Selection and Validation of Suitable Reference Genes for RT-qPCR Analysis in the Rare Aquatic Firefly Aquatica leii (Coleoptera: Lampyridae)
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Insects
2.2. Sample Preparation
2.2.1. Developmental Stage
2.2.2. Tissue
2.2.3. dsRNA Injection
2.3. RNA Extraction and cDNA Synthesis
2.4. Candidate Reference Genes and Primer Design
2.5. Quantitative Reverse Transcription-Polymerase Chain Reaction
2.6. Stability Evaluation of Candidate Reference Genes
2.7. Validation of a Selected Reference Gene
3. Results
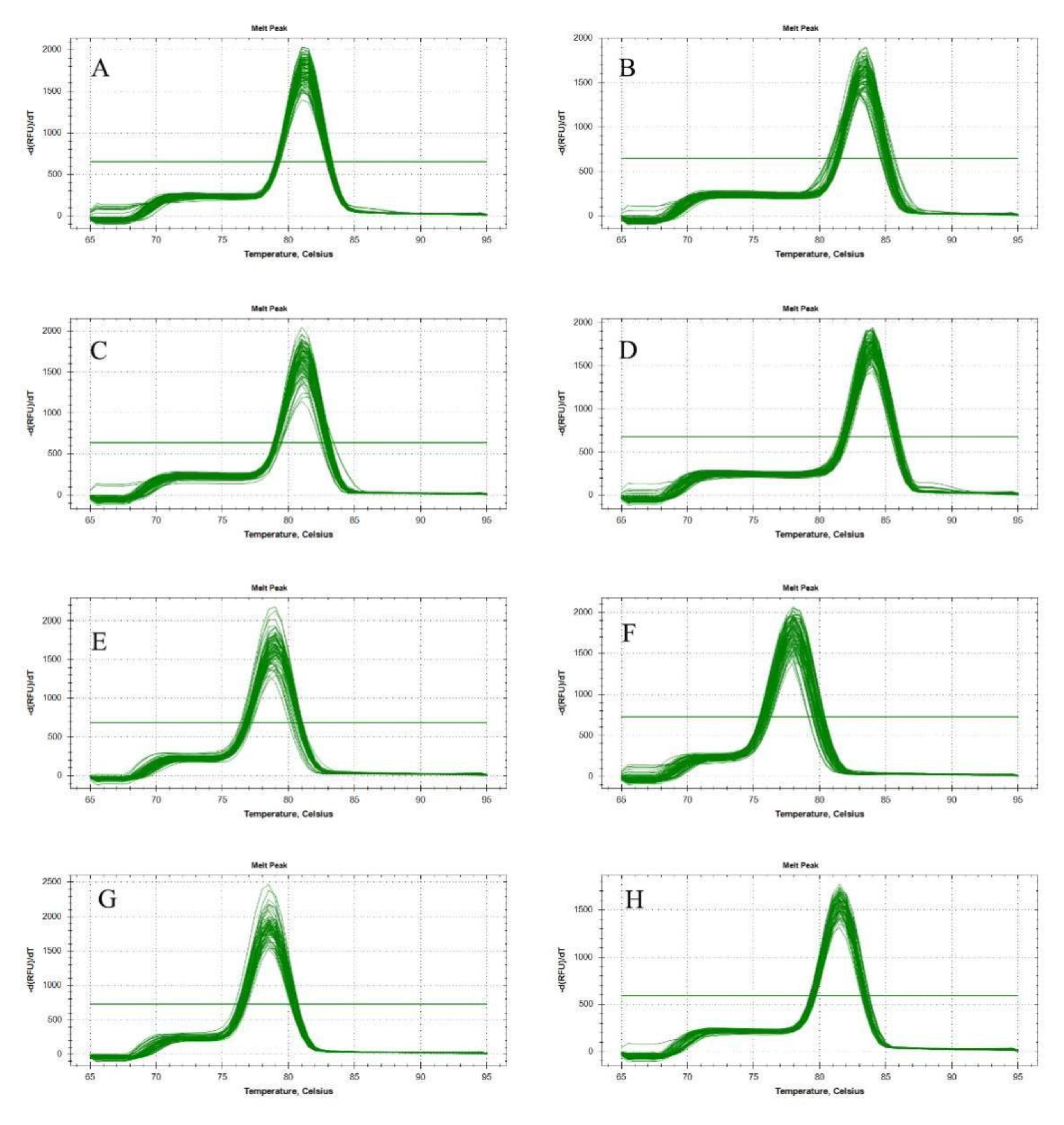
3.1. PCR Amplification of Candidate Reference Genes
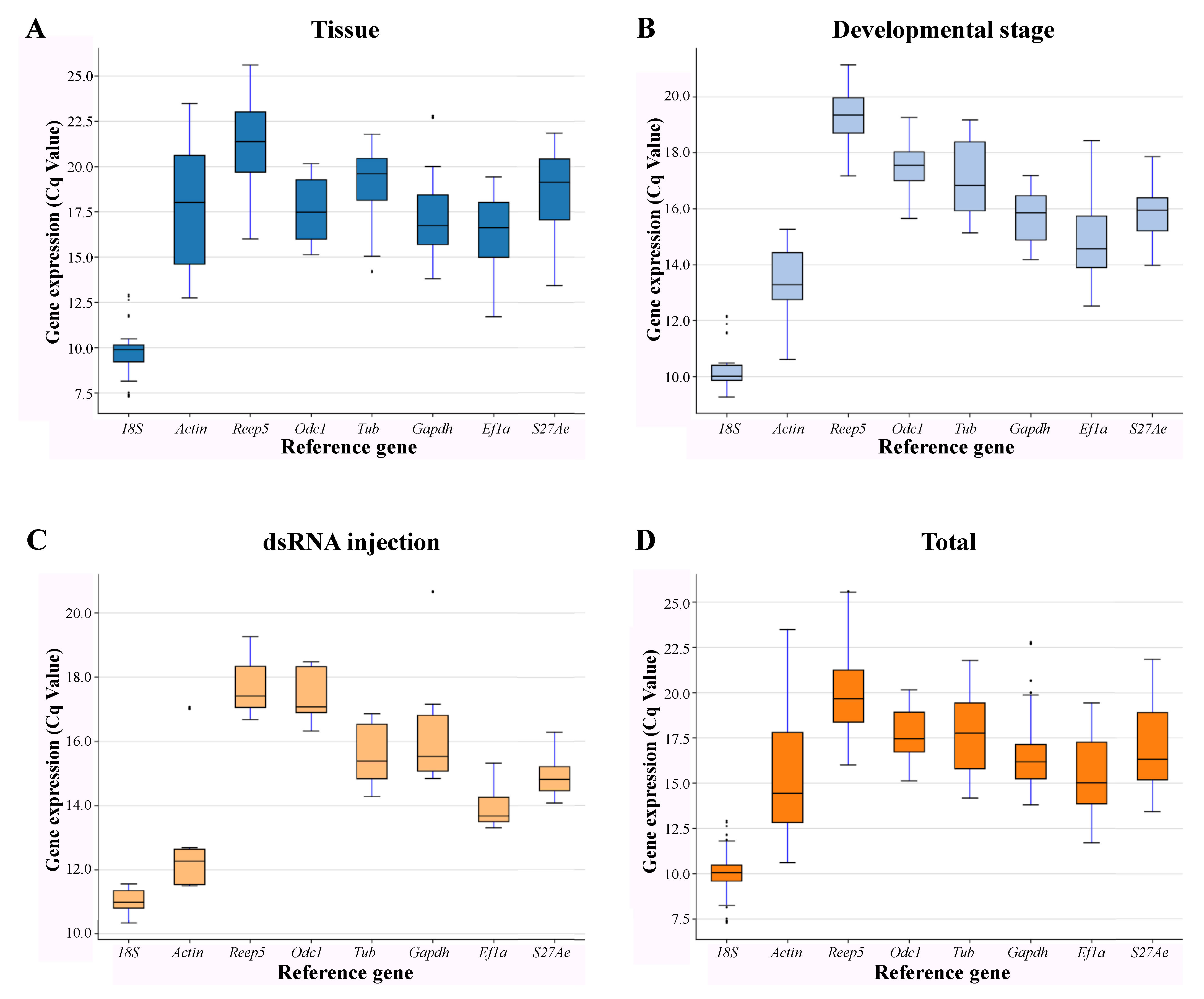
3.2. Expression Profiling of Candidate Reference Genes in A. leii
3.3. Expression Stability and Ranking of Candidate Reference Genes
3.4. Validation of the Recommended Candidate Reference Genes
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| LO | light organ |
| LLO | larval light organ |
| H-T | head and thorax |
| FB | fat body |
| T | testis |
| OV | ovary |
| RB | rest of body |
| L3 | 3rd-instar larvae |
| L5 | 5th-instar larvae |
| P1 | 1d- pupae |
| P3 | 3d- pupae |
| P5 | 5d- pupae |
Appendix A
| Gene | Protein | Function |
|---|---|---|
| 18S | 18S ribosomal RNA | 18S rRNA is the structural RNA for the small component of eukaryotic cytoplasmic ribosomes, and thus one of the basic components of all eukaryotic cells. |
| Actin | Actin, cytoplasmic 2 | Actins are highly conserved proteins that are involved in various types of cell motility and are ubiquitously expressed in all eukaryotic cells. |
| Reep5 | Receptor accessory protein 5 | The protein is a member of the REEP family, which generally facilitate intracellular trafficking through alterations to the endoplasmic reticulum, and which have the ability to enhance activity of G-protein coupled receptors. |
| Odc1 | Ornithine decarboxylase 1 | Catalyzes the first and rate-limiting step of polyamine biosynthesis that converts ornithine into putrescine, which is the precursor for the polyamines, spermidine and spermine. Polyamines are essential for cell proliferation and are implicated in cellular processes, ranging from DNA replication to apoptosis. |
| Tub | Tubulin beta chain | Tubulin in molecular biology can refer either to the tubulin protein superfamily of globular proteins, or one of the member proteins of that superfamily. α- and β-tubulins polymerize into microtubules, a major component of the eukaryotic cytoskeleton. |
| Gapdh | Glyceraldehyde-3-phosphate dehydrogenase | The protein is an enzyme of ~37kDa that catalyzes the sixth step of glycolysis and thus serves to break down glucose for energy and carbon molecules. |
| Ef1a | Elongation factor 1-alpha | This gene is responsible for the enzymatic delivery of aminoacyl tRNAs to the ribosome. |
| S27Ae | Ubiquitin-40 S ribosomal protein S27Ae | This gene encodes a fusion protein consisting of ubiquitin at the N terminus and ribosomal protein S27a at the C terminus. Ubiquitin is a highly conserved protein that has a major role in targeting cellular proteins for degradation by the 26S proteosome. Ribosomal protein S27a is a component of the 40S subunit of the ribosome |
| Fru | PREDICTED: Sex determination protein fruitless | The protein acts as a transcriptional regulator. Part of the somatic sex determination hierarchy |
| Luc | Luciferase | The protein anticipates the process of bioluminescence in fireflies. |


References
- Fu, X.H.; Ballantyne, L. Luciola leii sp. nov., a new species of aquatic firefly (Coleoptera: Lampyridae: Luciolinae) from mainland China. Can. Entomol. 2006, 138, 339–347. [Google Scholar]
- Fu, X.H.; Meyer Rochow, V.B. An investigation into the morphological and behavioral adaptations of the aquatic larvae of Aquatica leii (Coleoptera: Lampyridae) to prey upon freshwater snails that serve as intermediate hosts for the liver fluke. Biocontrol 2012, 62, 127–134. [Google Scholar] [CrossRef]
- Sato, K.; Goto, J.; Yamamoto, D. Sex mysteries of the fly courtship master regulator fruitless. Front Behav. Neurosci. 2019, 13, 1–6. [Google Scholar] [CrossRef]
- Citri, A.; Pang, Z.P.; Südhof, T.C.; Wernig, M.; Malenka, R.C. Comprehensive qPCR profiling of gene expression in single neuronal cells. Nat. Protoc. 2011, 7, 118–127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garson, J.; Grant, P.; Ayliffe, U.; Ferns, R.; Tedder, R. Real-time PCR quantitation of hepatitis B virus DNA using automated sample preparation and murine cytomegalovirus internal control. J. Virol. Methods 2005, 126, 207–213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ginzinger, D.G. Gene quantification using real-time quantitative PCR: An emerging technology hits the mainstream. Exp. Hematol. 2002, 30, 503–512. [Google Scholar] [CrossRef]
- Radonic, A.; Thulke, S.; Mackay, I.M.; Landt, O.; Siegert, W.; Nitsche, A. Guideline to reference gene selection for quantitative realtime PCR. Biochem. Biophys. Res. Commun. 2014, 313, 856–862. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.Y.; Li, Z.L.; Lu, M.X.; Cao, S.S.; Du, Y.Z. Selection of valid reference genes for quantitative real-time PCR in Cotesiachilonis (Hymenoptera: Braconidae) exposed to different temperatures. PLoS ONE 2019, 14, e0226139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sang, W.; He, L.; Wang, X.P.; Zhu, S.K.; Lei, C.L. Evaluation of Reference Genes for RT-qPCR in Tribolium castaneum (Coleoptera: Tenebrionidae) Under UVB Stress. Environ. Entomol. 2015, 44, 418–425. [Google Scholar] [CrossRef] [PubMed]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pfaffl, M.; Tichopad, A.; Prgomet, C.; Neuvians, T. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [PubMed]
- Silver, N.; Best, S.; Jiang, J.; Thein, S.L. Selection of housekeeping genes for gene expression studies inhuman reticulocytes using real-time PCR. BMC Mol. Biol. 2006, 7, 33–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, F.; Xiao, P.; Chen, D.; Xu, L.; Zhang, B. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012, 80, 75–78. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.H. An Illustrated Handbook of Chinese Fireflies, 1st ed.; The Commercial Press: Beijing, China, 2014; pp. 1–167. [Google Scholar]
- Lewis, S.M.; Cratsley, C.K. Flash signal evolution, mate choice, and predation in fireflies. Annu. Rev. Entomol. 2008, 53, 293–321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilson, T.; Hastings, J.W. Bioluminescence: Living Lights, Lights for Living, 1st ed.; Harvard University Press: Cambridge, MA, USA; London, UK, 2013; pp. 1–185. [Google Scholar]
- Owens, A.C.S.; Meyer-Rochow, V.B.; Yang, E.-C. Short- and mid-wavelength artificial light influences the flash signals of Antiquatica ficta fireflies (Coleoptera: Lampyridae). PLoS ONE 2018, 13, e0191576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, X.H.; Li, J.J.; Tian, Y.; Quan, W.P.; Zhang, S.; Liu, Q.; Liang, F.; Zhu, X.L.; Zhang, L.S.; Wang, D.P.; et al. Long-read sequence assembly of the firefly Pyrocoelia pectoralis genome. Gigascience 2017, 6, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fallon, T.R.; Lower, S.E.; Chang, C.H.; Bessho-Uehara, M.; Martin, G.J.; Bewick, A.J.; Behringer, M.; Debat, H.J.; Wong, I.; Day, J.C.; et al. Firefly genomes illuminate parallel origins of bioluminescence in beetles. Elife 2018, 7, 1–146. [Google Scholar] [CrossRef] [PubMed]
- Oba, Y.; Konishi, K.; Yano, D.; Shibata, H.; Kato, D.; Shirai, T. Resurrecting the ancient glow of the fireflies. Sci. Adv. 2020, 6, eabc5705. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.J.; Zheng, H.L.; Liu, Y.Y.; Li, H.W.; Jiang, Y.H.; Lin, L.B.; Deng, X.Y.; Zhang, Q.L. Selection of reference genes for quantitative real-time PCR in Aquatica leii (Coleoptera: Lampyridae) under five different experimental conditions. Front. Physiol. 2020, 11. [Google Scholar] [CrossRef]


| Access Number | Primer Name | Primer Seq (5′-3′) | Primer TM (°C) | Length (bp) | |
|---|---|---|---|---|---|
| 18S (18S ribosomal RNA) | MT899427 | 18S F | TCTTAACCGAGTGTCCAGGC | 56.43 | 158 |
| 18S R | CATTACCTCTGTGCGTTCCA | 54.91 | |||
| Actin (Actin, cytoplasmic 2) | MT899429 | Actin F | GTATCTCACACCGTCCCCAT | 55.83 | 143 |
| Actin R | CTTTCAGCGGTGGTTGTGAA | 56.02 | |||
| Reep5 (Receptor accessory protein 5) | MT899435 | Reep5 F | ACCTACCGATTTCAATGGATCTC | 58.04 | 119 |
| Reep5 R | GCTTTGCCGCTTCATTTTGG | 59.21 | |||
| Odc1 (Ornithine decarboxylase 1) | MT899431 | Odc1 F | AGACGCTGAGTGGATTTTGC | 58.84 | 101 |
| Odc1 R | CGTCCACATAATCCAGCACG | 59.07 | |||
| Tub (Tubulin beta chain) | MT899434 | Tub F | GTACGTTCGGGTCCATTTGG | 58.92 | 114 |
| Tub R | GACCAATTCAGCACCTTCGG | 59.2 | |||
| Gapdh (Glyceraldehyde-3-phosphate dehydrogenase) | MT899430 | Gapdh F | ATCATTCCAGCAGCAACAGG | 58.53 | 188 |
| Gapdh R | CCTTCGGCAGCTTCCTTTAC | 58.91 | |||
| Ef1a (Elongation factor 1-alpha) | MT899428 | Ef1a F | ATGGTTGTCGTCTTTGCACC | 59.05 | 141 |
| Ef1a R | ACGACGCAATTCCTTAACGG | 58.93 | |||
| S27Ae (Ubiquitin-40S ribosomal protein S27Ae) | MT899432 | S27Ae F | TCCACCTGATCAACAACGTT | 57.37 | 124 |
| S27Ae R | AGCACCACCTCGAAGTCTAA | 58.37 | |||
| Fru (fruitless) | MT899433 | Fru F | TCGCAAAACCTTCTTCCGAT | 57.82 | 106 |
| Fru R | GCACTTCCGTTGTTTCGTCT | 59.06 | |||
| Luc | MT990933 | Luc F | GGAGATATTGGGTATTACGATG | ds RNA primer | 378 |
| Luc R | CATCTTTGCATTTGGTTTCTTG | ||||
| Gfp | AAA27722.1 | GFP F | CTACGGCGTGCAGTGCTTCAGC | ds RNA primer | 350 |
| GFP F | AGTGGTCGGCGAGCTGCACGCTG |
| Candidate Genes | ΔCt Method | BestKeeper | NormFinder | geNorm | Recommended Genes | ||||
|---|---|---|---|---|---|---|---|---|---|
| Average of STDEV | Ranking | Stability | Ranking | Stability | Ranking | Stability | Ranking | ||
| Tissues | |||||||||
| 18S | 2.424 | 7 | 0.900 | 1 | 2.021 | 7 | 1.477 | 6 | Ef1a & Odc1 |
| Actin | 3.457 | 8 | 2.915 | 8 | 3.292 | 8 | 1.972 | 7 | |
| Reep5 | 1.780 | 5 | 2.110 | 7 | 1.136 | 6 | 1.012 | 4 | |
| Odc1 | 1.559 | 3 | 1.549 | 2 | 0.325 | 1 | 0.886 | 3 | |
| Tub | 1.550 | 2 | 1.697 | 3 | 0.739 | 2 | 0.684 | 2 | |
| Gapdh | 1.850 | 6 | 1.748 | 4 | 1.008 | 5 | 1.193 | 5 | |
| Ef1a | 1.549 | 1 | 1.859 | 5 | 0.835 | 3 | 0.538 | 1 | |
| S27Ae | 1.604 | 4 | 2.109 | 6 | 0.994 | 4 | 0.538 | 1 | |
| Developmental stages | |||||||||
| 18S | 1.648 | 8 | 0.507 | 1 | 1.437 | 8 | 1.264 | 7 | S27Ae & Reep5 |
| Actin | 1.436 | 7 | 0.959 | 6 | 1.170 | 6 | 1.136 | 6 | |
| Reep5 | 1.022 | 2 | 0.779 | 4 | 0.407 | 2 | 0.479 | 1 | |
| Odc1 | 1.074 | 3 | 0.684 | 2 | 0.491 | 3 | 0.789 | 3 | |
| Tub | 1.270 | 5 | 1.218 | 7 | 0.975 | 5 | 0.665 | 2 | |
| Gapdh | 1.264 | 4 | 0.808 | 5 | 0.910 | 4 | 1.041 | 5 | |
| Ef1a | 1.423 | 6 | 1.240 | 8 | 1.191 | 7 | 0.896 | 4 | |
| S27Ae | 0.976 | 1 | 0.689 | 3 | 0.216 | 1 | 0.479 | 1 | |
| dsRNA injection | |||||||||
| 18S | 1.344 | 6 | 0.323 | 1 | 1.046 | 6 | 0.744 | 5 | Odc1 & Tub |
| Actin | 1.894 | 8 | 1.386 | 8 | 1.749 | 8 | 1.234 | 7 | |
| Reep5 | 1.118 | 5 | 0.734 | 5 | 0.853 | 5 | 0.586 | 4 | |
| Odc1 | 0.941 | 2 | 0.686 | 4 | 0.126 | 1 | 0.251 | 1 | |
| Tub | 0.929 | 1 | 0.742 | 6 | 0.126 | 2 | 0.251 | 1 | |
| Gapdh | 1.658 | 7 | 1.319 | 7 | 1.418 | 7 | 1.014 | 6 | |
| Ef1a | 0.948 | 3 | 0.513 | 2 | 0.454 | 3 | 0.492 | 2 | |
| S27Ae | 1.042 | 4 | 0.570 | 3 | 0.724 | 4 | 0.552 | 3 | |
| Total | |||||||||
| 18S | 2.581 | 7 | 0.752 | 1 | 2.270 | 7 | 1.621 | 6 | Tub & S27Ae |
| Actin | 3.133 | 8 | 2.827 | 8 | 2.916 | 8 | 1.999 | 7 | |
| Reep5 | 1.683 | 4 | 1.834 | 5 | 0.919 | 4 | 0.839 | 2 | |
| Odc1 | 1.762 | 5 | 1.093 | 2 | 0.950 | 5 | 1.173 | 4 | |
| Tub | 1.646 | 1 | 1.926 | 7 | 0.864 | 1 | 0.781 | 1 | |
| Gapdh | 1.880 | 6 | 1.377 | 3 | 1.078 | 6 | 1.334 | 5 | |
| Ef1a | 1.660 | 3 | 1.700 | 4 | 0.902 | 3 | 0.922 | 3 | |
| S27Ae | 1.649 | 2 | 1.901 | 6 | 0.875 | 2 | 0.781 | 1 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, X.; Meyer-Rochow, V.B. Selection and Validation of Suitable Reference Genes for RT-qPCR Analysis in the Rare Aquatic Firefly Aquatica leii (Coleoptera: Lampyridae). Insects 2021, 12, 359. https://doi.org/10.3390/insects12040359
Fu X, Meyer-Rochow VB. Selection and Validation of Suitable Reference Genes for RT-qPCR Analysis in the Rare Aquatic Firefly Aquatica leii (Coleoptera: Lampyridae). Insects. 2021; 12(4):359. https://doi.org/10.3390/insects12040359
Chicago/Turabian StyleFu, Xinhua, and Victor Benno Meyer-Rochow. 2021. "Selection and Validation of Suitable Reference Genes for RT-qPCR Analysis in the Rare Aquatic Firefly Aquatica leii (Coleoptera: Lampyridae)" Insects 12, no. 4: 359. https://doi.org/10.3390/insects12040359
APA StyleFu, X., & Meyer-Rochow, V. B. (2021). Selection and Validation of Suitable Reference Genes for RT-qPCR Analysis in the Rare Aquatic Firefly Aquatica leii (Coleoptera: Lampyridae). Insects, 12(4), 359. https://doi.org/10.3390/insects12040359







