(Epi)Genetic Mechanisms Underlying the Evolutionary Success of Eusocial Insects
Abstract
:Simple Summary
Abstract
1. Introduction
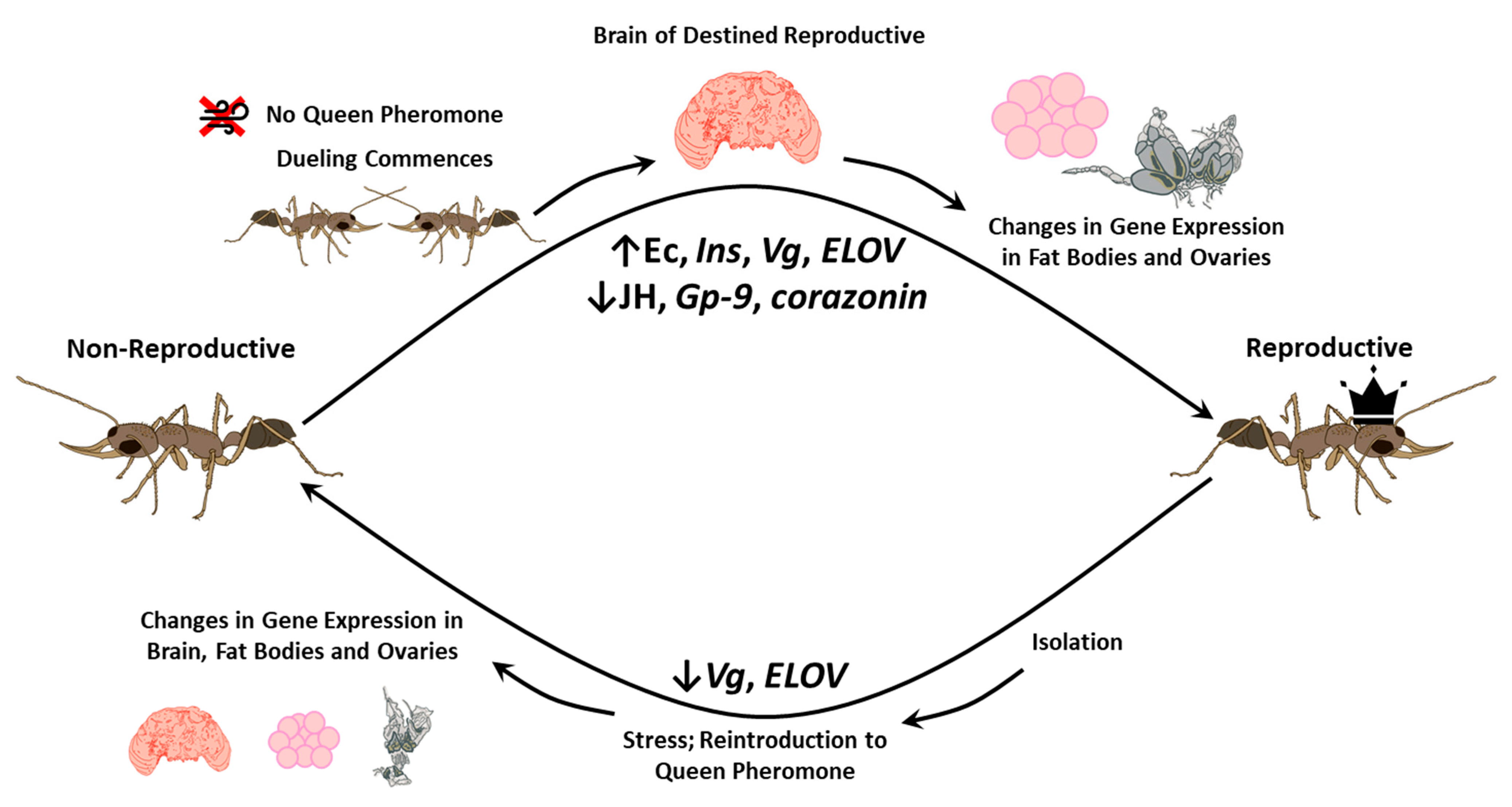
2. Caste Determination, Plasticity, and Caste-Specific Behavior
3. Reproduction and Juvenile Development
4. Age-Dependent Behavior, Aging, and Longevity
5. Social Communication
6. Neural Tissue and Functionality
7. Transgenerational Epigenetic Inheritance
8. Eusocial Insects as Models
9. Conclusions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Wilson, E.O. The Insect Societies; Belknap Press: Cambridge, MA, USA, 1971. [Google Scholar]
- Hamilton, W.D. The genetical evolution of social behaviour. J. Theor. Biol. 1964, 7, 1–16. [Google Scholar] [CrossRef]
- McCarrey, J.R. Epigenetic mechanisms regulating gene expression. In Introduction to Bioinformatics: A Theoretical and Practical Approach; Krawetz, S.A., Womble, D.D., Eds.; Humana Press: Totowa, NJ, USA, 2003; pp. 123–139. [Google Scholar]
- Waddington, C.H. Canalization of development and the inheritance of acquired characters. Nature 1942, 150, 563–565. [Google Scholar] [CrossRef]
- Waddington, C.H. The epigenotype. Endeavour 1942, 1, 18–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hebbes, T.R.; Thorne, A.W.; Crane-Robinson, C. A direct link between core histone acetylation and transcriptionally active chromatin. Embo J. 1988, 7, 1395–1402. [Google Scholar] [CrossRef] [PubMed]
- Brownell, J.E.; Zhou, J.; Ranalli, T.; Kobayashi, R.; Edmondson, D.G.; Roth, S.Y.; Allis, C.D. Tetrahymena histone acetyltransferase a: A homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell 1996, 84, 843–851. [Google Scholar] [CrossRef] [Green Version]
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef] [PubMed]
- Creyghton, M.P.; Cheng, A.W.; Welstead, G.G.; Kooistra, T.; Carey, B.W.; Steine, E.J.; Hanna, J.; Lodato, M.A.; Frampton, G.M.; Sharp, P.A.; et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA 2010, 107, 21931. [Google Scholar] [CrossRef] [Green Version]
- Marmorstein, R.; Roth, S.Y. Histone acetyltransferases: Function, structure, and catalysis. Curr. Opin. Genet. Dev. 2001, 11, 155–161. [Google Scholar] [CrossRef]
- Milazzo, G.; Mercatelli, D.; Di Muzio, G.; Triboli, L.; De Rosa, P.; Perini, G.; Giorgi, F.M. Histone deacetylases (HDACs): Evolution, specificity, role in transcriptional complexes, and pharmacological actionability. Genes 2020, 11, 556. [Google Scholar] [CrossRef]
- Yoshida, M.; Horinouchi, S.; Beppu, T. Trichostatin A and trapoxin: Novel chemical probes for the role of histone acetylation in chromatin structure and function. BioEssays 1995, 17, 423–430. [Google Scholar] [CrossRef] [PubMed]
- Dokmanovic, M.; Clarke, C.; Marks, P.A. Histone deacetylase inhibitors: Overview and perspectives. Mol. Cancer Res. 2007, 5, 981. [Google Scholar] [CrossRef] [Green Version]
- Razin, A.; Riggs, A. DNA methylation and gene function. Science 1980, 210, 604–610. [Google Scholar] [CrossRef]
- Holliday, R.; Pugh, J.E. DNA modification mechanisms and gene activity during development. Science 1975, 187, 226–232. [Google Scholar] [CrossRef] [PubMed]
- Klose, R.J.; Bird, A.P. Genomic DNA methylation: The mark and its mediators. Trends Biochem. Sci. 2006, 31, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Lyko, F. The DNA methyltransferase family: A versatile toolkit for epigenetic regulation. Nat. Rev. Genet. 2018, 19, 81–92. [Google Scholar] [CrossRef]
- Jin, B.; Robertson, K.D. DNA methyltransferases, DNA damage repair, and cancer. In Epigenetic Alterations in Oncogenesis; Karpf, A.R., Ed.; Springer: New York, NY, USA, 2013; pp. 3–29. [Google Scholar]
- Svedružić, Ž.M. Dnmt1: Structure and function. In Progress in Molecular Biology and Translational Science; Cheng, X., Blumenthal, R.M., Eds.; Academic Press: Cambridge, MA, USA, 2011; Volume 101, pp. 221–254. [Google Scholar]
- Chédin, F. The DNMT3 family of mammalian de novo DNA methyltransferases. In Progress in Molecular Biology and Translational Science; Cheng, X., Blumenthal, R.M., Eds.; Academic Press: Cambridge, MA, USA, 2011; Volume 101, pp. 255–285. [Google Scholar]
- Frye, M.; Harada, B.T.; Behm, M.; He, C. RNA modifications modulate gene expression during development. Science 2018, 361, 1346–1349. [Google Scholar] [CrossRef] [Green Version]
- Roignant, J.-Y.; Soller, M. m6A in mRNA: An ancient mechanism for fine-tuning gene expression. Trends Genet. 2017, 33, 380–390. [Google Scholar] [CrossRef] [PubMed]
- Chuang, J.C.; Jones, P.A. Epigenetics and microRNAs. Pediatric Res. 2007, 61, 24–29. [Google Scholar] [CrossRef]
- Wei, J.-W.; Huang, K.; Yang, C.; Kang, C.-S. Non-coding RNAs as regulators in epigenetics (review). Oncol. Rep. 2017, 37, 3–9. [Google Scholar] [CrossRef] [Green Version]
- Allis, C.D.; Caparros, M.-L.; Jenuwein, T.; Reinberg, D. Epigenetics, 2nd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Habor, NY, USA, 2015. [Google Scholar]
- Bonasio, R. Emerging topics in epigenetics: Ants, brains, and noncoding RNAs. Ann. N. Y. Acad. Sci. 2012, 1260, 14–23. [Google Scholar] [CrossRef]
- Gordon, J.A.R.; Grandy, R.A.; Lian, J.B.; Stein, J.L.; van Wijnen, A.J.; Stein, G.S. Chromatin. In Brenner’s Encyclopedia of Genetics, 2nd ed.; Maloy, S., Hughes, K., Eds.; Academic Press: San Diego, CA, USA, 2013; pp. 538–541. [Google Scholar]
- Morgan, H.D.; Santos, F.; Green, K.; Dean, W.; Reik, W. Epigenetic reprogramming in mammals. Hum. Mol. Genet. 2005, 14, R47–R58. [Google Scholar] [CrossRef] [Green Version]
- Peeters, C. The occurrence of sexual reproduction among ant workers. Biol. J. Linn. Soc. 1991, 44, 141–152. [Google Scholar] [CrossRef]
- Campbell, R.A.A.; Turner, G.C. The mushroom body. Curr. Biol. 2010, 20, R11–R12. [Google Scholar] [CrossRef] [Green Version]
- Bonasio, R.; Zhang, G.; Ye, C.; Mutti, N.S.; Fang, X.; Qin, N.; Donahue, G.; Yang, P.; Li, Q.; Li, C.; et al. Genomic comparison of the ants Camponotus floridanus and Harpegnathos saltator. Science 2010, 329, 1068–1071. [Google Scholar] [CrossRef] [Green Version]
- Gospocic, J.; Shields, E.J.; Glastad, K.M.; Lin, Y.; Penick, C.A.; Yan, H.; Mikheyev, A.S.; Linksvayer, T.A.; Garcia, B.A.; Berger, S.L.; et al. The neuropeptide corazonin controls social behavior and caste identity in ants. Cell 2017, 170, 748–759.e12. [Google Scholar] [CrossRef]
- Opachaloemphan, C.; Mancini, G.; Konstantinides, N.; Parikh, A.; Mlejnek, J.; Yan, H.; Reinberg, D.; Desplan, C. Early behavioral and molecular events leading to caste switching in the ant Harpegnathos. Genes Dev. 2021, 35, 410–424. [Google Scholar] [CrossRef] [PubMed]
- Chandra, V.; Fetter-Pruneda, I.; Oxley, P.R.; Ritger, A.L.; McKenzie, S.K.; Libbrecht, R.; Kronauer, D.J.C. Social regulation of insulin signaling and the evolution of eusociality in ants. Science 2018, 361, 398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Azevedo, S.V.; Hartfelder, K.; Amdam, G.V. Insulin-like peptides (AmILP1 and AmILP2) differentially affect female caste development in the honey bee (Apis mellifera L.). J. Exp. Biol. 2013, 216, 4347–4357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corona, M.; Velarde, R.A.; Remolina, S.; Moran-Lauter, A.; Wang, Y.; Hughes, K.A.; Robinson, G.E. Vitellogenin, juvenile hormone, insulin signaling, and queen honey bee longevity. Proc. Natl. Acad. Sci. USA 2007, 104, 7128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, M.; Liu, Y.; Wen, T.; Liu, W.; Gao, Q.; Zhao, J.; Xiong, Z.; Wang, Z.; Jiang, W.; Yu, Y.; et al. Chromatin accessibility and transcriptome landscapes of Monomorium pharaonis brain. Sci. Data 2020, 7, 217. [Google Scholar] [CrossRef]
- Nijhout, H.F.; Wheeler, D.E. Juvenile hormone and the physiological basis of insect polymorphisms. Q. Rev. Biol. 1982, 57, 109–133. [Google Scholar] [CrossRef]
- Wheeler, D.E. Developmental and physiological determinants of caste in social Hymenoptera: Evolutionary implications. Am. Nat. 1986, 128, 13–34. [Google Scholar] [CrossRef]
- Winston, M.L. The Biology of the Honey Bee; Harvard University Press: Cambridge, MA, USA, 1987. [Google Scholar]
- Whitfield, C.W.; Cziko, A.M.; Robinson, G.E. Gene expression profiles in the brain predict behavior in individual honey bees. Science 2003, 302, 296–299. [Google Scholar] [CrossRef] [PubMed]
- Liebig, J.; Hölldobler, B.; Peeters, C. Are ant workers capable of colony foundation? Naturwissenschaften 1998, 85, 133–135. [Google Scholar] [CrossRef]
- Penick, C.A.; Liebig, J.; Brent, C.S. Reproduction, dominance, and caste: Endocrine profiles of queens and workers of the ant Harpegnathos saltator. J. Comp. Physiol. A 2011, 197, 1063. [Google Scholar] [CrossRef] [PubMed]
- Hölldobler, B.; Wilson, E.O. The Ants; Belknap Press: Cambridge, MA, USA, 1990. [Google Scholar]
- Quadrana, L.; Colot, V. Plant transgenerational epigenetics. Annu. Rev. Genet. 2016, 50, 467–491. [Google Scholar] [CrossRef] [PubMed]
- Heard, E.; Martienssen, R.A. Transgenerational epigenetic inheritance: Myths and mechanisms. Cell 2014, 157, 95–109. [Google Scholar] [CrossRef] [Green Version]
- Bošković, A.; Rando, O.J. Transgenerational epigenetic inheritance. Annu. Rev. Genet. 2018, 52, 21–41. [Google Scholar] [CrossRef]
- Bonasio, R.; Tu, S.; Reinberg, D. Molecular signals of epigenetic states. Science 2010, 330, 612. [Google Scholar] [CrossRef] [Green Version]
- Yan, H.; Simola, D.F.; Bonasio, R.; Liebig, J.; Berger, S.L.; Reinberg, D. Eusocial insects as emerging models for behavioural epigenetics. Nat. Rev. Genet. 2014, 15, 677–688. [Google Scholar] [CrossRef]
- Opachaloemphan, C.; Yan, H.; Leibholz, A.; Desplan, C.; Reinberg, D. Recent advances in behavioral (epi)genetics in eusocial insects. Annu. Rev. Genet. 2018, 52, 489–510. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Liebig, J. Genetic basis of chemical communication in eusocial insects. Genes Dev. 2021, 35, 470–482. [Google Scholar] [CrossRef] [PubMed]
- Glastad, K.M.; Hunt, B.G.; Goodisman, M.A.D. Epigenetics in insects: Genome regulation and the generation of phenotypic diversity. Annu. Rev. Entomol. 2019, 64, 185–203. [Google Scholar] [CrossRef] [Green Version]
- Villagra, C.; Frías-Lasserre, D. Epigenetic molecular mechanisms in insects. Neotrop. Entomol. 2020, 49, 615–642. [Google Scholar] [CrossRef]
- Palli, S.R. Epigenetic regulation of post-embryonic development. Curr. Opin. Insect Sci. 2021, 43, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Toth, A.L.; Rehan, S.M. Molecular evolution of insect sociality: An eco-evo-devo perspective. Annu. Rev. Entomol. 2017, 62, 419–442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bewick, A.J.; Vogel, K.J.; Moore, A.J.; Schmitz, R.J. Evolution of DNA methylation across insects. Mol. Biol. Evol. 2016, 34, 654–665. [Google Scholar] [CrossRef] [Green Version]
- Bonasio, R.; Li, Q.; Lian, J.; Mutti, N.S.; Jin, L.; Zhao, H.; Zhang, P.; Wen, P.; Xiang, H.; Ding, Y.; et al. Genome-wide and caste-specific DNA methylomes of the ants Camponotus floridanus and Harpegnathos saltator. Curr. Biol. 2012, 22, 1755–1764. [Google Scholar] [CrossRef] [Green Version]
- Lyko, F.; Foret, S.; Kucharski, R.; Wolf, S.; Falckenhayn, C.; Maleszka, R. The honey bee epigenomes: Differential methylation of brain DNA in queens and workers. PLoS Biol. 2010, 8, e1000506. [Google Scholar] [CrossRef] [Green Version]
- Kucharski, R.; Maleszka, J.; Foret, S.; Maleszka, R. Nutritional control of reproductive status in honeybees via DNA methylation. Science 2008, 319, 1827–1830. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.F.; Wang, Y.; Zhang, W.X.; Liu, Z.G.; Xu, B.H.; Wang, H.F. Methionine as a methyl donor regulates caste differentiation in the European honey bee (Apis mellifera). Insect Sci. 2020. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Hou, L.; Zhu, D.; Xu, X.; An, S.; Wang, X. Identification and caste-dependent expression patterns of DNA methylation associated genes in Bombus terrestris. Sci. Rep. 2018, 8, 2332. [Google Scholar] [CrossRef] [PubMed]
- Marshall, H.; Lonsdale, Z.N.; Mallon, E.B. Methylation and gene expression differences between reproductive and sterile bumblebee workers. Evol. Lett. 2019, 3, 485–499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morandin, C.; Brendel, V.P.; Sundström, L.; Helanterä, H.; Mikheyev, A.S. Changes in gene DNA methylation and expression networks accompany caste specialization and age-related physiological changes in a social insect. Mol. Ecol. 2019, 28, 1975–1993. [Google Scholar] [CrossRef]
- Smith, C.R.; Mutti, N.S.; Jasper, W.C.; Naidu, A.; Smith, C.D.; Gadau, J. Patterns of DNA methylation in development, division of labor and hybridization in an ant with genetic caste determination. PLoS ONE 2012, 7, e42433. [Google Scholar] [CrossRef]
- Oldroyd, B.P.; Yagound, B. The role of epigenetics, particularly DNA methylation, in the evolution of caste in insect societies. Philos. Trans. R. Soc. B Biol. Sci. 2021, 376, 20200115. [Google Scholar] [CrossRef] [PubMed]
- Schulz, N.K.E.; Wagner, C.I.; Ebeling, J.; Raddatz, G.; Diddens-de Buhr, M.F.; Lyko, F.; Kurtz, J. Dnmt1 has an essential function despite the absence of CpG DNA methylation in the red flour beetle Tribolium Castaneum. Sci. Rep. 2018, 8, 16462. [Google Scholar] [CrossRef]
- Zhao, B.S.; Roundtree, I.A.; He, C. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 2017, 18, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Xiao, Y.; Li, Y.; Wang, X.; Qi, S.; Wang, Y.; Zhao, L.; Wang, K.; Peng, W.; Luo, G.-Z.; et al. RNA m6A modification functions in larval development and caste differentiation in honeybee (Apis mellifera). Cell Rep. 2021, 34, 108580. [Google Scholar] [CrossRef]
- Shinoda, T.; Itoyama, K. Juvenile hormone acid methyltransferase: A key regulatory enzyme for insect metamorphosis. Proc. Natl. Acad. Sci. USA 2003, 100, 11986. [Google Scholar] [CrossRef] [Green Version]
- Wojciechowski, M.; Lowe, R.; Maleszka, J.; Conn, D.; Maleszka, R.; Hurd, P.J. Phenotypically distinct female castes in honey bees are defined by alternative chromatin states during larval development. Genome Res. 2018, 28, 1532–1542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, C.; Rasheed, H.; Ran, Y.; Yang, X.; Xing, L.; Su, X. Transcriptome changes reveal the genetic mechanisms of the reproductive plasticity of workers in lower termites. BMC Genom. 2019, 20, 702. [Google Scholar] [CrossRef] [Green Version]
- Terrapon, N.; Li, C.; Robertson, H.M.; Ji, L.; Meng, X.; Booth, W.; Chen, Z.; Childers, C.P.; Glastad, K.M.; Gokhale, K.; et al. Molecular traces of alternative social organization in a termite genome. Nat. Commun. 2014, 5, 3636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheng, L.; Shields, E.J.; Gospocic, J.; Glastad, K.M.; Ratchasanmuang, P.; Berger, S.L.; Raj, A.; Little, S.; Bonasio, R. Social reprogramming in ants induces longevity-associated glia remodeling. Sci. Adv. 2020, 6, eaba9869. [Google Scholar] [CrossRef] [PubMed]
- Feldmeyer, B.; Elsner, D.; Foitzik, S. Gene expression patterns associated with caste and reproductive status in ants: Worker-specific genes are more derived than queen-specific ones. Mol. Ecol. 2014, 23, 151–161. [Google Scholar] [CrossRef]
- Paul, R.K.; Takeuchi, H.; Matsuo, Y.; Kubo, T. Gene expression of ecdysteroid-regulated gene E74 of the honeybee in ovary and brain. Insect Mol. Biol. 2005, 14, 9–15. [Google Scholar] [CrossRef]
- Martins, J.R.; Morais Franco Nunes, F.; Luz Paulino Simões, Z.; Maria Gentile Bitondi, M. A honeybee storage protein gene, hex 70a, expressed in developing gonads and nutritionally regulated in adult fat body. J. Insect Physiol. 2008, 54, 867–877. [Google Scholar] [CrossRef] [PubMed]
- Niu, D.; Zheng, H.; Corona, M.; Lu, Y.; Chen, X.; Cao, L.; Sohr, A.; Hu, F. Transcriptome comparison between inactivated and activated ovaries of the honey bee Apis Mellifera L. Insect Mol. Biol. 2014, 23, 668–681. [Google Scholar] [CrossRef]
- Grozinger, C.M.; Fan, Y.; Hoover, S.E.R.; Winston, M.L. Genome-wide analysis reveals differences in brain gene expression patterns associated with caste and reproductive status in honey bees (Apis mellifera). Mol. Ecol. 2007, 16, 4837–4848. [Google Scholar] [CrossRef]
- Judice, C.; Hartfelder, K.; Pereira, G.A.G. Caste-specific gene expression in the stingless bee Melipona quadrifasciata–are there common patterns in highly eusocial bees? Insectes Sociaux 2004, 51, 352–358. [Google Scholar] [CrossRef]
- Pereboom, J.J.M.; Jordan, W.C.; Sumner, S.; Hammond, R.L.; Bourke, A.F.G. Differential gene expression in queen–worker caste determination in bumble-bees. Proc. R. Soc. B Biol. Sci. 2005, 272, 1145–1152. [Google Scholar] [CrossRef]
- Sumner, S.; Pereboom, J.J.M.; Jordan, W.C. Differential gene expression and phenotypic plasticity in behavioural castes of the primitively eusocial wasp, Polistes canadensis. Proc. R. Soc. B Biol. Sci. 2006, 273, 19–26. [Google Scholar] [CrossRef] [Green Version]
- Hunt, B.G.; Goodisman, M.A.D. Evolutionary variation in gene expression is associated with dimorphism in eusocial Vespid wasps. Insect Mol. Biol. 2010, 19, 641–652. [Google Scholar] [CrossRef]
- Matsunami, M.; Nozawa, M.; Suzuki, R.; Toga, K.; Masuoka, Y.; Yamaguchi, K.; Maekawa, K.; Shigenobu, S.; Miura, T. Caste-specific microRNA expression in termites: Insights into soldier differentiation. Insect Mol. Biol. 2019, 28, 86–98. [Google Scholar] [CrossRef] [Green Version]
- Liu, F.; Peng, W.; Li, Z.; Li, W.; Li, L.; Pan, J.; Zhang, S.; Miao, Y.; Chen, S.; Su, S. Next-generation small RNA sequencing for microRNAs profiling in Apis mellifera: Comparison between nurses and foragers. Insect Mol. Biol. 2012, 21, 297–303. [Google Scholar] [CrossRef] [PubMed]
- Collins, D.H.; Mohorianu, I.; Beckers, M.; Moulton, V.; Dalmay, T.; Bourke, A.F.G. microRNAs associated with caste determination and differentiation in a primitively eusocial insect. Sci. Rep. 2017, 7, 45674. [Google Scholar] [CrossRef] [PubMed]
- Cardinal, S.; Danforth, B.N. The antiquity and evolutionary history of social behavior in bees. PLoS ONE 2011, 6, e21086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simola, D.F.; Ye, C.; Mutti, N.S.; Dolezal, K.; Bonasio, R.; Liebig, J.; Reinberg, D.; Berger, S.L. A chromatin link to caste identity in the carpenter ant Camponotus floridanus. Genome Res. 2013, 23, 486–496. [Google Scholar] [CrossRef] [Green Version]
- Simola, D.F.; Graham, R.J.; Brady, C.M.; Enzmann, B.L.; Desplan, C.; Ray, A.; Zwiebel, L.J.; Bonasio, R.; Reinberg, D.; Liebig, J.; et al. Epigenetic (re)programming of caste-specific behavior in the ant Camponotus floridanus. Science 2016, 351, aac6633. [Google Scholar] [CrossRef] [Green Version]
- Glastad, K.M.; Graham, R.J.; Ju, L.; Roessler, J.; Brady, C.M.; Berger, S.L. Epigenetic regulator CoREST controls social behavior in ants. Mol. Cell 2020, 77, 338–351.e336. [Google Scholar] [CrossRef]
- Fujioka, H.; Abe, M.S.; Fuchikawa, T.; Tsuji, K.; Shimada, M.; Okada, Y. Ant circadian activity associated with brood care type. Biol. Lett. 2017, 13, 20160743. [Google Scholar] [CrossRef]
- Fujioka, H.; Abe, M.S.; Okada, Y. Ant activity-rest rhythms vary with age and interaction frequencies of workers. Behav. Ecol. Sociobiol. 2019, 73, 30. [Google Scholar] [CrossRef]
- Libbrecht, R.; Nadrau, D.; Foitzik, S. A role of histone acetylation in the regulation of circadian rhythm in ants. iScience 2020, 23, 100846. [Google Scholar] [CrossRef] [Green Version]
- Etchegaray, J.-P.; Lee, C.; Wade, P.A.; Reppert, S.M. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature 2003, 421, 177–182. [Google Scholar] [CrossRef]
- Hosoda, H.; kato, K.; Asano, H.; Ito, M.; Kato, H.; Iwamoto, T.; Suzuki, A.; Masushige, S.; Kida, S. CBP/p300 is a cell type-specific modulator of CLOCK/BMAL1-mediated transcription. Mol. Brain 2009, 2, 34. [Google Scholar] [CrossRef] [Green Version]
- Mitaka, Y.; Tasaki, E.; Nozaki, T.; Fuchikawa, T.; Kobayashi, K.; Matsuura, K. Transcriptomic analysis of epigenetic modification genes in the termite Reticulitermes speratus. Insect Sci. 2020, 27, 202–211. [Google Scholar] [CrossRef] [PubMed]
- Kay, S.; Skowronski, D.; Hunt, B.G. Developmental DNA methyltransferase expression in the fire ant Solenopsis invicta. Insect Sci. 2018, 25, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Pozo, M.I.; Hunt, B.J.; Van Kemenade, G.; Guerra-Sanz, J.M.; Wäckers, F.; Mallon, E.B.; Jacquemyn, H. The effect of DNA methylation on bumblebee colony development. BMC Genom. 2021, 22, 73. [Google Scholar] [CrossRef] [PubMed]
- Kapheim, K.M.; Jones, B.M.; Pan, H.; Li, C.; Harpur, B.A.; Kent, C.F.; Zayed, A.; Ioannidis, P.; Waterhouse, R.M.; Kingwell, C.; et al. Developmental plasticity shapes social traits and selection in a facultatively eusocial bee. Proc. Natl. Acad. Sci. USA 2020, 117, 13615. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, R.; Yaguchi, H.; Maekawa, K. Histone modifying genes are involved in the molting period during soldier differentiation in Zootermopsis nevadensis. J. Insect Physiol. 2019, 117, 103892. [Google Scholar] [CrossRef] [PubMed]
- Cornette, R.; Gotoh, H.; Koshikawa, S.; Miura, T. Juvenile hormone titers and caste differentiation in the damp-wood termite Hodotermopsis sjostedti (Isoptera, Termopsidae). J. Insect Physiol. 2008, 54, 922–930. [Google Scholar] [CrossRef]
- Yan, H.; Bonasio, R.; Simola, D.F.; Liebig, J.; Berger, S.L.; Reinberg, D. DNA methylation in social insects: How epigenetics can control behavior and longevity. Annu. Rev. Entomol. 2015, 60, 435–452. [Google Scholar] [CrossRef] [PubMed]
- Kamakura, M. Royalactin induces queen differentiation in honeybees. Nature 2011, 473, 478–483. [Google Scholar] [CrossRef]
- Wang, K.; Liu, Z.G.; Pang, Q.; Zhang, W.W.; Chen, X.M.; Fan, R.L.; Yin, L.; Ji, T. Investigating the regulation of hypopharyngeal gland activity in honeybees (Apis mellifera carnica) under overwintering conditions via morphologic analysis combined with iTRAQ-based comparative proteomics. Ann. Entomol. Soc. Am. 2018, 111, 127–135. [Google Scholar] [CrossRef]
- Wang, K.; Liu, Z.G.; Lin, Z.G.; Yin, L.; Gao, F.C.; Chen, G.H.; Ji, T. Epigenetic modifications may regulate the activation of the hypopharyngeal gland of honeybees (Apis mellifera) during winter. Front. Genet. 2020, 11, 46. [Google Scholar] [CrossRef] [PubMed]
- Beros, S.; Scharf, I.; Lenhart, A.; Negroni, M.A.; Menzel, F.; Foitzik, S. Extreme lifespan extension in tapeworm-infected ant workers. Proc. R. Soc. B 2021. Submitted. [Google Scholar]
- Stoldt, M.; Klein, L.; Beros, S.; Butter, F.; Jongepier, E.; Feldmeyer, B.; Foitzik, S. Parasite presence induces gene expression changes in an ant host related to immunity and longevity. Genes 2021, 12, 95. [Google Scholar] [CrossRef]
- Carnes, M.U.; Campbell, T.; Huang, W.; Butler, D.G.; Carbone, M.A.; Duncan, L.H.; Harbajan, S.V.; King, E.M.; Peterson, K.R.; Weitzel, A.; et al. The genomic basis of postponed senescence in Drosophila melanogaster. PLoS ONE 2015, 10, e0138569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Penick, C.A.; Ghaninia, M.; Haight, K.L.; Opachaloemphan, C.; Yan, H.; Reinberg, D.; Liebig, J. Reversible plasticity in brain size, behaviour and physiology characterizes caste transitions in a socially flexible ant (Harpegnathos saltator). Proc. R. Soc. B Biol. Sci. 2021, 288, 20210141. [Google Scholar] [CrossRef]
- Smith, D.B.; Bernhardt, G.; Raine, N.E.; Abel, R.L.; Sykes, D.; Ahmed, F.; Pedroso, I.; Gill, R.J. Exploring miniature insect brains using micro-CT scanning techniques. Sci. Rep. 2016, 6, 21768. [Google Scholar] [CrossRef]
- Hartfelder, K.; Engels, W. Social insect polymorphism: Hormonal regulation of plasticity in development and reproduction in the honeybee. Curr. Top. Dev. Biol. 1998, 40, 45–77. [Google Scholar] [PubMed]
- Cardoso-Júnior, C.A.M.; Guidugli-Lazzarini, K.R.; Hartfelder, K. DNA methylation affects the lifespan of honey bee (Apis mellifera L.) workers–evidence for a regulatory module that involves vitellogenin expression but is independent of juvenile hormone function. Insect Biochem. Mol. Biol. 2018, 92, 21–29. [Google Scholar] [CrossRef]
- Ben-Shahar, Y.; Robichon, A.; Sokolowski, M.B.; Robinson, G.E. Influence of gene action across different time scales on behavior. Science 2002, 296, 741–744. [Google Scholar] [CrossRef]
- Liu, F.; Shi, T.; Qi, L.; Su, X.; Wang, D.; Dong, J.; Huang, Z.Y. lncRNA profile of Apis mellifera and its possible role in behavioural transition from nurses to foragers. BMC Genom. 2019, 20, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Weaver, I.C.G.; Cervoni, N.; Champagne, F.A.; D’Alessio, A.C.; Sharma, S.; Seckl, J.R.; Dymov, S.; Szyf, M.; Meaney, M.J. Epigenetic programming by maternal behavior. Nat. Neurosci. 2004, 7, 847–854. [Google Scholar] [CrossRef]
- Franklin, T.B.; Russig, H.; Weiss, I.C.; Gräff, J.; Linder, N.; Michalon, A.; Vizi, S.; Mansuy, I.M. Epigenetic transmission of the impact of early stress across generations. Biol. Psychiatry 2010, 68, 408–415. [Google Scholar] [CrossRef] [PubMed]
- Smedal, B.; Brynem, M.; Kreibich, C.D.; Amdam, G.V. Brood pheromone suppresses physiology of extreme longevity in honeybees. J. Exp. Biol. 2009, 212, 3795. [Google Scholar] [CrossRef] [Green Version]
- Huang, Z.Y.; Robinson, G.E. Honeybee colony integration: Worker-worker interactions mediate hormonally regulated plasticity in division of labor. Proc. Natl. Acad. Sci. USA 1992, 89, 11726–11729. [Google Scholar] [CrossRef] [Green Version]
- Huang, Z.-Y.; Robinson, G.E. Regulation of honey bee division of labor by colony age demography. Behav. Ecol. Sociobiol. 1996, 39, 147–158. [Google Scholar] [CrossRef]
- Eyer, M.; Dainat, B.; Neumann, P.; Dietemann, V. Social regulation of ageing by young workers in the honey bee, Apis mellifera. Exp. Gerontol. 2017, 87, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Amdam, G.V.; Rueppell, O.; Fondrk, M.K.; Page, R.E.; Nelson, C.M. The nurse’s load: Early-life exposure to brood-rearing affects behavior and lifespan in honey bees (Apis mellifera). Exp. Gerontol. 2009, 44, 467–471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amdam, G.V. Social context, stress, and plasticity of aging. Aging Cell 2011, 10, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Cardoso-Júnior, C.A.M.; Eyer, M.; Dainat, B.; Hartfelder, K.; Dietemann, V. Social context influences the expression of DNA methyltransferase genes in the honeybee. Sci. Rep. 2018, 8, 11076. [Google Scholar] [CrossRef]
- Yan, H.; Jafari, S.; Pask, G.; Zhou, X.; Reinberg, D.; Desplan, C. Evolution, developmental expression and function of odorant receptors in insects. J. Exp. Biol. 2020, 223, jeb208215. [Google Scholar] [CrossRef] [Green Version]
- Holman, L.; Trontti, K.; Helanterä, H. Queen pheromones modulate DNA methyltransferase activity in bee and ant workers. Biol. Lett. 2016, 12, 20151038. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Butler, C.G.; Fairey, E.M. The role of the queen in preventing oogenesis in worker honeybees. J. Apic. Res. 1963, 2, 14–18. [Google Scholar] [CrossRef]
- Hoover, S.E.R.; Keeling, C.I.; Winston, M.L.; Slessor, K.N. The effect of queen pheromones on worker honey bee ovary development. Naturwissenschaften 2003, 90, 477–480. [Google Scholar] [CrossRef] [PubMed]
- Vergoz, V.; Schreurs, H.A.; Mercer, A.R. Queen pheromone blocks aversive learning in young worker bees. Science 2007, 317, 384. [Google Scholar] [CrossRef]
- Fan, Y.; Richard, F.-J.; Rouf, N.; Grozinger, C.M. Effects of queen mandibular pheromone on nestmate recognition in worker honeybees, Apis mellifera. Anim. Behav. 2010, 79, 649–656. [Google Scholar] [CrossRef]
- Pankiw, T.; Huang, Z.Y.; Winston, M.L.; Robinson, G.E. Queen mandibular gland pheromone influences worker honey bee (Apis mellifera L.) foraging ontogeny and juvenile hormone titers. J. Insect Physiol. 1998, 44, 685–692. [Google Scholar] [CrossRef]
- Cardoso-Junior, C.A.M.; Ronai, I.; Hartfelder, K.; Oldroyd, B.P. Queen pheromone modulates the expression of epigenetic modifier genes in the brain of honeybee workers. Biol. Lett. 2020, 16, 20200440. [Google Scholar] [CrossRef] [PubMed]
- De Paula Junior, D.E.; de Oliveira, M.T.; Bruscadin, J.J.; Pinheiro, D.G.; Bomtorin, A.D.; Coelho Júnior, V.G.; Moda, L.M.R.; Simões, Z.L.P.; Barchuk, A.R. Caste-specific gene expression underlying the differential adult brain development in the honeybee Apis mellifera. Insect Mol. Biol. 2021, 30, 42–56. [Google Scholar] [CrossRef]
- Goldstein, A.Y.N.; Jan, Y.-N.; Luo, L. Function and regulation of Tumbleweed (RacGAP50C) in neuroblast proliferation and neuronal morphogenesis. Proc. Natl. Acad. Sci. USA 2005, 102, 3834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tejedor, F.; Zhu, X.R.; Kaltenbach, E.; Ackermann, A.; Baumann, A.; Canal, I.; Heisenberg, M.; Fischbach, K.F.; Pongs, O. Minibrain: A new protein kinase family involved in postembryonic neurogenesis in Drosophila. Neuron 1995, 14, 287–301. [Google Scholar] [CrossRef] [Green Version]
- Mutti, N.S.; Dolezal, A.G.; Wolschin, F.; Mutti, J.S.; Gill, K.S.; Amdam, G.V. IRS and TOR nutrient-signaling pathways act via juvenile hormone to influence honey bee caste fate. J. Exp. Biol. 2011, 214, 3977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biergans, S.D.; Claudianos, C.; Reinhard, J.; Galizia, C.G. DNA methylation mediates neural processing after odor learning in the honeybee. Sci. Rep. 2017, 7, 43635. [Google Scholar] [CrossRef] [Green Version]
- Biergans, S.D.; Claudianos, C.; Reinhard, J.; Galizia, C.G. DNA methylation adjusts the specificity of memories depending on the learning context and promotes relearning in honeybees. Front. Mol. Neurosci. 2016, 9, 82. [Google Scholar] [CrossRef] [Green Version]
- Biergans, S.D.; Jones, J.C.; Treiber, N.; Galizia, C.G.; Szyszka, P. DNA methylation mediates the discriminatory power of associative long-term memory in honeybees. PLoS ONE 2012, 7, e39349. [Google Scholar] [CrossRef]
- Biergans, S.D.; Giovanni Galizia, C.; Reinhard, J.; Claudianos, C. DNMTs and TET target memory-associated genes after appetitive olfactory training in honey bees. Sci. Rep. 2015, 5, 16223. [Google Scholar] [CrossRef] [Green Version]
- Zachepilo, T.G.; Lopatina, N.G. Methylation of histone H3 by lysine 4 in neurons of the mushroom bodies of the honeybee brain during memory formation. Cell Tissue Biol. 2020, 14, 270–274. [Google Scholar] [CrossRef]
- Alaux, C.; Sinha, S.; Hasadsri, L.; Hunt, G.J.; Guzmán-Novoa, E.; DeGrandi-Hoffman, G.; Uribe-Rubio, J.L.; Southey, B.R.; Rodriguez-Zas, S.; Robinson, G.E. Honey bee aggression supports a link between gene regulation and behavioral evolution. Proc. Natl. Acad. Sci. USA 2009, 106, 15400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cingolani, P.; Cao, X.; Khetani, R.S.; Chen, C.-C.; Coon, M.; Sammak, A.a.; Bollig-Fischer, A.; Land, S.; Huang, Y.; Hudson, M.E.; et al. Intronic non-CG DNA hydroxymethylation and alternative mRNA splicing in honey bees. BMC Genom. 2013, 14, 666. [Google Scholar] [CrossRef] [Green Version]
- Chandrasekaran, S.; Ament, S.A.; Eddy, J.A.; Rodriguez-Zas, S.L.; Schatz, B.R.; Price, N.D.; Robinson, G.E. Behavior-specific changes in transcriptional modules lead to distinct and predictable neurogenomic states. Proc. Natl. Acad. Sci. USA 2011, 108, 18020. [Google Scholar] [CrossRef] [Green Version]
- Rittschof, C.C.; Robinson, G.E. Manipulation of colony environment modulates honey bee aggression and brain gene expression. Genesbrainand Behav. 2013, 12, 802–811. [Google Scholar] [CrossRef] [Green Version]
- Rittschof, C.C.; Bukhari, S.A.; Sloofman, L.G.; Troy, J.M.; Caetano-Anollés, D.; Cash-Ahmed, A.; Kent, M.; Lu, X.; Sanogo, Y.O.; Weisner, P.A.; et al. Neuromolecular responses to social challenge: Common mechanisms across mouse, stickleback fish, and honey bee. Proc. Natl. Acad. Sci. USA 2014, 111, 17929–17934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li-Byarlay, H.; Rittschof, C.C.; Massey, J.H.; Pittendrigh, B.R.; Robinson, G.E. Socially responsive effects of brain oxidative metabolism on aggression. Proc. Natl. Acad. Sci. USA 2014, 111, 12533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shpigler, H.Y.; Saul, M.C.; Murdoch, E.E.; Cash-Ahmed, A.C.; Seward, C.H.; Sloofman, L.; Chandrasekaran, S.; Sinha, S.; Stubbs, L.J.; Robinson, G.E. Behavioral, transcriptomic and epigenetic responses to social challenge in honey bees. Genesbrain Behav. 2017, 16, 579–591. [Google Scholar] [CrossRef] [Green Version]
- Herb, B.R.; Shook, M.S.; Fields, C.J.; Robinson, G.E. Defense against territorial intrusion is associated with DNA methylation changes in the honey bee brain. BMC Genom. 2018, 19, 216. [Google Scholar] [CrossRef] [Green Version]
- Kapheim, K.M.; Jones, B.M.; Søvik, E.; Stolle, E.; Waterhouse, R.M.; Bloch, G.; Ben-Shahar, Y. Brain microRNAs among social and solitary bees. R. Soc. Open Sci. 2020, 7, 200517. [Google Scholar] [CrossRef]
- Itakura, S.; Hattori, K.; Umezawa, K. Identification and expression analysis of microRNAs in worker caste termites of Coptotermes formosanus and Reticulitermes speratus. J. Asia-Pac. Entomol. 2018, 21, 388–393. [Google Scholar] [CrossRef]
- Keleman, K.; Rajagopalan, S.; Cleppien, D.; Teis, D.; Paiha, K.; Huber, L.A.; Technau, G.M.; Dickson, B.J. Comm sorts robo to control axon guidance at the Drosophila midline. Cell 2002, 110, 415–427. [Google Scholar] [CrossRef] [Green Version]
- Rendić, D.; Sharrow, M.; Katoh, T.; Overcarsh, B.; Nguyen, K.; Kapurch, J.; Aoki, K.; Wilson, I.B.H.; Tiemeyer, M. Neural-specific α3-fucosylation of N-linked glycans in the Drosophila embryo requires fucosyltransferase A and influences developmental signaling associated with O-glycosylation. Glycobiology 2010, 20, 1353–1365. [Google Scholar] [CrossRef] [Green Version]
- Gómez-Skarmeta, J.-L.; del Corral, R.D.; de la Calle-Mustienes, E.; Ferrés-Marcó, D.; Modolell, J. Araucan and caupolican, two members of the novel iroquois complex, encode homeoproteins that control proneural and vein-forming genes. Cell 1996, 85, 95–105. [Google Scholar] [CrossRef] [Green Version]
- Mellerick, D.M.; Kassis, J.A.; Zhang, S.-D.; Odenwald, W.F. Castor encodes a novel zinc finger protein required for the development of a subset of CNS neurons in Drosophila. Neuron 1992, 9, 789–803. [Google Scholar] [CrossRef]
- Shields, E.J.; Sheng, L.; Weiner, A.K.; Garcia, B.A.; Bonasio, R. High-quality genome assemblies reveal long non-coding RNAs expressed in ant brains. Cell Rep. 2018, 23, 3078–3090. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Hore, T.A.; Reik, W. Reprogramming the methylome: Erasing memory and creating diversity. Cell Stem Cell 2014, 14, 710–719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guzman-Novoa, E.; Hunt, G.J.; Page, R.E., Jr.; Uribe-Rubio, J.L.; Prieto-Merlos, D.; Becerra-Guzman, F. Paternal effects on the defensive behavior of honeybees. J. Hered. 2005, 96, 376–380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Libbrecht, R.; Keller, L. Genetic compatability affects division of labor in the Argentine ant Linepithema humile. Evolution 2013, 67, 517–524. [Google Scholar] [CrossRef] [Green Version]
- Matsuura, K.; Mizumoto, N.; Kobayashi, K.; Nozaki, T.; Fujita, T.; Yashiro, T.; Fuchikawa, T.; Mitaka, Y.; Vargo, E.L. A genomic imprinting model of termite caste determination: Not genetic but epigenetic inheritance influences offspring caste fate. Am. Nat. 2018, 191, 677–690. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.; Li, G.; Li, C.; Zhang, J.; Wang, Q.; Simmons, D.K.; Chen, X.; Wijesena, N.; Zhu, W.; Wang, Z.; et al. Evolutionary transition between invertebrates and vertebrates via methylation reprogramming in embryogenesis. Natl. Sci. Rev. 2019, 6, 993–1003. [Google Scholar] [CrossRef]
- Harris, K.D.; Lloyd, J.P.B.; Domb, K.; Zilberman, D.; Zemach, A. DNA methylation is maintained with high fidelity in the honey bee germline and exhibits global non-functional fluctuations during somatic development. Epigenetics Chromatin 2019, 12, 62. [Google Scholar] [CrossRef] [PubMed]
- Yagound, B.; Duncan, M.; Chapman, N.C.; Oldroyd, B.P. Subfamily-dependent alternative reproductive strategies in worker honeybees. Mol. Ecol. 2017, 26, 6938–6947. [Google Scholar] [CrossRef] [PubMed]
- Yagound, B.; Smith, N.M.A.; Buchmann, G.; Oldroyd, B.P.; Remnant, E.J. Unique DNA methylation profiles are associated with cis-variation in honey bees. Genome Biol. Evol. 2019, 11, 2517–2530. [Google Scholar] [CrossRef] [PubMed]
- Yagound, B.; Remnant, E.J.; Buchmann, G.; Oldroyd, B.P. Intergenerational transfer of DNA methylation marks in the honey bee. Proc. Natl. Acad. Sci. USA 2020, 117, 32519–32527. [Google Scholar] [CrossRef]
- Bewick, A.J.; Sanchez, Z.; McKinney, E.C.; Moore, A.J.; Moore, P.J.; Schmitz, R.J. Dnmt1 is essential for egg production and embryo viability in the large milkweed bug, Oncopeltus fasciatus. Epigenet. Chromatin 2019, 12, 6. [Google Scholar] [CrossRef]
- Zhang, J.; Xing, Y.; Li, Y.; Yin, C.; Ge, C.; Li, F. DNA methyltransferases have an essential role in female fecundity in brown planthopper, Nilaparvata lugens. Biochem. Biophys. Res. Commun. 2015, 464, 83–88. [Google Scholar] [CrossRef]
- Zwier, M.V.; Verhulst, E.C.; Zwahlen, R.D.; Beukeboom, L.W.; van de Zande, L. DNA methylation plays a crucial role during early Nasonia development. Insect Mol. Biol. 2012, 21, 129–138. [Google Scholar] [CrossRef] [Green Version]
- Yan, H.; Opachaloemphan, C.; Mancini, G.; Yang, H.; Gallitto, M.; Mlejnek, J.; Leibholz, A.; Haight, K.; Ghaninia, M.; Huo, L.; et al. An engineered orco mutation produces aberrant social behavior and defective neural development in ants. Cell 2017, 170, 736–747 e739. [Google Scholar] [CrossRef]
- Sieber, K.; Saar, M.; Opachaloemphan, C.; Gallitto, M.; Yang, H.; Yan, H. Embryo injections for CRISPR-mediated mutagenesis in the ant Harpegnathos saltator. JoVE 2021, 168, e61930. [Google Scholar]
- Trible, W.; Olivos-Cisneros, L.; McKenzie, S.K.; Saragosti, J.; Chang, N.C.; Matthews, B.J.; Oxley, P.R.; Kronauer, D.J.C. orco mutagenesis causes loss of antennal lobe glomeruli and impaired social behavior in ants. Cell 2017, 170, 727–735.e10. [Google Scholar] [CrossRef] [PubMed]
- Chiu, Y.K.; Hsu, J.C.; Chang, T.; Huang, Y.C.; Wang, J. Mutagenesis mediated by CRISPR/Cas9 in the red imported fire ant, Solenopsis invicta. Insectes Sociaux 2020, 67, 317–326. [Google Scholar] [CrossRef]
- Kohno, H.; Suenami, S.; Takeuchi, H.; Sasaki, T.; Kubo, T. Production of knockout mutants by CRISPR/Cas9 in the European honeybee, Apis mellifera L. Zool. Sci. 2016, 33, 505–512. [Google Scholar] [CrossRef]
- Değirmenci, L.; Geiger, D.; Rogé Ferreira, F.L.; Keller, A.; Krischke, B.; Beye, M.; Steffan-Dewenter, I.; Scheiner, R. CRISPR/Cas 9-mediated mutations as a new tool for studying taste in honeybees. Chem. Senses 2020, 45, 655–666. [Google Scholar] [CrossRef]
- Hu, X.F.; Zhang, B.; Liao, C.H.; Zeng, Z.J. High-efficiency CRISPR/Cas9-mediated gene editing in honeybee (Apis mellifera) embryos. G3 Genes|Genomes|Genet. 2019, 9, 1759. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kohno, H.; Kubo, T. mKast is dispensable for normal development and sexual maturation of the male European honeybee. Sci. Rep. 2018, 8, 11877. [Google Scholar] [CrossRef]
- Roth, A.; Vleurinck, C.; Netschitailo, O.; Bauer, V.; Otte, M.; Kaftanoglu, O.; Page, R.E.; Beye, M. A genetic switch for worker nutrition-mediated traits in honeybees. PLoS Biol. 2019, 17, e3000171. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Traniello, I.M.; Rana, S.; Cash-Ahmed, A.C.; Sankey, A.L.; Yang, C.; Robinson, G.E. Neurodevelopmental and transcriptomic effects of CRISPR/Cas9-induced somatic orco mutation in honey bees. J. Neurogenet. 2021. [Google Scholar] [CrossRef]
- Schulte, C.; Theilenberg, E.; Müller-Borg, M.; Gempe, T.; Beye, M. Highly efficient integration and expression of piggyBac-derived cassettes in the honeybee (Apis mellifera). Proc. Natl. Acad. Sci. USA 2014, 111, 9003–9008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Otte, M.; Netschitailo, O.; Kaftanoglu, O.; Wang, Y.; Page, R.E., Jr.; Beye, M. Improving genetic transformation rates in honeybees. Sci. Rep. 2018, 8, 16534. [Google Scholar] [CrossRef]
- Holtzman, L.; Gersbach, C.A. Editing the epigenome: Reshaping the genomic landscape. Annu. Rev. Genom. Hum. Genet. 2018, 19, 43–71. [Google Scholar] [CrossRef]
- Nakamura, M.; Gao, Y.; Dominguez, A.A.; Qi, L.S. CRISPR technologies for precise epigenome editing. Nat. Cell Biol. 2021, 23, 11–22. [Google Scholar] [CrossRef] [PubMed]


| Term | Definition | Reference(s) |
|---|---|---|
| Inclusive fitness | A measurement of fitness in which the success of an animal is derived from the summation of an animal’s own reproductive fitness and of cooperative or altruistic behaviors exhibited by genetically similar individuals. | [2] |
| Eusociality | The highest degree of sociality exhibited by animals. Distinguished by overlapping generations in a colony, cooperative brood care, and division of labor. | [1] |
| Epigenetics | The study of changes in traits unrelated to changes in the genetic code. Such traits are mitotically heritable (through cell division). | [3,4,5] |
| Histone modification | The addition of an acetyl group, methyl group, phosphate group, or ubiquitin protein to histone proteins. | [6,7,8] |
| H3K27ac | Acetylation of histone H3 on lysine 27, a histone modification associated with transcriptional activation. | [9] |
| HAT | Histone acetyltransferase that transfers acetyl groups to lysine amino acids. | [7,10] |
| HDAC | Histone deacetylase for removal of acetyl groups from histones. | [11] |
| HDACi | Histone deacetylase inhibitors. | [12,13] |
| DNA methylation | Addition of a methyl group to a cytosine nucleotide. | [14,15,16] |
| DNMT family | The DNA methyltransferase family of proteins that are responsible for catalyzing DNA methylation. | [17,18] |
| DNMT1 | The maintenance DNA methyltransferase. | [19] |
| DNMT3 | The de novo DNA methyltransferase. | [20] |
| N6-methyladenosine | A form of RNA methylation, which has functions in RNA regulation. | [21,22] |
| miRNAs | microRNAs are non-coding RNAs of around 22 nucleotides in length. They suppress translation by binding to mRNA. | [23] |
| lncRNAs | Long non-coding RNAs are non-coding RNAs longer than 200 nucleotides. They have variable functions. | [24,25,26] |
| Chromatin | A complex of DNA and histone proteins which may be modified to be condensed or relaxed, thereby suppressing or promoting gene expression. | [27] |
| Epigenetic reprogramming | Erasure and rewriting of histone marks and DNA methylation. | [28] |
| Gamergate | A pseudoqueen: lack of queen pheromone in the colony induces workers to achieve reproductive status. | [29] |
| Mushroom body | The region of the insect brain responsible for olfactory and visual learning and memory functions. | [30] |
| IGF | Homolog of insulin-like growth factor in mammals, also called Ilp-1 in Apis mellifera and Ilp-2 in Harpegnathos saltator. | [31,32,33] |
| Ins | Homolog of mammalian insulin, also called Ilp-1 in Harpegnathos saltator, Ilp-2 in Apis mellifera and Ooceraea biroi, and LIRP in Monomorium pharaonis. | [31,32,33,34,35,36,37] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sieber, K.R.; Dorman, T.; Newell, N.; Yan, H. (Epi)Genetic Mechanisms Underlying the Evolutionary Success of Eusocial Insects. Insects 2021, 12, 498. https://doi.org/10.3390/insects12060498
Sieber KR, Dorman T, Newell N, Yan H. (Epi)Genetic Mechanisms Underlying the Evolutionary Success of Eusocial Insects. Insects. 2021; 12(6):498. https://doi.org/10.3390/insects12060498
Chicago/Turabian StyleSieber, Kayli R., Taylor Dorman, Nicholas Newell, and Hua Yan. 2021. "(Epi)Genetic Mechanisms Underlying the Evolutionary Success of Eusocial Insects" Insects 12, no. 6: 498. https://doi.org/10.3390/insects12060498
APA StyleSieber, K. R., Dorman, T., Newell, N., & Yan, H. (2021). (Epi)Genetic Mechanisms Underlying the Evolutionary Success of Eusocial Insects. Insects, 12(6), 498. https://doi.org/10.3390/insects12060498






