Genome-Wide Identification of Neuropeptides and Their Receptors in an Aphid Endoparasitoid Wasp, Aphidius gifuensi
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Insect Rearing
2.2. RNA Sequencing and Gene Identification
2.3. Phylogenetic Analysis
2.4. Expression Analysis
3. Results
3.1. Transcriptome Sequencing, Assembly, and Annotation
3.2. Identification of Neuropeptide Precursors
3.3. Neuropeptide Receptors in A. gifuensis
3.4. Expression Profiles of Neuropeptides and Their Receptors
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schoofs, L.; De Loof, A.; Van Hiel, M.B. Neuropeptides as regulators of behavior in insects. Annu. Rev. Ѐntomol. 2017, 62, 35–52. [Google Scholar] [CrossRef] [PubMed]
- Van Wielendaele, P.; Wynant, N.; Dillen, S.; Zels, S.; Badisco, L.; Broeck, J.V. Neuropeptide F regulates male reproductive processes in the desert locust, Schistocerca gregaria. Insect Biochem. Mol. Biol. 2013, 43, 252–259. [Google Scholar] [CrossRef] [PubMed]
- Ragionieri, L.; Predel, R. The neuropeptidome of Carabus (Coleoptera, Adephaga: Carabidae). Insect Biochem. Mol. Biol. 2019, 118, 103309. [Google Scholar] [CrossRef] [PubMed]
- Hou, L.; Yang, P.; Jiang, F.; Liu, Q.; Wang, X.; Kang, L. The neuropeptide F/nitric oxide pathway is essential for shaping locomotor plasticity underlying locust phase transition. eLife 2017, 6. [Google Scholar] [CrossRef] [PubMed]
- Ons, S. Neuropeptides in the regulation of Rhodnius prolixus physiology. J. Insect Physiol. 2017, 97, 77–92. [Google Scholar] [CrossRef]
- Van Wielendaele, P.; Dillen, S.; Zels, S.; Badisco, L.; Broeck, J.V. Regulation of feeding by Neuropeptide F in the desert locust, Schistocerca gregaria. Insect Biochem. Mol. Biol. 2013, 43, 102–114. [Google Scholar] [CrossRef]
- Okada, Y.; Katsuki, M.; Okamoto, N.; Fujioka, H.; Okada, K. A specific type of insulin-like peptide regulates the conditional growth of a beetle weapon. PLoS Biol. 2019, 17, e3000541. [Google Scholar] [CrossRef]
- Hou, L.; Jiang, F.; Yang, P.; Wang, X.; Kang, L. Molecular characterization and expression profiles of neuropeptide precursors in the migratory locust. Insect Biochem. Mol. Biol. 2015, 63, 63–71. [Google Scholar] [CrossRef]
- Audsley, N.; Down, R.E. G protein coupled receptors as targets for next generation pesticides. Insect Biochem. Mol. Biol. 2015, 67, 27–37. [Google Scholar] [CrossRef]
- Ons, S.; Lavore, A.; Sterkel, M.; Wulff, J.P.; Sierra, I.; Martínez-Barnetche, J.; Rodriguez, M.H.; Rivera-Pomar, R. Identification of G protein coupled receptors for opsines and neurohormones in Rhodnius prolixus. Genomic and transcriptomic analysis. Insect Biochem. Mol. Biol. 2016, 69, 34–50. [Google Scholar] [CrossRef]
- Jaeger, W.C.; Armstrong, S.P.; Hill, S.J.; Pfleger, K.D.G. Biophysical detection of diversity and bias in GPCR function. Front. Endocrinol. 2014, 5. [Google Scholar] [CrossRef]
- Li, C.; Song, X.; Chen, X.; Liu, X.; Sang, M.; Wu, W.; Yun, X.; Hu, X.; Li, B. Identification and comparative analysis of G protein-coupled receptors in Pediculus humanus humanus. Genomics 2014, 104, 58–67. [Google Scholar] [CrossRef]
- Harmar, A.J. Family-B G-protein-coupled receptors. Genome Biol. 2001, 2. [Google Scholar] [CrossRef]
- Van Hiel, B.; Vandersmissen, H.P.; Van Loy, T.; Broeck, J.V. An evolutionary comparison of leucine-rich repeat containing G protein-coupled receptors reveals a novel LGR subtype. Peptides 2012, 34, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Vogel, K.; Brown, M.R.; Strand, M.R. Ovary ecdysteroidogenic hormone requires a receptor tyrosine kinase to activate egg formation in the mosquito Aedes aegypti. Proc. Natl. Acad. Sci. USA 2015, 112, 5057–5062. [Google Scholar] [CrossRef] [PubMed]
- Rewitz, K.F.; Yamanaka, N.; Gilbert, L.I.; O’Connor, M.B. The insect neuropeptide ptth activates receptor tyrosine kinase torso to initiate metamorphosis. Science 2009, 326, 1403–1405. [Google Scholar] [CrossRef] [PubMed]
- Bai, H.; Zhu, F.; Shah, K.; Palli, S.R. Large-scale RNAi screen of G protein-coupled receptors involved in larval growth, molting and metamorphosis in the red flour beetle. BMC Genom. 2011, 12, 388. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Ma, Z.; Kang, L. Two dopamine receptors play different roles in phase change of the migratory locust. Front. Behav. Neurosci. 2015, 9. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Ma, Z.; Du, B.; Li, T.; Li, W.; Xu, L.; He, J.; Kang, L. Dop1 enhances conspecific olfactory attraction by inhibiting miR-9a maturation in locusts. Nat. Commun. 2018, 9, 1193. [Google Scholar] [CrossRef] [PubMed]
- Kang, X.L.; Zhang, J.Y.; Wang, D.; Zhao, Y.M.; Han, X.L.; Wang, J.X.; Zhao, X.F. The steroid hormone 20-hydroxyecdysone binds to dopamine receptor to repress lepidopteran insect feeding and promote pupation. PLoS Genet. 2019, 15, e1008331. [Google Scholar] [CrossRef] [PubMed]
- Shahid, S.; Shi, Y.; Yang, C.; Li, J.J.; Ali, M.Y.; Smagghe, G.; Liu, T.X. CCHamide2-receptor regulates feeding behavior in the pea aphid, Acyrthosiphon pisum. Peptide 2021, 143, 170596. [Google Scholar] [CrossRef]
- Shi, Y.; Nachman, R.J.; Gui, S.; Piot, N.; Kaczmarek, K.; Zabrocki, J.; Dow, J.A.T.; Davies, S.; Smagghe, G. Efficacy and biosafety assessment of neuropeptide CAPA analogues against the peach-potato aphid (Myzus persicae). Insect Sci. 2021. [Google Scholar] [CrossRef]
- Garland, S.L. Are GPCRs still a source of new targets? J. Biomol. Screen 2013, 18, 947–966. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yang, D.; Zhou, Q.; Labroska, V.; Qin, S.; Darbalaei, S.; Wu, Y.; Yuliantie, E.; Xie, L.; Tao, H.; Cheng, J.; et al. G protein-coupled receptors: Structure- and function-based drug discovery. Signal Transduct. Target. Ther. 2021, 6, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Li, T.; Wang, Y.; Liu, S. G-protein coupled receptors (GPCRs) in insects—A potential target for new insecticide development. Molecules 2021, 26, 2993. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Teng, Z.W.; Gu, G.X.; Qi, Y.X.; Guo, L.; Xiao, S.; Wang, F.; Fang, Q.; Wang, F.; Song, Q.S.; et al. Genome-wide characterization and transcriptomic analyses of neuropeptides and their receptors in an endoparasitoid wasp, Pteromalus puparum. Arch. Insect Biochem. Physiol. 2020, 103, e21625. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.; Xiong, S.; Xu, G.; Ye, X.; Yao, H.; Wang, F.; Fang, Q.; Song, Q.; Ye, G. Identification of Neuropeptides and Their Receptors in the Ectoparasitoid, Habrobracon hebetor. Front. Physiol. 2020, 11, 575655. [Google Scholar] [CrossRef] [PubMed]
- Hauser, F.; Neupert, S.; Williamson, M.; Predel, R.; Tanaka, Y.; Grimmelikhuijzen, C.J. Genomics and peptidomics of neuropeptides and protein hormones present in the parasitic wasp Nasonia vitripennis. J. Proteome Res. 2010, 9, 5296. [Google Scholar] [CrossRef]
- Kang, Z.W.; Liu, F.H.; Liu, X.; Yu, W.B.; Tan, X.L.; Zhang, S.Z.; Tian, H.G.; Liu, T.X. The potential coordination of the heat-shock proteins and antioxidant enzyme genes of Aphidius gifuensis in response to thermal stress. Front. Physiol. 2017, 8. [Google Scholar] [CrossRef]
- Pan, M.Z.; Liu, T.X. Suitability of three aphid species for Aphidius gifuensis (Hymenoptera: Braconidae): Parasitoid performance varies with hosts of origin. Biol. Control 2014, 69, 90–96. [Google Scholar] [CrossRef]
- Yang, S.; Wei, J.N.; Yang, S.Y.; Kuang, R.P. Current status and future trends of augmentative release of Aphidius gifuensis for control of Myzus persicae in China’s Yunnan province. J. Entomol. Res. Soc. 2011, 13, 87–99. [Google Scholar]
- Kang, Z.W.; Liu, F.H.; Tan, X.L.; Zhang, Z.F.; Zhu, J.Y.; Tian, H.G.; Liu, T.X. Infection of powdery mildew reduces the fitness of grain aphids (Sitobion avenae) through restricted nutrition and induced defense response in wheat. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Kang, Z.W.; Liu, F.H.; Zhang, Z.F.; Tian, H.G.; Liu, T.X. Volatile β-Ocimene can regulate developmental performance of peach aphid Myzus persicae through activation of defense responses in Chinese cabbage Brassica pekinensis. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Pan, M.Z.; Wei, Y.Y.; Wang, F.R.; Liu, T.X. Influence of plant species on biological control effectiveness of Myzus persicae by Aphidius gifuensis. Crop. Prot. 2020, 135, 105223. [Google Scholar] [CrossRef]
- Pan, M.Z.; Cao, H.H.; Liu, T.X. Effects of winter wheat cultivars on the life history traits and olfactory response of Aphidius gifuensis. BioControl 2014, 59, 539–546. [Google Scholar] [CrossRef]
- Pan, M.Z.; Liu, T.X.; Nansen, C. Avoidance of parasitized host by female wasps of Aphidius gifuensis (Hymenoptera: Braconidae): The role of natal rearing effects and host availability? Insect Sci. 2018, 25, 1035–1044. [Google Scholar] [CrossRef]
- Fan, J.; Zhang, Q.; Xu, Q.; Xue, W.; Han, Z.; Sun, J.; Chen, J. Differential expression analysis of olfactory genes based on a combination of sequencing platforms and behavioral investigations in Aphidius gifuensis. Front. Physiol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Du, Z.; Tian, L.; Zhang, L.; Huang, Z.; Wei, S.; Song, F.; Cai, W.; Yu, Y.; Yang, H.; et al. Chromosome-level genome assembly of the aphid parasitoid Aphidius gifuensis using Oxford Nanopore sequencing and Hi-C technology. Mol. Ecol. Resour. 2020, 21, 941–954. [Google Scholar] [CrossRef]
- Kang, Z.W.; Tian, H.G.; Liu, F.H.; Liu, X.; Jing, X.F.; Liu, T.X. Identification and expression analysis of chemosensory receptor genes in an aphid endoparasitoid Aphidius gifuensis. Sci. Rep. 2017, 7, 3939. [Google Scholar] [CrossRef]
- Kang, Z.W.; Liu, F.H.; Pang, R.P.; Tian, H.G.; Liu, T.X. Effect of sublethal doses of imidacloprid on the biological performance of aphid endoparasitoid Aphidius gifuensis (Hymenoptera: Aphidiidae) and influence on its related gene expression. Front. Physiol. 2018, 9, 1729. [Google Scholar] [CrossRef]
- Tasman, K.; Hidalgo, S.; Zhu, B.; Rands, S.A.; Hodge, J.J.L. Neonicotinoids disrupt memory, circadian behaviour and sleep. Sci. Rep. 2021, 11, 2061. [Google Scholar] [CrossRef] [PubMed]
- Broeck, J.V. Neuropeptides and their precursors in the fruitfly, Drosophila melanogaster. Peptides 2001, 22, 241–254. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Rozewicki, J.; Li, S.; Amada, K.M.; Standley, D.M.; Katoh, K. MAFFT-DASH: Integrated protein sequence and structural alignment. Nucleic Acids Res. 2019, 47, W5–W10. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef]
- Kang, Z.W.; Liu, F.H.; Xu, Y.Y.; Cheng, J.H.; Lin, X.L.; Jing, X.F.; Tian, H.G.; Liu, T.X. Identification of candidate odorant-degrading enzyme genes in the antennal transcriptome of Aphidius gifuensis. Ѐntomol. Res. 2020, 51, 36–54. [Google Scholar] [CrossRef]
- Hummon, A.B.; Richmond, T.A.; Verleyen, P.; Baggerman, G.; Huybrechts, J.; Ewing, M.A.; Vierstraete, E.; Rodriguez-Zas, S.L.; Schoofs, L.; Robinson, G.E.; et al. From the genome to the proteome: Uncovering peptides in the Apis Brain. Science 2006, 314, 647–649. [Google Scholar] [CrossRef]
- Roller, L.; Yamanaka, N.; Watanabe, K.; Daubnerová, I.; Žitňan, D.; Kataoka, H.; Tanaka, Y. The unique evolution of neuropeptide genes in the silkworm Bombyx mori. Insect Biochem. Mol. Biol. 2008, 38, 1147–1157. [Google Scholar] [CrossRef]
- Li, B.; Predel, R.; Neupert, S.; Hauser, F.; Tanaka, Y.; Cazzamali, G.; Williamson, M.; Arakane, Y.; Verleyen, P.; Schoofs, L.; et al. Genomics, transcriptomics, and peptidomics of neuropeptides and protein hormones in the red flour beetle Tribolium castaneum. Genome Res. 2007, 18, 113–122. [Google Scholar] [CrossRef]
- Tanaka, Y.; Suetsugu, Y.; Yamamoto, K.; Noda, H.; Shinoda, T. Transcriptome analysis of neuropeptides and G-protein coupled receptors (GPCRs) for neuropeptides in the brown planthopper Nilaparvata lugens. Peptides 2014, 53, 125–133. [Google Scholar] [CrossRef]
- Zhang, H.; Bai, J.; Huang, S.; Liu, H.; Lin, J.; Hou, Y. Neuropeptides and G-Protein coupled receptors (GPCRs) in the red palm weevil Rhynchophorus ferrugineus Olivier (Coleoptera: Dryophthoridae). Front. Physiol. 2020, 11, 159. [Google Scholar] [CrossRef] [PubMed]
- Roller, L.; Čižmár, D.; Bednár, B.; Žitňan, D. Expression of RYamide in the nervous and endocrine system of Bombyx mori. Peptides 2016, 80, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Collin, C.; Hauser, F.; Krogh-Meyer, P.; Hansen, K.K.; de Valdivia, E.G.; Williamson, M.; Grimmelikhuijzen, C.J. Identification of the Drosophila and Tribolium receptors for the recently discovered insect RYamide neuropeptides. Biochem. Biophys. Res. Commun. 2011, 412, 578–583. [Google Scholar] [CrossRef] [PubMed]
- Ida, T.; Takahashi, T.; Tominaga, H.; Sato, T.; Kume, K.; Ozaki, M.; Hiraguchi, T.; Maeda, T.; Shiotani, H.; Terajima, S.; et al. Identification of the novel bioactive peptides dRYamide-1 and dRYamide-2, ligands for a neuropeptide Y-like receptor in Drosophila. Biochem. Biophys. Res. Commun. 2011, 410, 872–877. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Veenstra, J.A.; Perrimon, N. Control of lipid metabolism by tachykinin in Drosophila. Cell Rep. 2014, 9, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Van Loy, T.; Vandersmissen, H.P.; Poels, J.; Van Hiel, B.; Verlinden, H.; Broeck, J.V. Tachykinin-related peptides and their receptors in invertebrates: A current view. Peptides 2010, 31, 520–524. [Google Scholar] [CrossRef] [PubMed]
- Gui, S.H.; Jiang, H.B.; Xu, L.; Pei, Y.X.; Liu, X.Q.; Smagghe, G.; Wang, J.J. Role of a tachykinin-related peptide and its receptor in modulating the olfactory sensitivity in the oriental fruit fly, Bactrocera dorsalis (Hendel). Insect Biochem. Mol. Biol. 2016, 80, 71–78. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, X.; Wang, Z.; Chen, T.; Zhou, S.; Chen, J.; Pang, L.; Ye, X.; Shi, M.; Huang, J.; et al. Symbiotic bracovirus of a parasite manipulates host lipid metabolism via tachykinin signaling. PLos Pathog. 2021, 17, e1009365. [Google Scholar] [CrossRef]
- Qi, Y.X.; Jin, M.; Ni, X.Y.; Ye, G.Y.; Lee, Y.; Huang, J. Characterization of three serotonin receptors from the small white butterfly, Pieris rapae. Insect Biochem. Mol. Biol. 2017, 87, 107–116. [Google Scholar] [CrossRef]
- Gassias, E.; Durand, N.; Demondion, E.; Bourgeois, T.; Aguilar, P.; Bozzolan, F.; Debernard, S. A critical role for Dop1-mediated dopaminergic signaling in the plasticity of behavioral and neuronal responses to sex pheromone in a moth. J. Exp. Biol. 2019, 222. [Google Scholar] [CrossRef]
- Finetti, L.; Pezzi, M.; Civolani, S.; Calò, G.; Scapoli, C.; Bernacchia, G. Characterization of Halyomorpha halys TAR1 reveals its involvement in (E)-2-decenal pheromone perception. J. Exp. Biol. 2021, 224. [Google Scholar] [CrossRef]
- Ma, Z.; Guo, X.; Lei, H.; Li, T.; Hao, S.; Kang, L. Octopamine and tyramine respectively regulate attractive and repulsive behavior in locust phase changes. Sci. Rep. 2015, 5, srep08036. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Li, L.; Yang, P.; Ma, Z. Calmodulin as a downstream gene of octopamine-OAR α1 signalling mediates olfactory attraction in gregarious locusts. Insect Mol. Biol. 2016, 26, 1–12. [Google Scholar] [CrossRef]
- Ravi, P.; Trivedi, D.; Hasan, G. FMRFa receptor stimulated Ca2+ signals alter the activity of flight modulating central dopaminergic neurons in Drosophila melanogaster. PLoS Genet. 2018, 14, e1007459. [Google Scholar] [CrossRef]
- Kiss, B.; Szlanka, T.; Zvara, A.; Zurovec, M.; Sery, M.; Kakaš, S.; Ramasz, B.; Hegedüs, Z.; Lukacsovich, T.; Puskás, L.; et al. Selective elimination/RNAi silencing of FMRF-related peptides and their receptors decreases the locomotor activity in Drosophila melanogaster. Gen. Comp. Endocrinol. 2013, 191, 137–145. [Google Scholar] [CrossRef]
- Krupp, J.J.; Billeter, J.-C.; Wong, A.; Choi, H.C.; Nitabach, M.N.; Levine, J.D. Pigment-dispersing factor modulates pheromone production in clock cells that influence mating in Drosophila. Neuron 2013, 79, 54–68. [Google Scholar] [CrossRef]
- Ayub, M.; Hermiz, M.; Lange, A.B.; Orchard, I. SIFamide influences feeding in the Chagas disease vector, Rhodnius prolixus. Front. Neurosci. 2020, 14, 134. [Google Scholar] [CrossRef]
- Sellami, A.; Veenstra, J.A. SIFamide acts on fruitless neurons to modulate sexual behavior in Drosophila melanogaster. Peptides 2015, 74, 50–56. [Google Scholar] [CrossRef] [PubMed]
- Corta, E.; Bakkali, A.; A Berrueta, L.; Gallo, B.; Vicente, F. Kinetics and mechanism of amitraz hydrolysis in aqueous media by HPLC and GC-MS. Talanta 1999, 48, 189–199. [Google Scholar] [CrossRef]
- Finetti, L.; Roeder, T.; Calò, G.; Bernacchia, G. The insect type 1 tyramine receptors: From structure to behavior. Insects 2021, 12, 315. [Google Scholar] [CrossRef]
- Meyer, J.M.; Ejendal, K.F.K.; Avramova, L.V.; Garland-Kuntz, E.E.; Giraldo-Calderón, G.I.; Brust, T.F.; Watts, V.J.; Hill, C.A. A “genome-to-lead” approach for insecticide discovery: Pharmacological characterization and screening of Aedes aegypti D1-like dopamine receptors. PLos Negl. Trop. Dis. 2012, 6, e1478. [Google Scholar] [CrossRef] [PubMed]

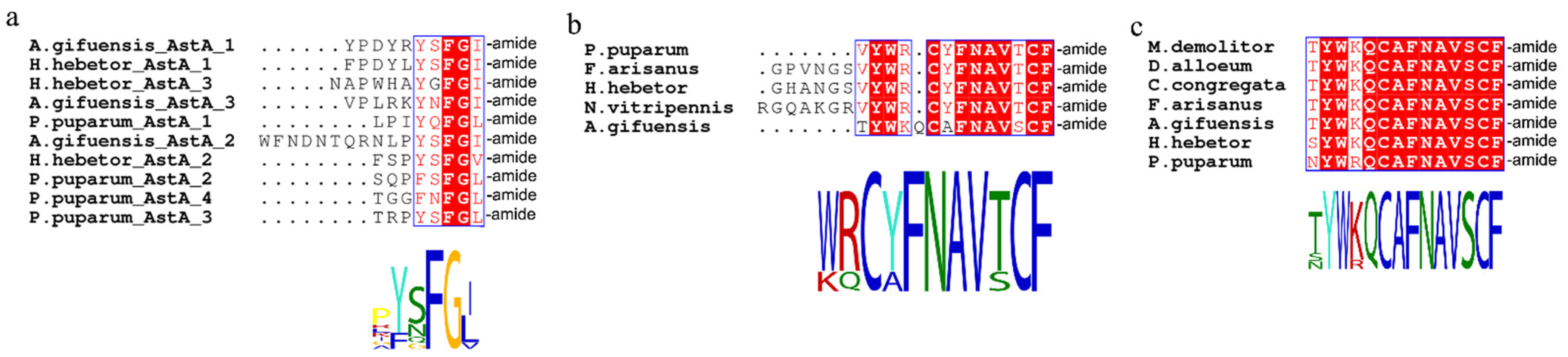
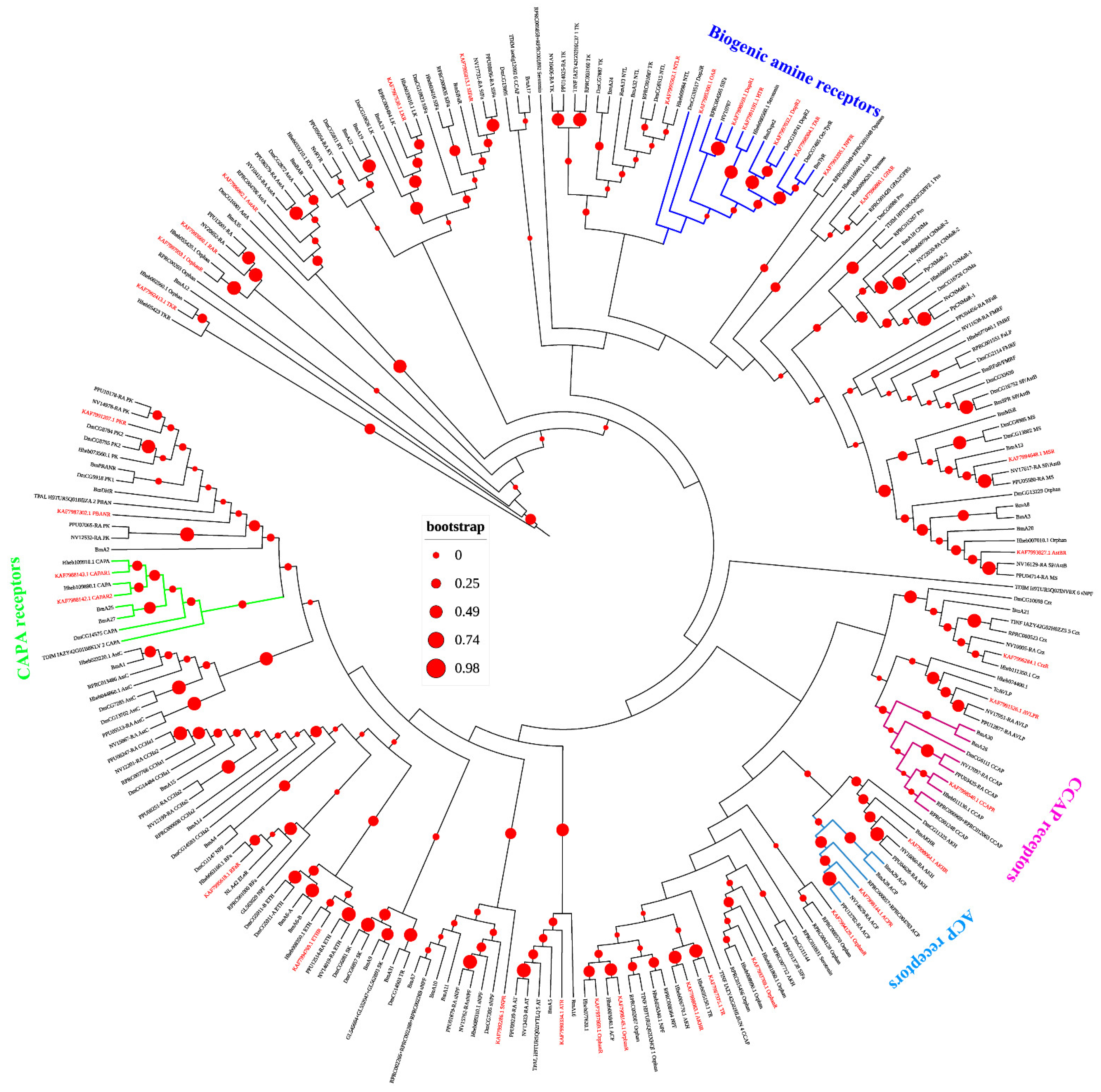
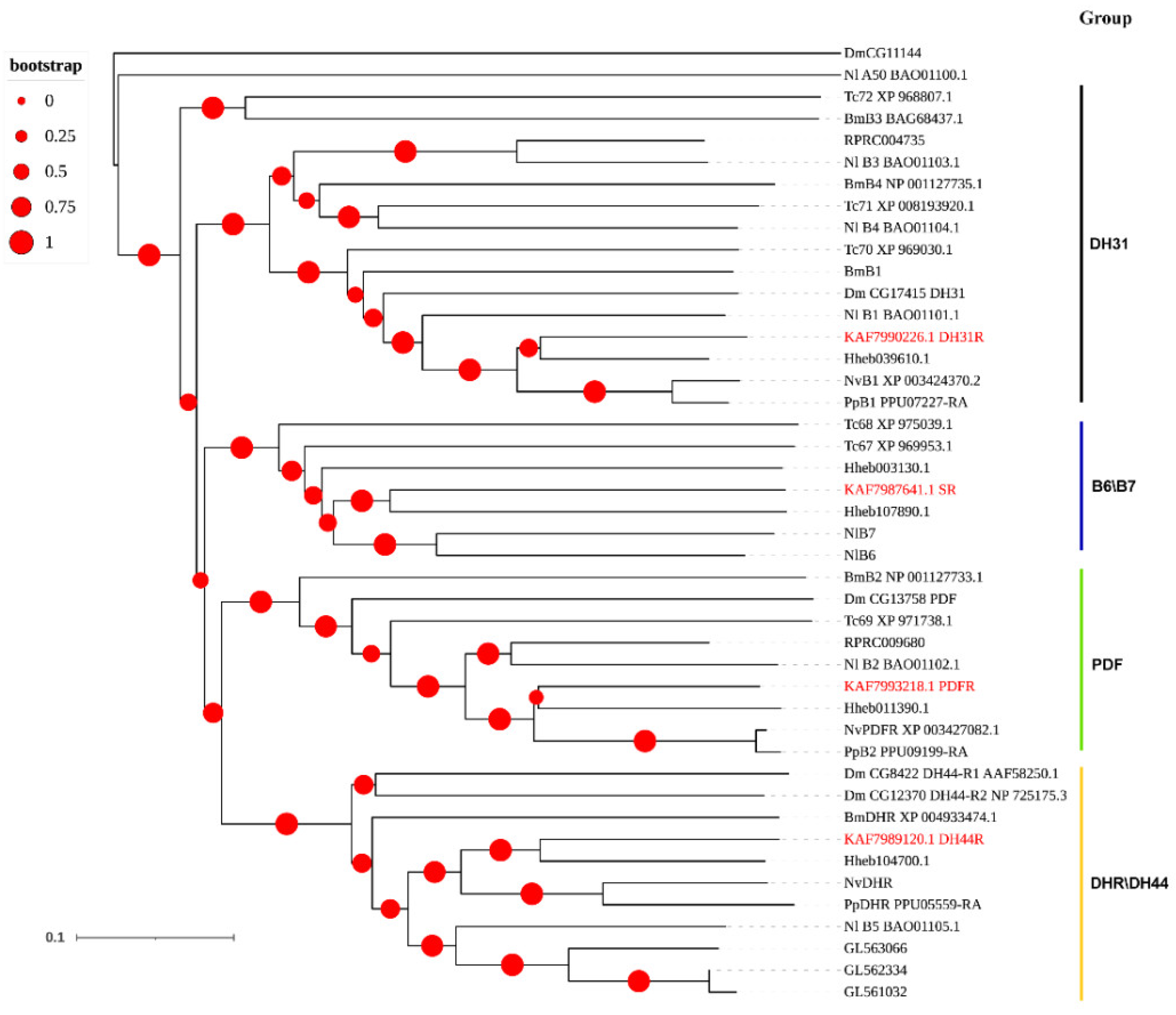
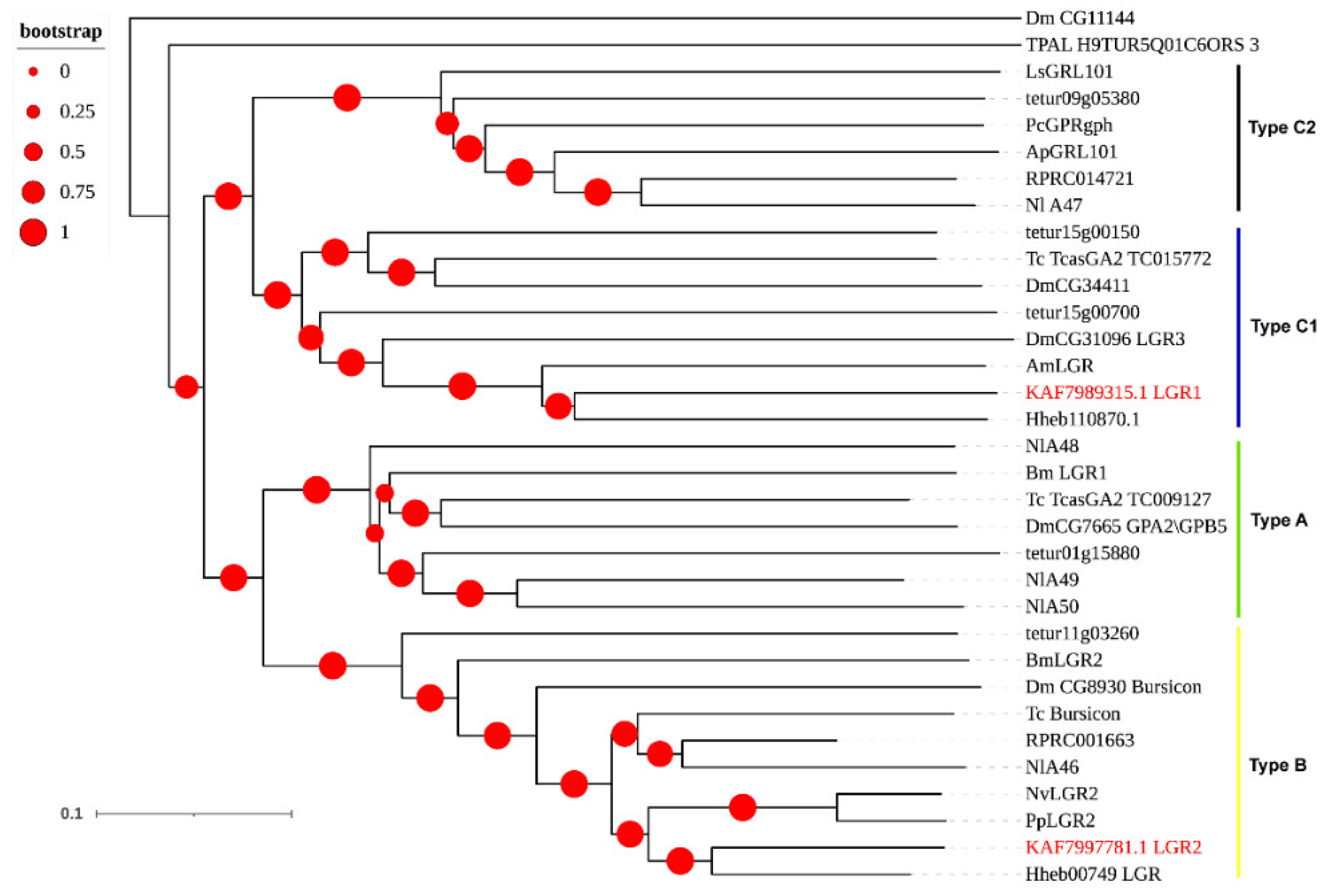
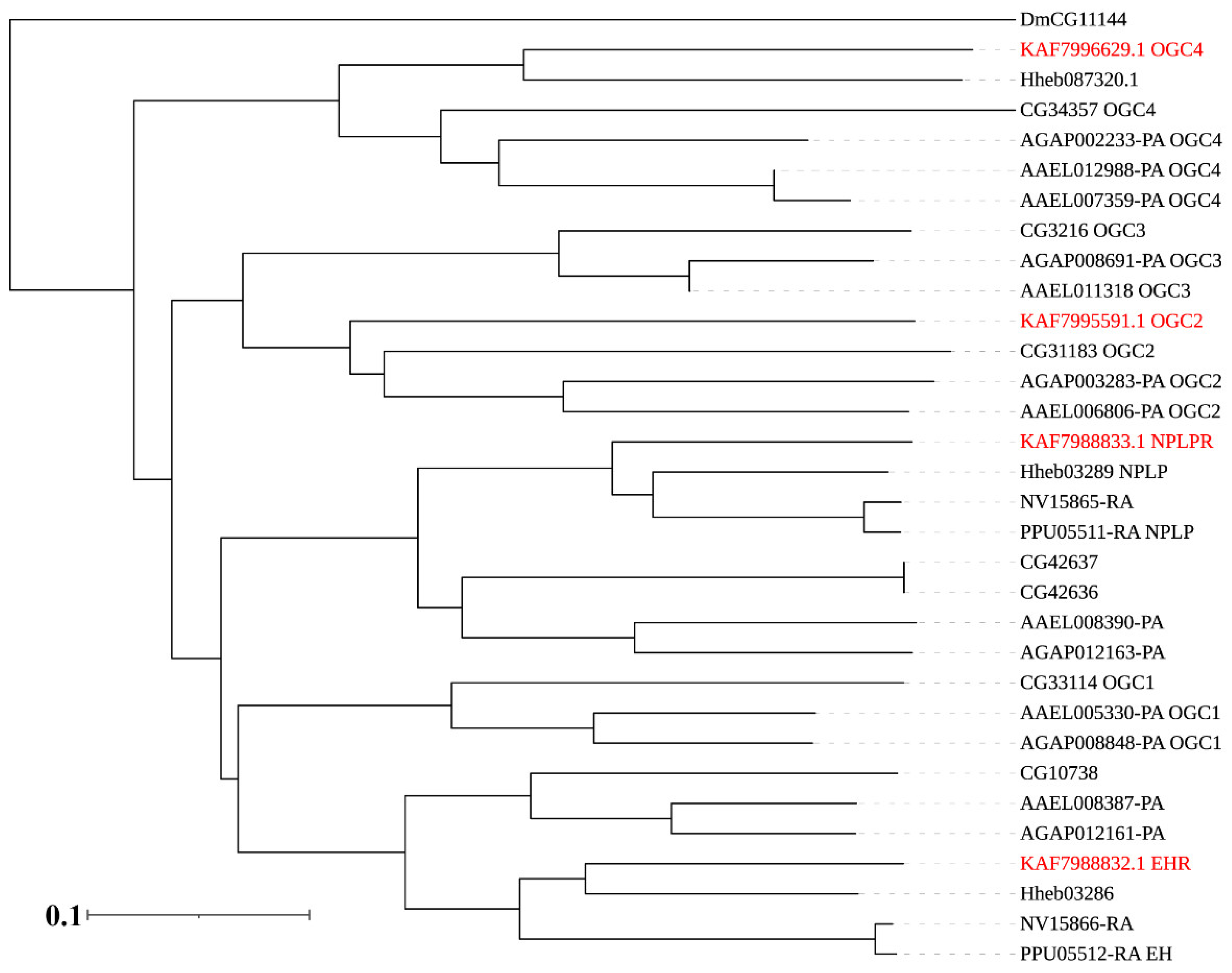
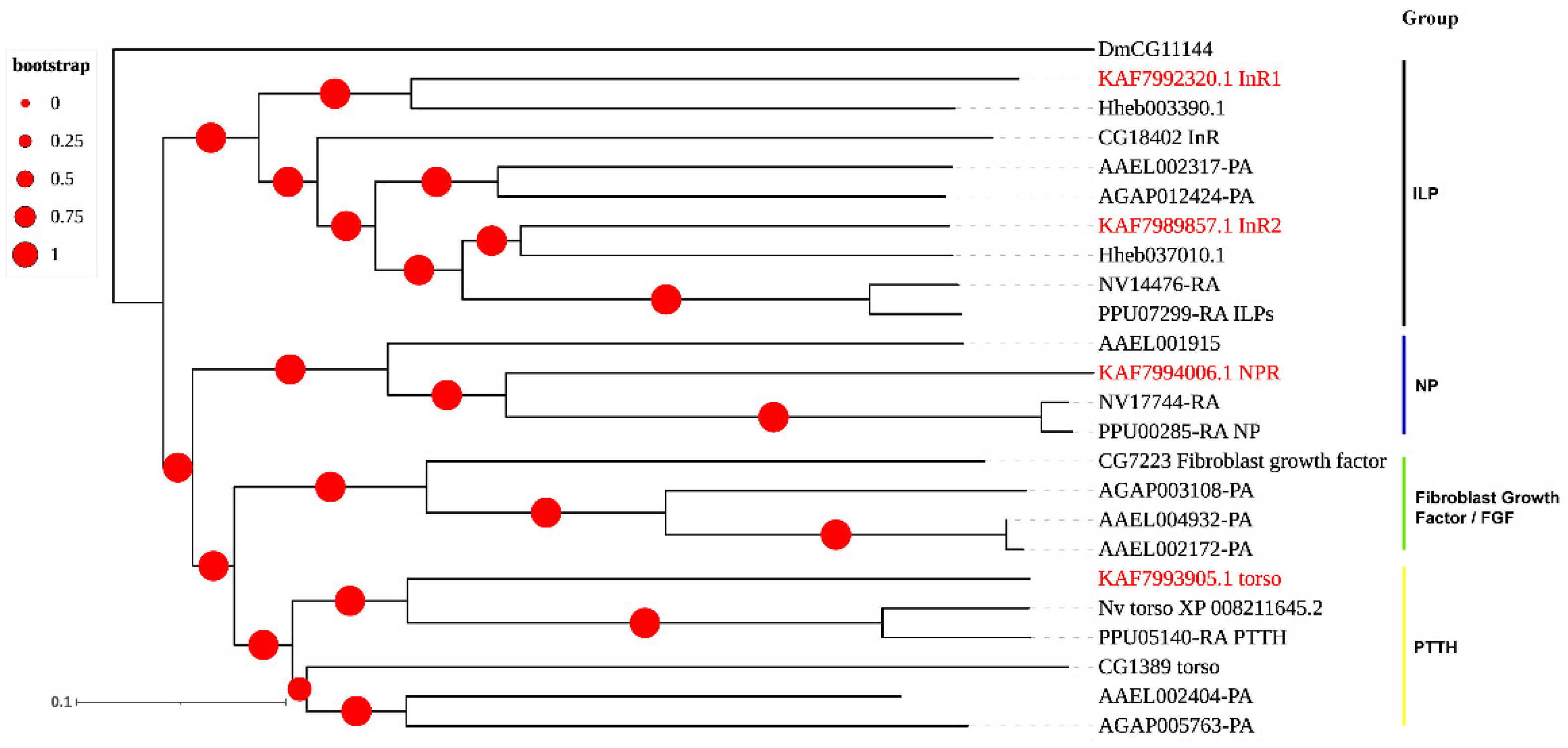
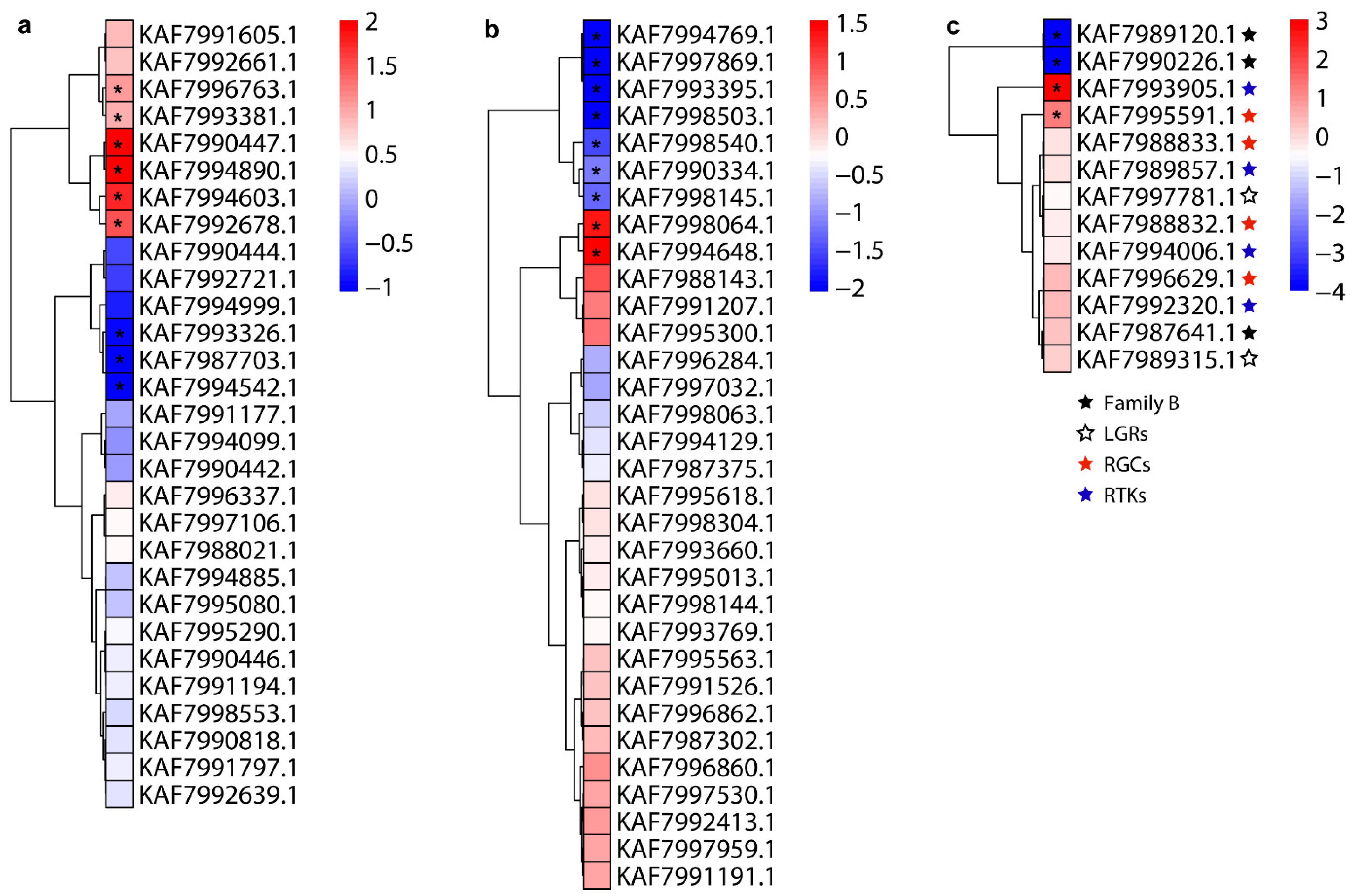
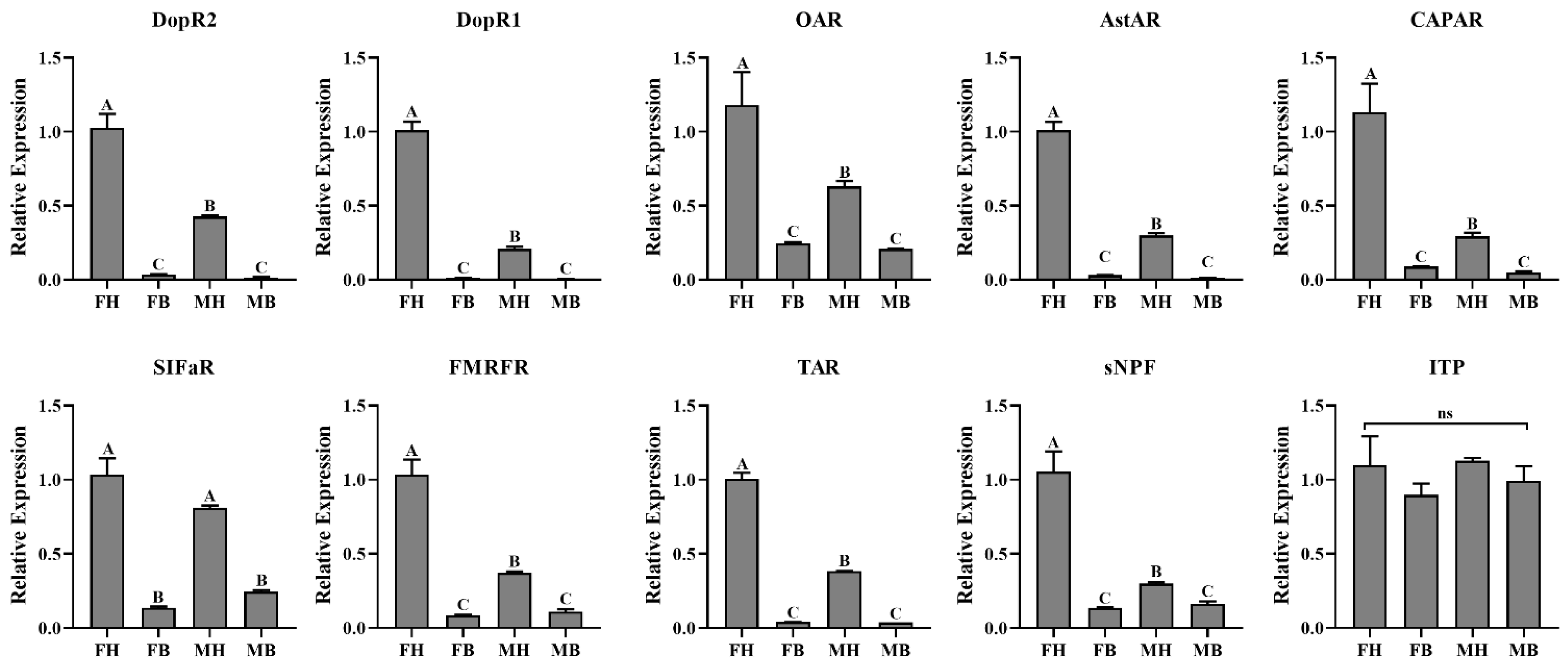
| Peptide Name | Gene ID | Acronym | Protein (AA) | Assigned Receptor ID |
|---|---|---|---|---|
| Allatostatin A | KAF7998325.1 | AstA | 181 | KAF7996862.1 |
| Allatostatin CC | KAF7990447.1 | AstCC | 136 | nd 1 |
| Allatostatin CCC | KAF7990446.1 | AstCCC | 90 | nd |
| Bursicon alpha | KAF7994885.1 | Burα | 147 | KAF7997781.1 |
| Bursicon beta | KAF7994885.1 | Burβ | 123 | KAF7997781.1 |
| CAPA splicing variant a | KAF7994999.1 | CAPA | 103 | KAF7988142.1 KAF7988143.1 |
| Crustacean cardioactive peptide | KAF7995080.1 | CCAP | 130 | KAF7998540.1 |
| CNMamide | KAF7990444.1 | CNMa | 152 | nd |
| Corazonin | KAF7991797.1 | Crz | 122 | KAF7996284.1 |
| Diuretic hormone 31 | KAF7996337.1 | DH31 | 110 | KAF7990226.1 |
| Diuretic hormone 44 | KAF7994603.1 | DH44 | 198 | KAF7989120.1 |
| Eclosion hormone | KAF7992721.1 | EH | 85 | KAF7988832.1 |
| Ecdysis triggering hormone | KAF7993382.1 | ETH | 168 | KAF7994769.1 |
| FMRFamide | KAF7993326.1 | FMRF | 305 | nd |
| IDLSRF-like peptide | KAF7991605.1 | IDLSRF | 201 | nd |
| Insulin-like peptide 1 | KAF7990199.1 | ILP1 | 645 | KAF7992320.1 KAF7989857.1 |
| Elevenin | KAF7994890.1 | Ele | 151 | nd |
| Inotocin | KAF7991194.1 | Inotocin | 143 | nd |
| ITG-like peptide | KAF7998553.1 | ITG | 237 | nd |
| Ion transport peptide | KAF7996763.1 | ITP | 225 | nd |
| Leucokinin | KAF7987704.1 | LK | 218 | KAF7997530.1 |
| Myosupressin | KAF7994099.1 | MS | 99 | KAF7989857.1 |
| Natalisin | KAF7987703.1 | NTL | 296 | KAF7995563.1 |
| Neuroparsin | KAF7991177.1 | NP | 130 | KAF7994006.1 |
| Neuropeptide F 2 | KAF7997106.1 | NPF2 | 123 | KAF7993395.1 |
| NVP-like putative neuropeptide | KAF7992678.1 | NVP | 735 | nd |
| Neuropeptide-like precursor | KAF7992639.1 | NPLP | 526 | KAF7988833.1 |
| Orcokinin-A | KAF7990442.1 | OKA | 154 | nd |
| Pheromone biosynthesis activating Neuropeptide/hugin-pyrokinin | KAF7995290.1 | PBAN | 158 | KAF7987302.1 KAF7991207.1 |
| Pigment dispersing factor | KAF7994542.1 | 88 | KAF7993218.1 | |
| Proctolin | KAF7992386.1 | Pro | 220 | nd |
| Prothoracicotropic hormone | KAF7990443.1 | PTTH | 113 | KAF7993905.1 |
| Short neuropeptide F | KAF7990818.1 | sNPF | 95 | KAF7992286.1 |
| SIFamide | KAF7993381.1 | SIFa | 123 | KAF7995013.1 |
| Trissin | KAF7988021.1 | Tris | 76 | nd |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kong, X.; Li, Z.-X.; Gao, Y.-Q.; Liu, F.-H.; Chen, Z.-Z.; Tian, H.-G.; Liu, T.-X.; Xu, Y.-Y.; Kang, Z.-W. Genome-Wide Identification of Neuropeptides and Their Receptors in an Aphid Endoparasitoid Wasp, Aphidius gifuensi. Insects 2021, 12, 745. https://doi.org/10.3390/insects12080745
Kong X, Li Z-X, Gao Y-Q, Liu F-H, Chen Z-Z, Tian H-G, Liu T-X, Xu Y-Y, Kang Z-W. Genome-Wide Identification of Neuropeptides and Their Receptors in an Aphid Endoparasitoid Wasp, Aphidius gifuensi. Insects. 2021; 12(8):745. https://doi.org/10.3390/insects12080745
Chicago/Turabian StyleKong, Xue, Zhen-Xiang Li, Yu-Qing Gao, Fang-Hua Liu, Zhen-Zhen Chen, Hong-Gang Tian, Tong-Xian Liu, Yong-Yu Xu, and Zhi-Wei Kang. 2021. "Genome-Wide Identification of Neuropeptides and Their Receptors in an Aphid Endoparasitoid Wasp, Aphidius gifuensi" Insects 12, no. 8: 745. https://doi.org/10.3390/insects12080745
APA StyleKong, X., Li, Z.-X., Gao, Y.-Q., Liu, F.-H., Chen, Z.-Z., Tian, H.-G., Liu, T.-X., Xu, Y.-Y., & Kang, Z.-W. (2021). Genome-Wide Identification of Neuropeptides and Their Receptors in an Aphid Endoparasitoid Wasp, Aphidius gifuensi. Insects, 12(8), 745. https://doi.org/10.3390/insects12080745






