is the first report of mitogenome sequences from the genus Atkinsoniella of subfamily Cicadellinae. The lengths of these two mitogenomes were 15,621 and 15,895 bp, for A. grahami and A. xanthonota, respectively. Comparison with other previously reported mitogenomes of Cicadellinae showed that all had a similar structural characteristics and nucleotide compositions. All of the Cicadellinae mitogenomes were highly conserved in holistic organization, exhibiting the same gene order as hypothetical ancestral insects, and all mitogenomes were composed of 37 typically encoded genes and a control region, except for Cuerna sp. (KX437741) and Cicadella viridis (KY752061), which lacked trnV, s-rRNA, and a control region, due to the incomplete mitogenomes sequences. These two newly sequenced mitogenomes of genus Atkinsoniella can provide valuable data for future studies of phylogenetic relationships of Cicadellinae.
Maximum likelihood and Bayesian inference analyses among the major lineages based on the concatenated alignments of AA, PCG12, and PCG12RNA indicated that each subfamily of leafhoppers (Cicadellinae) and treehoppers (Membracidae) was recovered as monophyletic group, and that the treehoppers originated from paraphyletic Cicadellidae, as per previous reports. The analyses produced a well-resolved framework for the relationships within each subfamily. The relationships within subfamily Cicadellinae, Typhlocybinae, Mileewinae, and Coelidiinae were stable in all phylogenetic trees with high support. However, a few deep nodes received low or moderate support values and the phylogenetic relationship among subfamilies was not well resolved, which may have been restricted by the limited sampling molecular data in this study. Perhaps, more mitogenomic taxon sampling, richer molecular data, and a combined approach of mitogenomes and nuclear markers would elucidate the unresolved relationships among these subfamilies and help to understand the phylogenetic and evolutionary relationships within Cicadellidae.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/insects12040338/s1: Figure S1. Phylogenetic trees inferred by maximum likelihood (ML) based on the amino acids (AA) from 73 species. Numbers on each node correspond to the bootstrap support values (BS) for 10,000 replicates; Figure S2. Phylogenetic trees inferred by maximum likelihood (ML) based on the 1st and 2nd codon positions of the protein-coding genes (PCG12) from 73 species. Numbers on each node correspond to the bootstrap support values (BS) for 10,000 replicates; Figure S3. Phylogenetic trees inferred by Bayesian inference (BI) based on the 1st and 2nd codon positions of the protein-coding genes (PCG12) from 73 species. Numbers on each node correspond to the Bayesian posterior probability (PP) for 100 million generations; Figure S4. Phylogenetic trees inferred by maximum likelihood (ML) based on the 1st and 2nd codon position of the protein coding genes and rRNA genes (PCG12RNA) from 67 species. Numbers on each node correspond to the bootstrap support values (BS) for 10,000 replicates; Figure S5. Phylogenetic trees inferred by Bayesian inference (BI) based on the 1st and 2nd code position of the protein-coding genes and rRNA genes (PCG12RNA) from 67 species. Numbers on each node correspond to the Bayesian posterior probability (PP) for 100 million generations; Table S1. The starting partitions used to initiate the PartitionFinder analysis; Table S2. The Start codons and stop codons of each protein coding gene in nine Cicadellinae mitogenomes; Table S3. Partition strategies and evolutionary models of AA, PCG12 and PCG12RNA datasets used in the phylogenetic analysis.
Author Contributions
Conceptualization, M.-F.Y. and Y.J.; species identification, M.-F.Y. and Y.J.; methodology and experiments, Y.J., H.-X.L. and X.-F.Y.; data analysis, Y.J. and H.-X.L.; writing—original draft preparation, Y.J.; writing—review and editing, Y.J., H.-X.L., X.-F.Y. and M.-F.Y.; visualization, Y.J. and H.-X.L.; supervision, M.-F.Y.; project administration, M.-F.Y.; funding acquisition, M.-F.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Program of Excellent Innovation Talents, Guizhou Province, China ([2016]-4022); the National Natural Science Foundation of China (31672335); the International Cooperation Base for Insect Evolutionary Biology and Pest Control ([2016]-5802); the Guizhou Provincial Department of Education Youth Science and Technology Talents Growth Project (Qian Jiao He KY word [2018]103); the Introducing Talent Research Program for Guizhou University (Gui Da Ren Ji He word [2016]70); the National Nature Science Foundation of Guizhou University Cultivating program (Qian Ke He Platform Talent [2017]5788-32); the Graduate Innovation Fund, Guizhou Province, China (Qianjiaohe YJSCXJH(2019)023).
Institutional Review Board Statement
Not applicable.
Data Availability Statement
Date available on request.
Acknowledgments
The authors would like to thank Feng Zhang (Nanjing Agricultural University, Nanjing, China) for providing assistance with software analysis, and Hongli He and Jiajia Wang for collecting specimens.
Conflicts of Interest
All of the authors declare no conflict of interest.
References
- Naveed, H.; Zhang, Y.L. Newly recorded leafhoppers of the subfamily Cicadellinae (Hemiptera: Cicadellidae) with description of a new species from Pakistan. Zootaxa 2018, 4504, 285–295. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.F.; Meng, Z.H.; Li, Z.Z. Hemiptera: Cicadellidae (II): Cicadellinae.Fauna Sinica: Insecta Vol. 67; Science Press: Beijing, China, 2017; p. 637. ISBN 978-7-03-052950-3. (In Chinese) [Google Scholar]
- Redak, R.A.; Purcell, A.H.; Lopes, J.R.; Blua, M.J.; Iii, R.F.M.; Andersen, P.C. The biology of xylem fluid-feeding insect vectors of Xylella fastidiosa and their relation to disease epidemiology. Annu. Rev. Èntomol. 2004, 49, 243–270. [Google Scholar] [CrossRef] [PubMed]
- Kleina, H.T.; Kudlawiec, K.; Esteves, M.B.; DalBó, M.A.; Oliveira, T.D.P.; Maluta, N.; Lopes, J.R.S.; May-De-Mio, L.L. Settling and feeding behavior of sharpshooter vectors of Xylella fastidiosa on plum genotypes resistant to leaf scald disease. Eur. J. Plant Pathol. 2020, 158, 633–644. [Google Scholar] [CrossRef]
- Hopkins, D.L.; Purcell, A.H. Xylella fastidiosa: Cause of Pierce’s disease of grapevine and other emergent diseases. Plant Dis. 2002, 86, 1056–1066. [Google Scholar] [CrossRef]
- Krugner, R.; Sisterson, M.S.; Backus, E.A.; Burbank, L.P.; Redak, R.A. Sharpshooters: A review of what moves Xylella fastidiosa. Austral Èntomol. 2019, 58, 248–267. [Google Scholar] [CrossRef]
- Boore, J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef]
- Cameron, S.L. Insect mitochondrial genomics: Implications for evolution and phylogeny. Annu. Rev. Èntomol. 2014, 59, 95–117. [Google Scholar] [CrossRef] [PubMed]
- Wolstenholme, D.R. Animal mitochondrial DNA: Structure and evolution. Int. Rev. Cytol. 1992, 141, 173–216. [Google Scholar] [CrossRef]
- Curole, J.P.; Kocher, T.D. Mitogenomics: Digging deeper with complete mitochondrial genomes. Trends Ecol. Evol. 1999, 14, 394–398. [Google Scholar] [CrossRef]
- Yu, J.N.; Azuma, N.; Abe, S. Genetic differentiation between collections of hatchery and wild masu salmon (Oncorhynchus masou) inferred from mitochondrial and microsatellite DNA analyses. Environ. Biol. Fishes 2011, 94, 259–271. [Google Scholar] [CrossRef]
- Lin, C.P.; Danforth, B.N. How do insect nuclear and mitochondrial gene substitution patterns differ? Insights from Bayesian analyses of combined datasets. Mol. Phylogenetics Evol. 2004, 30, 686–702. [Google Scholar] [CrossRef]
- Yu, P.F.; Li, Q.; Wang, M.X.; Cheng, P.; Lin, C.; Han, B.Y. Analysis of complete mitochondrial genome and phylogenetic relationship of Bothrogonia ferruginea. J. Agric. Biotechnol. 2019, 27, 1246–1258. (In Chinese) [Google Scholar] [CrossRef]
- Xu, X.L.; Yan, B.; Yu, X.F.; Yang, M.F. The complete mitochondrial genome of Bothrogonia qiongana (Hemiptera: Cicadellidae) with phylogenetic analyses. Mitochondrial DNA Part B 2020, 5, 2750–2751. [Google Scholar] [CrossRef] [PubMed]
- Song, N.; Cai, W.Z.; Li, H. Insufficient power of mitogenomic data in resolving the auchenorrhynchan monophyly. Zool. J. Linn. Soc. 2017, 183, 776–790. [Google Scholar] [CrossRef]
- Zhong, L.K.; Li, H.X.; Yu, X.F.; Yang, M.F. Complete mitochondrial genome sequence of Cicadella viridis (Hemiptera: Cicadellidae: Cicadellinae). Mitochondrial DNA Part B 2019, 4, 1287–1288. [Google Scholar] [CrossRef]
- Zhong, L.K.; Yang, M.F.; Yu, X.F. The mitochondrial genome of Cofana yasumatsui (Hemiptera: Cicadellidae: Cicadellinae). Mitochondrial DNA Part B 2020, 5, 1075–1076. [Google Scholar] [CrossRef]
- He, H.L.; Li, H.X.; Yang, M.F. Complete mitochondrial genome sequence of Mileewa albovittata (Hemiptera: Cicadellidae: Mileewinae). Mitochondrial DNA Part B 2019, 4, 740–741. [Google Scholar] [CrossRef]
- He, H.L.; Yang, M.F. The mitogenome of Mileewa margheritae (Hemiptera: Cicadellidae: Mileewinae). Mitochondrial DNA Part B 2020, 5, 3163–3164. [Google Scholar] [CrossRef] [PubMed]
- He, H.L.; Yang, M.F. Characterization and phylogenetic analysis of the mitochondrial genome of Mileewa ponta (Hemiptera: Cicadellidae: Mileewinae). Mitochondrial DNA Part B 2020, 5, 2976–2977. [Google Scholar] [CrossRef]
- Li, D.F.; Dai, R.H. The complete mitochondrial genome of Tituria pyramidata (Hemiptera: Cicadellidae: Ledrinae) from China. Mitochondrial DNA Part B 2020, 5, 1757–1758. [Google Scholar] [CrossRef]
- Wang, J.J.; Li, D.F.; Li, H.; Yang, M.F.; Dai, R.H. Structural and phylogenetic implications of the complete mitochondrial genome of Ledra auditura. Sci. Rep. 2019, 9, 15746. [Google Scholar] [CrossRef] [PubMed]
- Song, N.; Cai, W.Z.; Li, H. Deep-level phylogeny of Cicadomorpha inferred from mitochondrial genomes sequenced by NGS. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef]
- Zhou, N.; Wang, M.; Cui, L.; Chen, X.; Han, B. Complete mitochondrial genome of Empoasca vitis (Hemiptera: Cicadellidae). Mitochondrial DNA 2016, 27, 1052–1053. [Google Scholar] [CrossRef]
- Wang, J.J.; Yang, M.F.; Dai, R.H.; Li, H. Complete mitochondrial genome of Evacanthus heimianus (Hemiptera: Cicadellidae: Evacanthinae) from China. Mitochondrial DNA Part B 2018, 4, 284–285. [Google Scholar] [CrossRef]
- Wang, J.J.; Yang, M.F.; Dai, R.H.; Li, H.; Wang, X.Y. Characterization and phylogenetic implications of the complete mitochondrial genome of Idiocerinae (Hemiptera: Cicadellidae). Int. J. Biol. Macromol. 2018, 120, 2366–2372. [Google Scholar] [CrossRef] [PubMed]
- Dai, R.H.; Wang, J.J.; Yang, M.F. The complete mitochondrial genome of the leafhopper Idioscopus clypealis (Hemiptera: Cicadellidae: Idiocerinae). Mitochondrial DNA Part B 2018, 3, 32–33. [Google Scholar] [CrossRef]
- Choudhary, J.S.; Naaz, N.; Das, B.; Bhatt, B.P.; Prabhakar, C.S. Complete mitochondrial genome of Idioscopus nitidulus (Hemiptera: Cicadellidae). Mitochondrial DNA Part B 2018, 3, 191–192. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.J.; Dai, R.H.; Li, H.; Zhan, H.P. Characterization of the complete mitochondrial genome of Japanagallia spinosa and Durgades nigropicta (Hemiptera: Cicadellidae: Megophthalminae). Biochem. Syst. Ecol. 2017, 74, 33–41. [Google Scholar] [CrossRef]
- Wang, J.J.; Wu, Y.F.; Yang, M.F.; Dai, R.H. The phylogenetic implications of the mitochondrial genomes of Macropsis notata and Oncopsis nigrofasciata. Front. Genet. 2020, 11, 1–9. [Google Scholar] [CrossRef]
- Wang, J.J.; Li, H.; Dai, R.H. Complete mitochondrial genome of Taharana fasciana (Insecta, Hemiptera: Cicadellidae) and comparison with other Cicadellidae insects. Genetica 2017, 145, 593–602. [Google Scholar] [CrossRef]
- Wang, X.Y.; Wang, J.J.; Fan, Z.H.; Dai, R.H. Complete mitogenome of Olidiana ritcheriina (Hemiptera: Cicadellidae) and phylogeny of Cicadellidae. PeerJ 2019, 7, e8072. [Google Scholar] [CrossRef]
- Wang, J.J.; Wu, Y.F.; Dai, R.H.; Yang, M.F. Comparative mitogenomes of six species in the subfamily Iassinae (Hemiptera: Cicadellidae) and phylogenetic analysis. Int. J. Biol. Macromol. 2020, 149, 1294–1303. [Google Scholar] [CrossRef]
- Du, Y.M.; Zhang, C.N.; Dietrich, C.H.; Zhang, Y.L.; Dai, W. Characterization of the complete mitochondrial genomes of Maiestas dorsalis and Japananus hyalinus (Hemiptera: Cicadellidae) and comparison with other Membracoidea. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef]
- Wang, J.J.; Xing, J.C. Complete mitochondrial genome of Abrus expansivus (Hemiptera: Cicadellidae: Deltocephalinae) from China. Mitochondrial DNA Part B 2019, 4, 197–198. [Google Scholar] [CrossRef]
- Mao, M.; Yang, X.S.; Bennett, G. The complete mitochondrial genome of Macrosteles quadrilineatus (Hemiptera: Cicadellidae). Mitochondrial DNA Part B 2017, 2, 173–175. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.M.; Dietrich, C.H.; Dai, W. Complete mitochondrial genome of Macrosteles quadrimaculatus (Matsumura) (Hemiptera: Cicadellidae: Deltocephalinae) with a shared tRNA rearrangement and its phylogenetic implications. Int. J. Biol. Macromol. 2019, 122, 1027–1034. [Google Scholar] [CrossRef]
- Du, Y.M.; Dai, W.; Dietrich, C.H. Mitochondrial genomic variation and phylogenetic relationships of three groups in the genus Scaphoideus (Hemiptera: Cicadellidae: Deltocephalinae). Sci. Rep. 2017, 7, 16908. [Google Scholar] [CrossRef][Green Version]
- Wu, Y.F.; Dai, R.H.; Zhan, H.P.; Qu, L. Complete mitochondrial genome of Drabescoides nuchalis (Hemiptera: Cicadellidae). Mitochondrial DNA Part A 2015, 27, 3626–3627. [Google Scholar] [CrossRef]
- Yu, P.F.; Wang, M.X.; Cui, L.; Chen, X.X.; Han, B.Y. The complete mitochondrial genome of Tambocerus sp. (Hemiptera: Cicadellidae). Mitochondrial DNA Part A 2015, 28, 133–134. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Liang, A.P. Complete DNA sequence of the mitochondrial genome of the treehopper Leptobelus gazella (Membracoidea: Hemiptera). Mitochondrial DNA Part A 2016, 27, 3318–3319. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Leavengood, J.M.; Chapman, E.G.; Burkhardt, D.; Song, F.; Jiang, P.; Liu, J.P.; Zhou, X.G.; Cai, W.Z. Mitochondrial phylogenomics of Hemiptera reveals adaptive innovations driving the diversification of true bugs. Proc. R. Soc. B Biol. Sci. 2017, 284, 20171223. [Google Scholar] [CrossRef] [PubMed]
- Mao, M.; Yang, X.S.; Bennett, G. The complete mitochondrial genome of Entylia carinata (Hemiptera: Membracidae). Mitochondrial DNA Part B 2016, 1, 662–663. [Google Scholar] [CrossRef] [PubMed]
- Liang, A.P.; Gao, J.; Zhao, X. Characterization of the complete mitochondrial genome of the treehopper Darthula hardwickii (Hemiptera: Aetalionidae). Mitochondrial DNA Part A 2015, 27, 3291–3292. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Bu, C.P.; Wipfler, B.; Liang, A.P. Comparative analysis of the mitochondrial genomes of Callitettixini Spittlebugs (Hemiptera: Cercopidae) confirms the overall high evolutionary speed of the AT-Rich region but reveals the presence of short conservative elements at the tribal level. PLoS ONE 2014, 9, e109140. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.Y.; Hasegawa, H.; Cooley, J.R.; Simon, C.; Yoshimura, J.; Cai, W.Z.; Sota, T.; Li, H. Mitochondrial genomics reveals shared phylogeographic patterns and demographic history among three periodical Cicada species groups. Mol. Biol. Evol. 2019, 36, 1187–1200. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.M.; Kuoh, C.L. Eight new species and a new record species of the genus Atkinsoniella from China (Homoptera: Cicadellidae). J. Anhui Agric. Univ. 1993, 20, 7–17. (In Chinese) [Google Scholar] [CrossRef]
- Yang, M.F.; Li, Z.Z. Two new species of Atkinsoniella from Sichuan Province, China (Homoptera: Cicadellidae). Acta Entomol. Sin. 2002, 45, 40–42. (In Chinese) [Google Scholar] [CrossRef]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2017, 45, e18. [Google Scholar] [CrossRef]
- Meng, G.L.; Li, Y.Y.; Yang, C.T.; Liu, S.L. MitoZ: A toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 2019, 47, e63. [Google Scholar] [CrossRef]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef]
- Lowe, T.M.; Chan, P.P. tRNAscan-SE On-line: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016, 44, W54–W57. [Google Scholar] [CrossRef] [PubMed]
- Greiner, S.; Lehwark, P.; Bock, R. Organellar genome DRAW (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef]
- Perna, N.T.; Kocher, T.D. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 1995, 41, 353–358. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef]
- Ranwez, V.; Harispe, S.; Delsuc, F.; Douzery, E.J.P. MACSE: Multiple alignment of coding sequences accounting for frameshifts and stop codons. PLoS ONE 2011, 6, e22594. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zhou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2019, 20, 348–355. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef]
- Talavera, G.; Castresana, J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef] [PubMed]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New Methods for Selecting Partitioned Models of Evolution for Molecular and Morphological Phylogenetic Analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating Maximum-Likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.Q.; Huang, Y.X.; Bartlett, C.R.; Zhou, F.M.; Meng, R.; Qin, D.Z. Characterization of the complete mitochondrial genomes of two species of the genus Aphaena Guérin-Méneville (Hemiptera: Fulgoridae) and its phylogenetic implications. Int. J. Biol. Macromol. 2019, 141, 29–40. [Google Scholar] [CrossRef]
- Huang, W.J.; Zhang, Y.L. Characterization of two complete mitochondrial genomes of Ledrinae (Hemiptera: Cicadellidae) and phylogenetic analysis. Insects 2020, 11, 609. [Google Scholar] [CrossRef]
- Xu, D.L.; Yu, T.H.; Zhang, Y.L. Characterization of the complete mitochondrial genome of Drabescus ineffectus and Roxasellana stellata (Hemiptera: Cicadellidae: Deltocephalinae: Drabescini) and their phylogenetic implications. Insects 2020, 11, 534. [Google Scholar] [CrossRef]
- Tang, J.; Huang, W.J.; Zhang, Y.L. The Complete mitochondrial genome of four Hylicinae (Hemiptera: Cicadellidae): Structural features and phylogenetic implications. Insects 2020, 11, 869. [Google Scholar] [CrossRef]
- Zhang, D.X.; Hewitt, G.M. Insect mitochondrial control region: A review of its structure, evolution and usefulness in evolutionary studies. Biochem. Syst. Ecol. 1997, 25, 99–120. [Google Scholar] [CrossRef]
- Bian, D.D.; Ye, W.T.; Dai, M.L.; Lu, Z.T.; Li, M.X.; Fang, Y.L.; Qu, J.W.; Su, W.J.; Li, F.C.; Sun, H.N.; et al. Phylogenetic relationships of Limacodidae and insights into the higher phylogeny of Lepidoptera. Int. J. Biol. Macromol. 2020, 159, 356–363. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.S.; Song, L.; Mao, J.H.; Shi, Y.X.; Wu, C.J.; Zhang, Y.X.; Huang, L.; Peng, W.F.; Liu, X.M. Complete mitochondrial genome of the soybean leaffolder, Omiodes indicata (Lepidoptera: Pyraloidea: Crambidae), and phylogenetic analysis for Pyraloidea. Int. J. Biol. Macromol. 2018, 115, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Xin, Z.Z.; Zhu, X.Y.; Zhao, X.M.; Wang, Y.; Tang, B.P.; Zhang, H.B.; Zhang, D.Z.; Zhou, C.L.; Liu, Q.N. The complete mitochondrial genome of Euproctis similis (Lepidoptera: Noctuoidea: Erebidae) and phylogenetic analysis. Int. J. Biol. Macromol. 2017, 105, 219–227. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.H.; Yang, T.T.; Liu, Y.; Zhang, H.B.; Tang, B.P.; Liu, Q.N.; Ma, Y.F. The complete mitochondrial genome of Sinna extrema (Lepidoptera: Nolidae) and its implications for the phylogenetic relationships of Noctuoidea species. Int. J. Biol. Macromol. 2019, 137, 317–326. [Google Scholar] [CrossRef]
- Zhu, X.Y.; Xin, Z.Z.; Wang, Y.; Zhang, H.B.; Zhang, D.Z.; Wang, Z.F.; Zhou, C.L.; Tang, B.P.; Liu, Q.N. The complete mitochondrial genome of Clostera anachoreta (Lepidoptera: Notodontidae) and phylogenetic implications for Noctuoidea species. Genomics 2017, 109, 221–226. [Google Scholar] [CrossRef]
- Dietrich, C.H.; Allen, J.M.; Lemmon, A.R.; Lemmon, E.M.; Takiya, D.M.; Evangelista, O.; Walden, K.K.O.; Grady, P.G.S.; Johnson, K.P. Anchored Hybrid Enrichment-Based Phylogenomics of Leafhoppers and Treehoppers (Hemiptera: Cicadomorpha: Membracoidea). Insect Syst. Divers. 2017, 1, 57–72. [Google Scholar] [CrossRef]
- Dietrich, C.; Rakitov, R.; Holmes, J.; Black, W. Phylogeny of the major lineages of Membracoidea (Insecta: Hemiptera: Cicadomorpha) based on 28S rDNA sequences. Mol. Phylogenet. Evol. 2001, 18, 293–305. [Google Scholar] [CrossRef]
- Skinner, R.K.; Dietrich, C.H.; Walden, K.K.O.; Gordon, E.; Sweet, A.D.; Podsiadlowski, L.; Petersen, M.; Simon, C.; Takiya, D.M.; Johnson, K.P. Phylogenomics of Auchenorrhyncha (Insecta: Hemiptera) using transcriptomes: Examining controversial relationships via degeneracy coding and interrogation of gene conflict. Syst. Èntomol. 2019, 45, 85–113. [Google Scholar] [CrossRef]
- Young, D.A. Taxonomic Study of the Cicadellinae (Homoptera: Cicadellidae) pt. 1: Proconiini. Bull. U. S. Natl. Mus. 1968, 261, 1–287. [Google Scholar] [CrossRef]
- Linnavuori, R.; Delong, D.M. Studies of the Neotropical Mileewaninae (Homoptera: Cicadellidae). J. Kans. Entomol. Soc. 1977, 50, 410–421. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).