Immune Response of Eastern Honeybee Worker to Nosema ceranae Infection Revealed by Transcriptomic Investigation
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Transcriptome Data Source
2.2. Confirmation of Effective Infection of Eastern Honeybee Workers by N. ceranae
2.3. Identification and Analysis of DEGs
2.4. GO Categorization and KEGG Pathway Enrichment Analysis of DEGs
2.5. Construction and Analysis of Immune-Associated Regulatory Network of DEGs
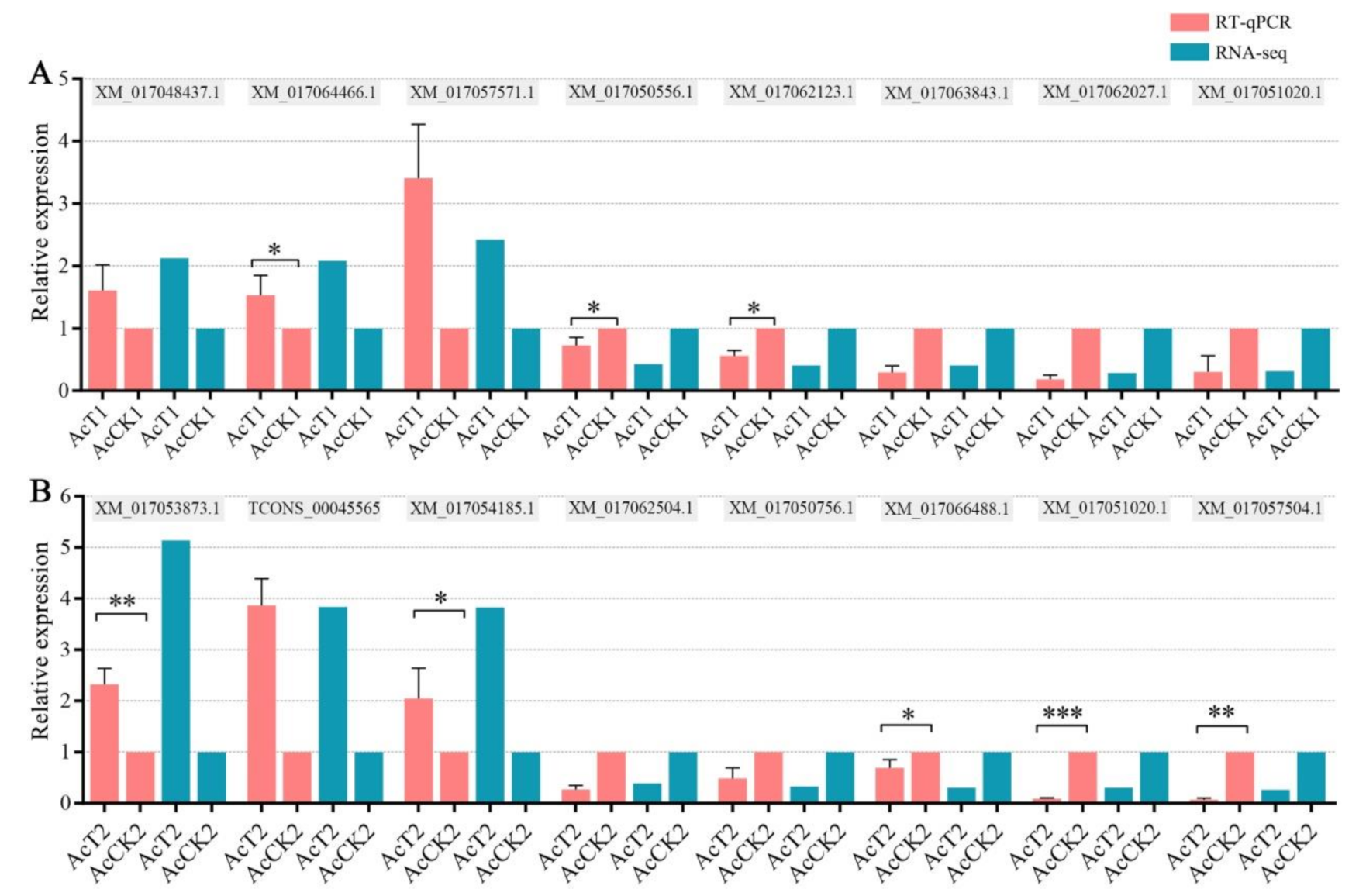
2.6. RT-qPCR Validation of DEGs
2.7. Statistical Analysis
3. Results
3.1. Effective Infection of A. c. cerana Worker by N. ceranae
3.2. DEGs Involved in A. c. cerana Workers’ Midgut Response to N. ceranae Infection
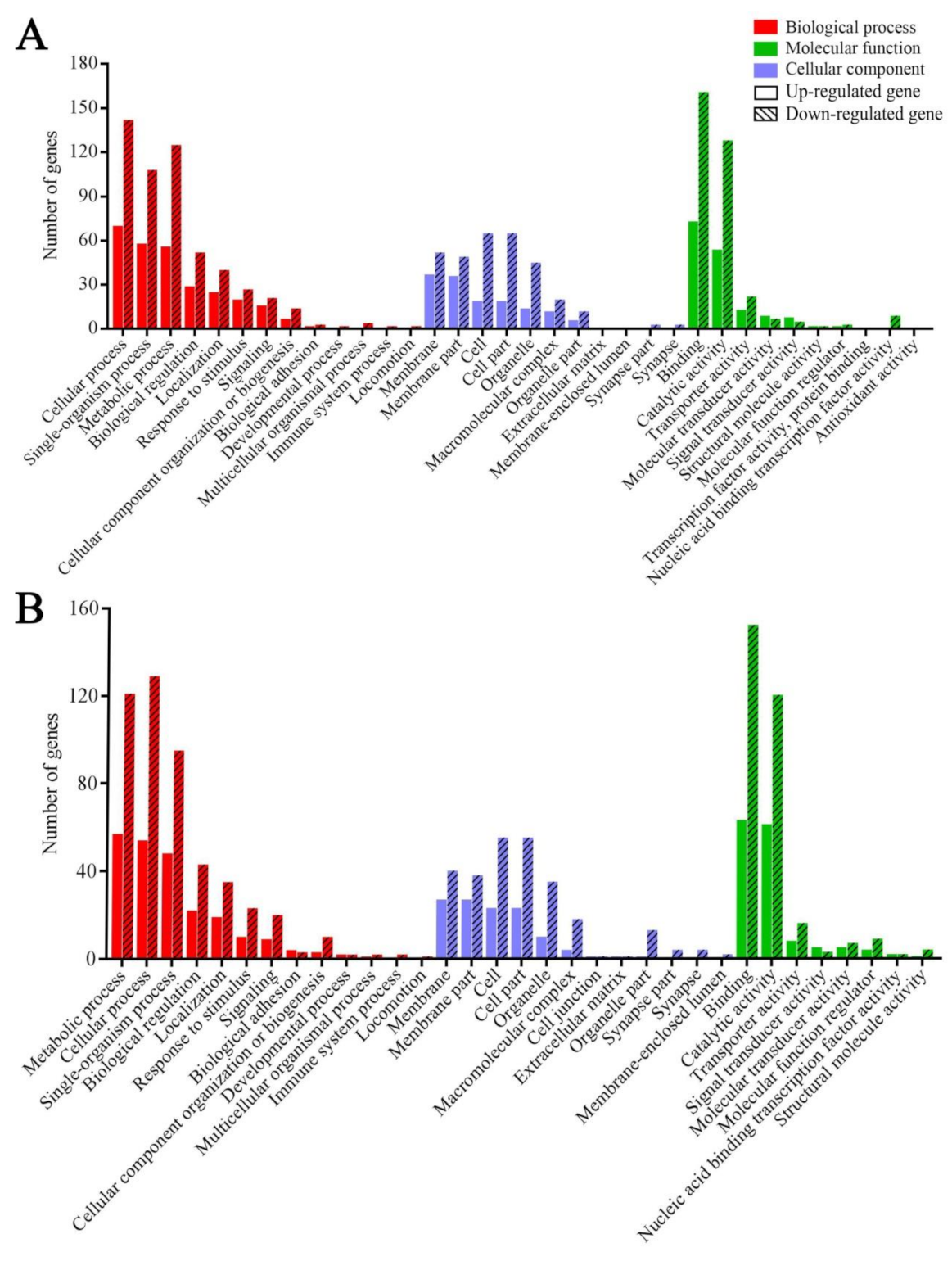
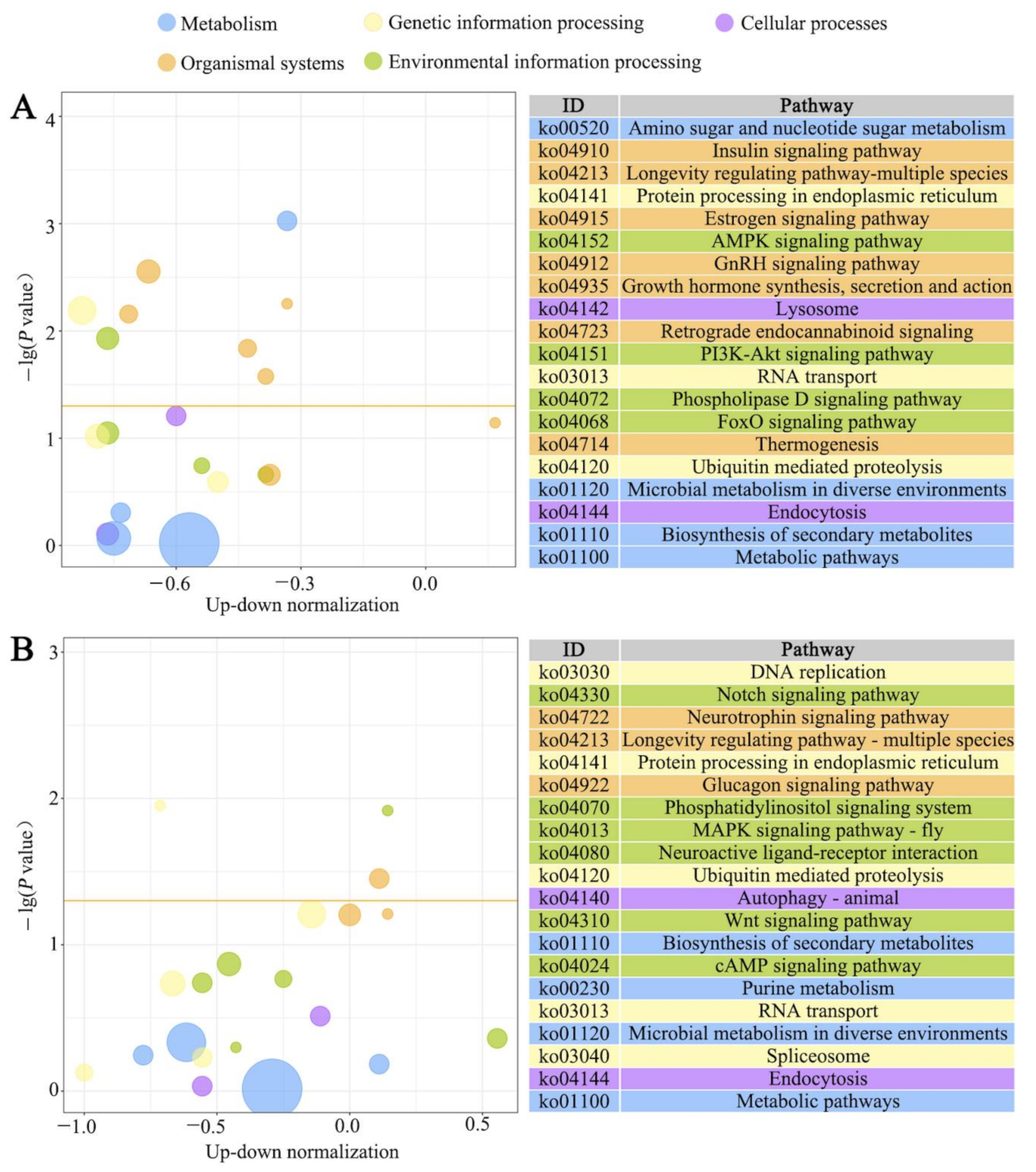
3.3. GO Terms and KEGG Pathways Enriched by Host DEGs
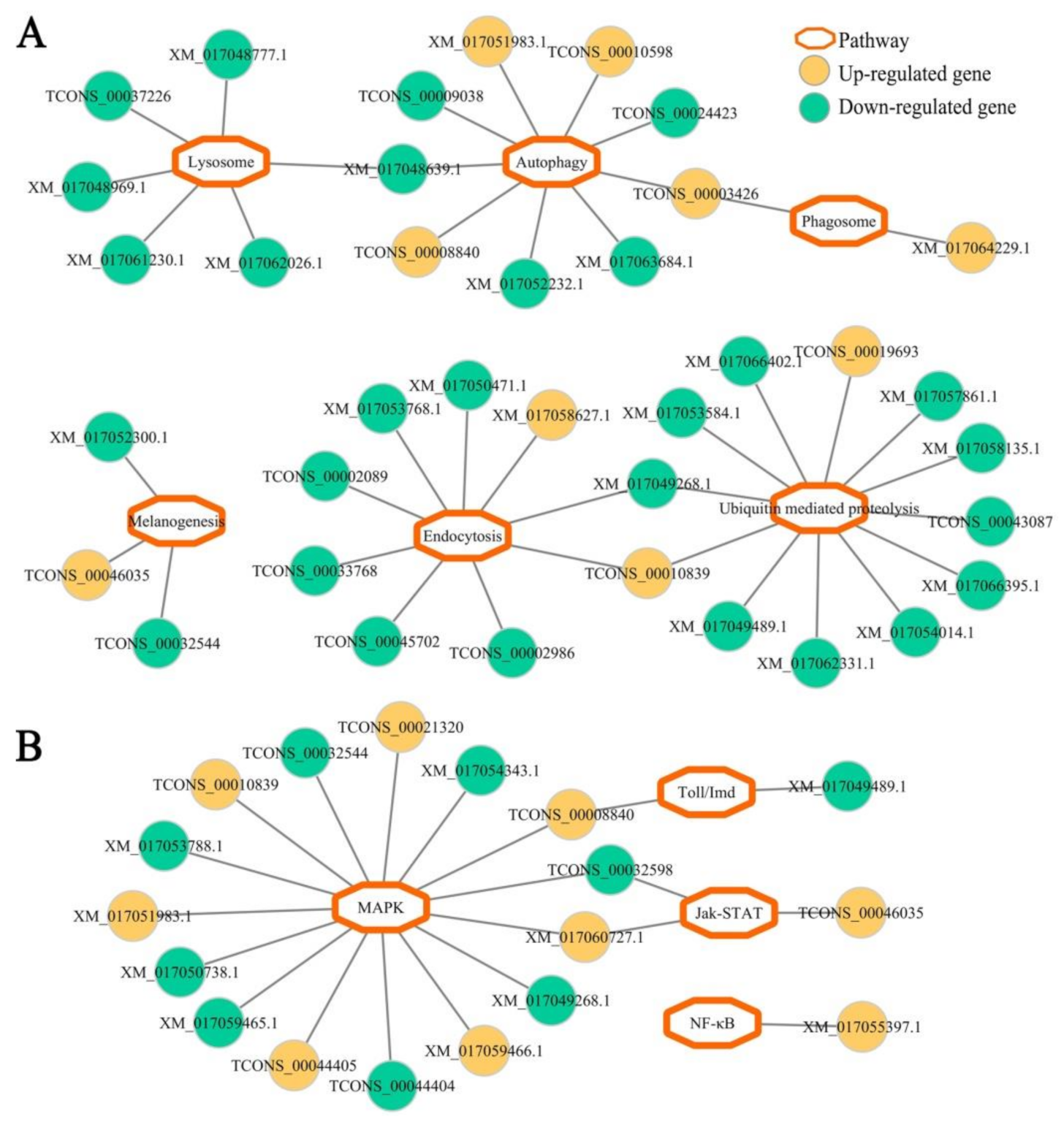
3.4. Host DEGs Relevant to Cellular and Humoral Immune Pathways
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Calderone, N.W. Insect pollinated crops, insect pollinators and US agriculture: Trend analysis of aggregate data for the period 1992–2009. PLoS ONE 2012, 7, e37235. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Xiao, Y.; Li, Y.; Wang, X.; Qi, S.; Wang, Y.; Zhao, L.; Wang, K.; Peng, W.; Luo, G.Z.; et al. RNA m6A modification functions in larval development and caste differentiation in honeybee (Apis mellifera). Cell Rep. 2021, 34, 108580. [Google Scholar] [CrossRef]
- Zayed, A.; Robinson, G.E. Understanding the relationship between brain gene expression and social behavior: Lessons from the honey bee. Annu. Rev. Genet. 2012, 46, 591–615. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.V.; Steinhauer, N.; Rennich, K.; Wilson, M.E.; Tarpy, D.R.; Caron, D.M. A national survey of managed honey bee 2013–2014 annual colony losses in the USA. Apidologie 2015, 46, 292–305. [Google Scholar] [CrossRef] [Green Version]
- Foret, S.; Kucharski, R.; Pellegrini, M.; Feng, S.; Jacobsen, S.E.; Robinson, G.E.; Maleszka, R. DNA methylation dynamics, metabolic fluxes, gene splicing, and alternative phenotypes in honey bees. Proc. Natl. Acad. Sci. USA 2012, 109, 4968–4973. [Google Scholar] [CrossRef] [Green Version]
- Li-Byarlay, H.; Li, Y.; Stroud, H.; Feng, S.; Newman, T.C.; Kaneda, M.; Hou, K.K.; Worley, K.C.; Elsik, C.G.; Wickline, S.A.; et al. RNA interference knockdown of DNA methyltransferase 3 affects gene alternative splicing in the honey bee. Proc. Natl. Acad. Sci. USA 2013, 110, 12750–12755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schulte, C.; Theilenberg, E.; Müller-Borg, M.; Gempe, T.; Beye, M. Highly efficient integration and expression of piggyBac-derived cassettes in the honeybee (Apis mellifera). Proc. Natl. Acad. Sci. USA. 2014, 111, 9003–9008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, X.F.; Zhang, B.; Liao, C.H.; Zeng, Z.J. High-efficiency CRISPR/Cas9-mediated gene editing in honeybee (Apis mellifera) embryos. G3 Genes Genom. Genet. 2019, 9, 1759–1766. [Google Scholar] [CrossRef] [Green Version]
- Higes, M.; García-Palencia, P.; Urbieta, A.; Nanetti, A.; Martín-Hernández, R. Nosema apis and Nosema ceranae tissue tropism in worker honey bees (Apis mellifera). Vet. Pathol. 2020, 57, 132–138. [Google Scholar] [CrossRef] [PubMed]
- Martín-Hernández, R.; Bartolomé, C.; Chejanovsky, N.; Le Conte, Y.; Dalmon, A.; Dussaubat, C.; García-Palencia, P.; Meana, A.; Pinto, M.A.; Soroker, V.; et al. Nosema ceranae in Apis mellifera: A 12 years postdetection perspective. Environ. Microbiol. 2018, 20, 1302–1329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fries, I.; Feng, F.; da Silva, A.; Slemenda, S.B.; Pieniazek, N.J. Nosema ceranae n. sp. (Microspora, Nosematidae), morphological and molecular characterization of a microsporidian parasite of the Asian honey bee Apis cerana (Hymenoptera, Apidae). Eur. J. Protistol. 1996, 32, 356–365. [Google Scholar] [CrossRef]
- Klee, J.; Besana, A.M.; Genersch, E.; Gisder, S.; Nanetti, A.; Tam, D.Q.; Chinh, T.X.; Puerta, F.; Ruz, J.M.; Kryger, P.; et al. Widespread dispersal of the microsporidian Nosema ceranae, an emergent pathogen of the western honey bee, Apis mellifera. J. Invertebr. Pathol. 2007, 96, 1–10. [Google Scholar] [CrossRef]
- Gisder, S.; Schüler, V.; Horchler, L.L.; Groth, D.; Genersch, E. Long-term temporal trends of Nosema spp. infection prevalence in Northeast Germany: Continuous spread of Nosema ceranae, an emerging pathogen of honey bees (Apis mellifera), but no general replacement of Nosema apis. Front. Cell. Infect Microbiol. 2017, 7, 301. [Google Scholar] [CrossRef] [Green Version]
- Paris, L.; El Alaoui, H.; Delbac, F.; Diogon, M. Effects of the gut parasite Nosema ceranae on honey bee physiology and behavior. Curr. Opin. Insect Sci. 2018, 26, 149–154. [Google Scholar] [CrossRef]
- Holt, H.L.; Aronstein, K.A.; Grozinger, C.M. Chronic parasitization by Nosema microsporidia causes global expression changes in core nutritional, metabolic and behavioral pathways in honey bee workers (Apis mellifera). BMC Genom. 2013, 14, 799. [Google Scholar] [CrossRef] [Green Version]
- Dussaubat, C.; Sagastume, S.; Gómez-Moracho, T.; Botías, C.; García-Palencia, P.; Martín-Hernández, R.; Le Conte, Y.; Higes, M. Comparative study of Nosema ceranae (Microsporidia) isolates from two different geographic origins. Vet. Microbiol. 2013, 162, 670–678. [Google Scholar] [CrossRef] [PubMed]
- García-Palencia, P.; Martín-Hernández, R.; González-Porto, A.; Marin, P.; Meana, A.; Higes, M. Natural infection by Nosema ceranae causes similar lesions as in experimentally infected caged-workers honey bees (Apis mellifera). J. Apic. Res. 2010, 49, 278–283. [Google Scholar] [CrossRef]
- Mayack, C.; Naug, D. Energetic stress in the honeybee Apis mellifera from Nosema ceranae infection. J. Invertebr. Pathol. 2009, 100, 185–188. [Google Scholar] [CrossRef] [PubMed]
- Higes, M.; Juarranz, Á.; Dias-Almeida, J.; Lucena, S.; Botías, C.; Meana, A.; García-Palencia, P.; Martín-Hernández, R. Apoptosis in the pathogenesis of Nosema ceranae (Microsporidia: Nosematidae) in honey bees (Apis mellifera). Environ. Microbiol. Rep. 2013, 5, 530–536. [Google Scholar] [CrossRef] [PubMed]
- Kurze, C.; Le Conte, Y.; Dussaubat, C.; Erler, S.; Kryger, P.; Lewkowski, O.; Müller, T.; Widder, M.; Moritz, R.F. Nosema tolerant honeybees (Apis mellifera) escape parasitic manipulation of apoptosis. PLoS ONE 2015, 10, e0140174. [Google Scholar] [CrossRef]
- Kurze, C.; Le Conte, Y.; Kryger, P.; Lewkowski, O.; Müller, T.; Moritz, R. Infection dynamics of Nosema ceranae in honey bee midgut and host cell apoptosis. J. Invertebr. Pathol. 2018, 154, 1–4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antúnez, K.; Martín-Hernández, R.; Prieto, L.; Meana, A.; Zunino, P.; Higes, M. Immune suppression in the honey bee (Apis mellifera) following infection by Nosema ceranae (Microsporidia). Environ. Microbiol. 2009, 11, 2284–2290. [Google Scholar] [CrossRef] [PubMed]
- Chaimanee, V.; Chantawannakul, P.; Chen, Y.; Evans, J.D.; Pettis, J.S. Differential expression of immune genes of adult honey bee (Apis mellifera) after inoculated by Nosema ceranae. J. Insect. Physiol. 2012, 58, 1090–1095. [Google Scholar] [CrossRef]
- Osta, M.A.; Christophides, G.K.; Vlachou, D.; Kafatos, F.C. Innate immunity in the malaria vector Anopheles gambiae: Comparative and functional genomics. J. Exp. Biol. 2004, 207, 2551–2563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Casteels-Josson, K.; Zhang, W.; Capaci, T.; Casteels, P.; Tempst, P. Acute transcriptional response of the honeybee peptide-antibiotics gene repertoire and required post-translational conversion of the precursor structures. J. Biol. Chem. 1994, 269, 28569–28575. [Google Scholar] [CrossRef]
- Garrido, P.M.; Porrini, M.P.; Antúnez, K.; Branchiccela, B.; Martínez-Noël, G.M.; Zunino, P.; Salerno, G.; Eguaras, M.J.; Ieno, E. Sublethal effects of acaricides and Nosema ceranae infection on immune related gene expression in honeybees. Vet. Res. 2016, 47, 51. [Google Scholar] [CrossRef] [Green Version]
- Sinpoo, C.; Paxton, R.J.; Disayathanoowat, T.; Krongdang, S.; Chantawannakul, P. Impact of Nosema ceranae and Nosema apis on individual worker bees of the two host species (Apis cerana and Apis mellifera) and regulation of host immune response. J. Insect Physiol. 2018, 105, 1–8. [Google Scholar] [CrossRef]
- Aufauvre, J.; Misme-Aucouturier, B.; Vigues, B.; Texier, C.; Delbac, F.; Blot, N. Transcriptome analyses of the honeybee response to Nosema ceranae and insecticides. PLoS ONE 2014, 9, e91686. [Google Scholar] [CrossRef]
- Dussaubat, C.; Brunet, J.L.; Higes, M.; Colbourne, J.K.; Lopez, J.; Choi, J.H.; MartinHernandez, R.; Botias, C.; Cousin, M.; McDonnell, C.; et al. Gut pathology and responses to the microsporidium Nosema ceranae in the honey bee Apis mellifera. PLoS ONE 2012, 7, e37017. [Google Scholar] [CrossRef] [Green Version]
- Fu, Z.M.; Chen, H.Z.; Liu, S.Y.; Zhu, Z.W.; Fan, X.X.; Fan, Y.C.; Wan, J.Q.; Zhang, L.; Xiong, C.L.; Xu, G.J.; et al. Immune responses of Apis mellifera ligustica to Nosema ceranae stress. Sci. Agric. Sin. 2019, 52, 3069–3082. [Google Scholar]
- Geng, S.H.; Zhou, D.D.; Fan, X.X.; Jiang, H.B.; Zhu, Z.W.; Wang, J.; Fan, Y.C.; Wang, X.R.; Xiong, C.L.; Zheng, Y.Z.; et al. Transcriptome analysis reveals the molecular mechanism underlying Nosema ceranae infection of Apis mellifera ligustica. Acta Entomol. Sin. 2020, 63, 294–308. [Google Scholar]
- Radlof, S.E.; Hepburn, C.; Hepburn, H.R.; Fuchs, S.; Hadisoesilo, S.; Tan, K.; Engel, M.S.; Kuznetsov, V. Population structure and classifcation of Apis cerana. Apidologie 2010, 41, 589–601. [Google Scholar] [CrossRef] [Green Version]
- Brown, M.J.F. Oldroyd Benjamin Siriwat Wongsiri Asian honey bees. Biology, Conservation, and Human Interactions. Harvard University Press Cambridge, Massachusetts Pp. Anim. Behav. 2006, 73, 553–554. [Google Scholar] [CrossRef]
- Peng, Y.S.; Fang, Y.Z.; Xu, S.Y.; Ge, L.S. The resistance mechanism of the Asian honey bee, Apis cerana Fabr., to an ectoparasitic mite, Varroa jacobsoni Oudemans. J. Invertebr. Pathol. 1987, 49, 54–60. [Google Scholar] [CrossRef]
- Xu, P.; Shi, M.; Chen, X.X. Antimicrobial peptide evolution in the Asiatic honey bee Apis cerana. PLoS ONE 2009, 4, e4239. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Qin, M.; Yang, L.; Song, Z.; Luo, L.; Bao, H.; Ma, Z.; Zhou, Z.; Xu, J. A genome-wide analysis of simple sequence repeats in Apis cerana and its development as polymorphism markers. Gene 2017, 599, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Ono, M.; Okada, I.; Sasaki, M. Heat protection by balling in the Japanese honeybee Apis cerana japonica as a defensive behavior against the hornet, Vespa simillima xanthoptera (Hymenoptera: Vespidae). Experientia 1987, 43, 1031–1034. [Google Scholar] [CrossRef]
- Chen, D.; Du, Y.; Chen, H.; Fan, Y.; Fan, X.; Zhu, Z.; Wang, J.; Xiong, C.; Zheng, Y.; Hou, C.; et al. Comparative identification of micrornas in Apis cerana cerana workers’ midguts in response to Nosema ceranae invasion. Insects 2019, 10, 258. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Evans, J.D.; Smith, I.B.; Pettis, J.S. Nosema ceranae is a long-present and wide-spread microsporidian infection of the European honey bee (Apis mellifera) in the United States. J. Invertebr. Pathol. 2008, 97, 186–188. [Google Scholar] [CrossRef]
- Benjeddou, M.; Leat, N.; Allsopp, M.; Davison, S. Detection of acute bee paralysis virus and black queen cell virus from honeybees by reverse transcriptase pcr. Appl. Environ. Microbiol. 2001, 67, 2384–2387. [Google Scholar] [CrossRef] [Green Version]
- Genersch, E. Development of a rapid and sensitive RT-PCR method for the detection of deformed wing virus, a pathogen of the honeybee (Apis mellifera). Vet. J. 2005, 169, 121–123. [Google Scholar] [CrossRef] [PubMed]
- Ribiere, M.; Triboulot, C.; Mathieu, L.; Aurieres, C.; Faucon, J.P.; Pepin, M. Molecular diagnosis of chronic bee paralysis virus infection. Apidologie 2002, 33, 339–351. [Google Scholar] [CrossRef]
- Stoltz, D.; Shen, X.R.; Boggis, C.; Sisson, G. Molecular diagnosis of Kashmir bee virus infection. J. Apic. Res. 1995, 34, 153–160. [Google Scholar] [CrossRef]
- Singh, R.; Levitt, A.L.; Rajotte, E.G.; Holmes, E.C.; Ostiguy, N.; Vanengelsdorp, D.; Lipkin, W.I.; Depamphilis, C.W.; Toth, A.L.; Cox-Foster, D.L. RNA viruses in hymenopteran pollinators: Evidence of inter-Taxa virus transmission via pollen and potential impact on non-Apis hymenopteran species. PLoS ONE 2010, 5, e14357. [Google Scholar] [CrossRef]
- Fu, Z.M.; Zhou, D.D.; Chen, H.Z.; Geng, S.H.; Chen, D.F.; Zheng, Y.Z.; Xiong, C.L.; Xu, G.J.; Zhang, X.; Guo, R. Analysis of highly expressed genes in Apis cerana cerana workers’ midguts responding to Nosema ceranae stress. J. Sichuan Univ. (Nat. Sci. Ed.) 2020, 57, 191–198. [Google Scholar]
- Forsgren, E.; Fries, I. Comparative virulence of Nosema ceranae and Nosema apis in individual European honey bees. Vet. Parasitol. 2010, 170, 212–217; [Google Scholar] [CrossRef]
- Chen, D.; Chen, H.; Du, Y.; Zhou, D.; Geng, S.; Wang, H.; Wan, J.; Xiong, C.; Zheng, Y.; Guo, R. Genome-wide identification of long non-coding RNAs and their regulatory networks involved in Apis mellifera ligustica response to Nosema ceranae infection. Insects 2019, 10, 245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, X.; Kim, H.; Zhong, S.; Chen, H.; Hu, Z.; Zhou, B. De novo transcriptome assembly for rudimentary leaves in Litchi chinesis Sonn. and identification of differentially expressed genes in response to reactive oxygen species. BMC Genom. 2014, 15, 805. [Google Scholar] [CrossRef] [Green Version]
- Ye, J.; Fang, L.; Zheng, H.; Zhang, Y.; Chen, J.; Zhang, Z.; Wang, J.; Li, S.; Li, R.; Bolund, L.; et al. WEGO: A web tool for plotting GO annotations. Nucleic Acids Res. 2006, 34, W293–W297. [Google Scholar] [CrossRef]
- Du, Y.; Zhou, D.D.; Wan, J.Q.; Lu, J.X.; Fan, X.X.; Fan, Y.C.; Chen, H.; Xiong, C.L.; Zheng, Y.Z.; Fu, Z.M.; et al. Profiling and regulation network of differentially expressed genes during the development process of Apis mellifera ligustica worker’s midgut. Sci. Agric. Sin. 2020, 53, 201–212. [Google Scholar]
- Su, G.; Morris, J.H.; Demchak, B.; Bader, G.D. Biological network exploration with Cytoscape 3. Curr. Protoc. Bioinform. 2014, 47, 8–13. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Anaysis of relative gene expression data using realtime quantitative PCR and the 2-ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Huang, Q.; Chen, Y.P.; Wang, R.W.; Cheng, S.; Evans, J.D. Host-parasite interactions and purifying selection in a microsporidian parasite of honey bees. PLoS ONE 2016, 11, e0147549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, W.F.; Solter, L.F. Comparative development and tissue tropism of Nosema apis and Nosema ceranae. J. Invertebr. Pathol. 2013, 113, 35–41. [Google Scholar] [CrossRef]
- Higes, M.; García-Palencia, P.; Martín-Hernández, R.; Meana, A. Experimental infection of Apis mellifera honeybees with Nosema ceranae (Microsporidia). J. Invertebr. Pathol. 2007, 94, 211–217. [Google Scholar] [CrossRef] [PubMed]
- Vidau, C.; Panek, J.; Texier, C.; Biron, D.G.; Belzunces, L.P.; Le Gall, M.; Broussard, C.; Delbac, F.; El Alaoui, H. Differential proteomic analysis of midguts from Nosema ceranae-infected honeybees reveals manipulation of key host functions. J. Invertebr. Pathol. 2014, 121, 89–96. [Google Scholar] [CrossRef]
- Panek, J.; Paris, L.; Roriz, D.; Mone, A.; Dubuffet, A.; Delbac, F.; Diogon, M.; El Alaoui, H. Impact of the microsporidian Nosema ceranae on the gut epithelium renewal of the honeybee, Apis mellifera. J. Invertebr. Pathol. 2018, 159, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Evans, J.D.; Aronstein, K.; Chen, Y.P.; Hetru, C.; Imler, J.L.; Jiang, H.; Kanost, M.; Thompson, G.J.; Zou, Z.; Hultmark, D. Immune pathways and defence mechanisms in honey bees Apis mellifera. Insect Mol. Biol. 2006, 15, 645–656. [Google Scholar] [CrossRef] [Green Version]
- Govind, S. Innate immunity in Drosophila: Pathogens and pathways. Insect Sci. 2008, 15, 29–43. [Google Scholar] [CrossRef]
- Aronstein, K.A.; Murray, K.D. Chalkbrood disease in honey bees. J. Invertebr. Pathol. 2010, 103, S20–S29. [Google Scholar] [CrossRef]
- Glinski, Z.; Jarosz, J. Infection and immunity in the honey bee Apis mellifera. Apiacta 2001, 36, 12–24. [Google Scholar]
- Glinski, Z.; Buczek, K. Response of the Apoidea to fungal infections. Apiacta 2003, 38, 183–189. [Google Scholar]
- Xu, Z.; Zhu, L.; Yang, Y.; Zhang, Y.; Lu, M.; Tao, L.; Xu, W. Bifenthrin induces DNA damage and autophagy in Spodoptera frugiperda (Sf9) insect cells. In Vitro Cell. Dev. Biol. Anim. 2021, 57, 264–271. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Klionsky, D.J. Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 2009, 43, 67–93. [Google Scholar] [CrossRef] [Green Version]
- Nakagawa, I.; Amano, A.; Mizushima, N.; Yamamoto, A.; Yamaguchi, H.; Kamimoto, T.; Nara, A.; Funao, J.; Nakata, M.; Tsuda, K.; et al. Autophagy defends cells against invading group a Streptococcus. Science 2004, 306, 1037–1040. [Google Scholar] [CrossRef]
- Orvedahl, A.; MacPherson, S.; Sumpter, R.J.; Tallóczy, Z.; Zou, Z.; Levine, B. Autophagy protects against Sindbis virus infection of the central nervous system. Cell Host Microbe. 2010, 7, 115–127. [Google Scholar] [CrossRef] [Green Version]
- Shelly, S.; Lukinova, N.; Bambina, S.; Berman, A.; Cherry, S. Autophagy is an essential component of Drosophila immunity against vesicular stomatitis virus. Immunity 2009, 30, 588–598. [Google Scholar] [CrossRef] [Green Version]
- Moy, R.H.; Cherry, S. Antimicrobial autophagy: A conserved innate immune response in Drosophila. J. Innate. Immun. 2013, 5, 444–455. [Google Scholar] [CrossRef] [PubMed]
- Eng, M.W.; van Zuylen, M.N.; Severson, D.W. Apoptosis-related genes control autophagy and influence DENV-2 infection in the mosquito vector, Aedes aegypti. Insect Biochem. Mol. Biol. 2016, 76, 70–83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paulus, G.L.; Xavier, R.J. Autophagy and checkpoints for intracellular pathogen defense. Curr. Opin. Gastroenterol. 2015, 31, 14–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, Y.; Bowman, J.W.; Jung, J.U. Autophagy during viral infection-a double-edged sword. Nat. Rev. Microbiol. 2018, 16, 341–354. [Google Scholar] [CrossRef]
- McBride, W.H.; Iwamoto, K.S.; Syljuasen, R.; Pervan, M.; Pajonk, F. The role of the ubiquitin/proteasome system in cellular responses to radiation. Oncogene 2003, 22, 5755–5773. [Google Scholar] [CrossRef] [Green Version]
- Kurze, C.; Mayack, C.; Hirche, F.; Stangl, G.I.; Le Conte, Y.; Kryger, P.; Moritz, R.F. Nosema spp. infections cause no energetic stress in tolerant honeybees. Parasitol. Res. 2016, 115, 2381–2388. [Google Scholar] [CrossRef] [Green Version]
- Dussaubat, C.; Maisonnasse, A.; Crauser, D.; Beslay, D.; Costagliola, G.; Soubeyrand, S.; Kretzchmar, A.; Le Conte, Y. Flight behavior and pheromone changes associated to Nosema ceranae infection of honey bee workers (Apis mellifera) in field conditions. J. Invertebr. Pathol. 2013, 113, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Wolf, S.; McMahon, D.P.; Lim, K.S.; Pull, C.D.; Clark, S.J.; Paxton, R.J.; Osborne, J.L. So near and yet so far: Harmonic radar reveals reduced homing ability of Nosema infected honeybees. PLoS ONE 2014, 9, e103989. [Google Scholar] [CrossRef] [Green Version]
- Kaur, G.; Iyer, L.M.; Burroughs, A.M.; Aravind, L. Bacterial death and TRADD-N domains help define novel apoptosis and immunity mechanisms shared by prokaryotes and metazoans. Elife 2021, 10, e70394. [Google Scholar] [CrossRef]
- Narayan, K. Insect defence: Its impact on microbial control of insect pests. Curr. Sci. 2004, 86, 800–814. [Google Scholar]
- Guo, R.; Chen, D.; Diao, Q.; Xiong, C.; Zheng, Y.; Hou, C. Transcriptomic investigation of immune responses of the Apis cerana cerana larval gut infected by Ascosphaera apis. J. Invertebr. Pathol. 2019, 166, 107210. [Google Scholar] [CrossRef]
- Stanley, D.; Miller, J.; Tunaz, H. Eicosanoid actions in insect immunity. J. Innate. Immun. 2009, 1, 282–290. [Google Scholar] [CrossRef]
- Yeh, J.X.; Park, E.; Schultz, K.L.W.; Griffin, D.E. NF-κB activation promotes alphavirus replication in mature neurons. J. Virol. 2019, 93, e01071-19. [Google Scholar] [CrossRef]
- Hayden, M.S.; Ghosh, S. NF-κB, the first quarter-century: Remarkable progress and outstanding questions. Genes. Dev. 2012, 26, 203–234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khor, S.; Cai, D. Control of lifespan and survival by Drosophila NF-κB signaling through neuroendocrine cells and neuroblasts. Aging (Albany NY) 2020, 12, 24604–24622. [Google Scholar] [CrossRef] [PubMed]
- Valanne, S.; Wang, J.H.; Rämet, M. The Drosophila Toll signaling pathway. J. Immunol. 2011, 186, 649–656. [Google Scholar] [CrossRef] [Green Version]
- Myllymäki, H.; Valanne, S.; Rämet, M. The Drosophila imd signaling pathway. J. Immunol. 2014, 192, 3455–3462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffmann, J.A.; Reichhart, J.M. Drosophila innate immunity: An evolutionary perspective. Nat. Immunol. 2002, 3, 121–126. [Google Scholar] [CrossRef]
- De Gregorio, E.; Spellman, P.T.; Tzou, P.; Rubin, G.M.; Lemaitre, B. The Toll and Imd pathways are the major regulators of the immune response in Drosophila. EMBO J. 2002, 21, 2568–2579. [Google Scholar] [CrossRef] [Green Version]
- Tanji, T.; Hu, X.; Weber, A.N.; Ip, Y.T. Toll and IMD pathways synergistically activate an innate immune response in Drosophila melanogaster. Mol. Cell. Biol. 2007, 27, 4578–4588. [Google Scholar] [CrossRef] [Green Version]
- Lin, C.C.; Chou, C.M.; Hsu, Y.L.; Lien, J.C.; Wang, Y.M.; Chen, S.T.; Tsai, S.C.; Hsiao, P.W.; Huang, C.J. Characterization of two mosquito STATs, AaSTAT and CtSTAT. Differential regulation of tyrosine phosphorylation and DNA binding activity by lipopolysaccharide treatment and by Japanese encephalitis virus infection. J. Biol. Chem. 2004, 279, 3308–3317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Guo, R.; Kumar, D.; Ma, H.; Liu, J.; Hu, X.; Cao, G.; Xue, R.; Gong, C. Identification gene expression and immune function of the novel Bm-STAT gene in virus-infected Bombyx mori. Gene 2016, 577, 82–88. [Google Scholar] [CrossRef] [PubMed]
- Dostert, C.; Jouanguy, E.; Irving, P.; Troxler, L.; Galiana-Arnoux, D.; Hetru, C.; Hoffmann, J.A.; Imler, J.L. The Jak-STAT signaling pathway is required but not sufficient for the antiviral response of drosophila. Nat. Immunol. 2005, 6, 946–953. [Google Scholar] [CrossRef] [PubMed]
- Agaisse, H.; Petersen, U.M.; Boutros, M.; Mathey-Prevot, B.; Perrimon, N. Signaling role of hemocytes in Drosophila JAK/STAT-dependent response to septic injury. Dev. Cell 2003, 5, 441–450. [Google Scholar] [CrossRef] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xing, W.; Zhou, D.; Long, Q.; Sun, M.; Guo, R.; Wang, L. Immune Response of Eastern Honeybee Worker to Nosema ceranae Infection Revealed by Transcriptomic Investigation. Insects 2021, 12, 728. https://doi.org/10.3390/insects12080728
Xing W, Zhou D, Long Q, Sun M, Guo R, Wang L. Immune Response of Eastern Honeybee Worker to Nosema ceranae Infection Revealed by Transcriptomic Investigation. Insects. 2021; 12(8):728. https://doi.org/10.3390/insects12080728
Chicago/Turabian StyleXing, Wenhao, Dingding Zhou, Qi Long, Minghui Sun, Rui Guo, and Limei Wang. 2021. "Immune Response of Eastern Honeybee Worker to Nosema ceranae Infection Revealed by Transcriptomic Investigation" Insects 12, no. 8: 728. https://doi.org/10.3390/insects12080728
APA StyleXing, W., Zhou, D., Long, Q., Sun, M., Guo, R., & Wang, L. (2021). Immune Response of Eastern Honeybee Worker to Nosema ceranae Infection Revealed by Transcriptomic Investigation. Insects, 12(8), 728. https://doi.org/10.3390/insects12080728






