Frequent Drivers, Occasional Passengers: Signals of Symbiont-Driven Seasonal Adaptation and Hitchhiking in the Pea Aphid, Acyrthosiphon pisum
Abstract
:Simple Summary
Abstract
1. Introduction
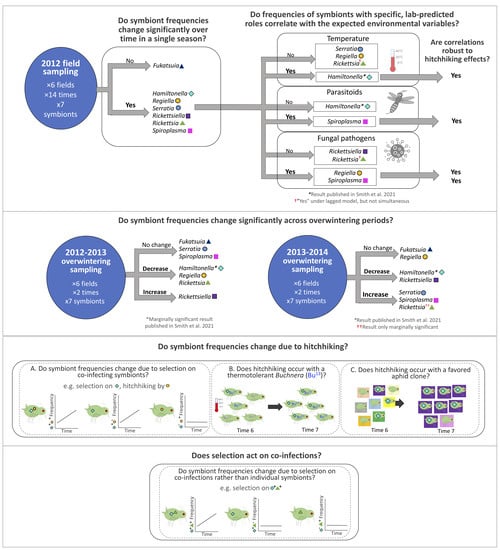
2. Methods
2.1. 2012 Aphid Sampling
2.2. Insect Counts and Climatic Measurements
2.3. Mortality Assays
2.4. Imputing Environmental Variables
2.5. Aphid Sampling across Overwintering Periods
2.6. Molecular Methods
2.7. Statistical Analyses—Change over Time in 2012 and across Overwintering Periods
2.8. Statistical Analyses—Environmental Correlates of 2012 Symbiont Dynamics—Simultaneous Analyses
2.9. Statistical Analyses—Environmental Correlates of 2012 Symbiont Dynamics—Simultaneous + Lagged Analyses
2.10. Hitchhiking Effects—Removal of Aphids with the Most Common Co-Infecting Symbiont Prior to Re-Assessing Symbiont vs. Environment Correlations
2.11. Hitchhiking Effects—Removing Co-Infected Aphids from Paired Timepoint Datasets, to Identify the Influence of Co-Infection on Symbiont Dynamics
2.12. Hitchhiking Effects—Plotting Symbiont Species vs. Symbiont (Co-)Infection Context Trendlines for Comparisons to Hypothesized Patterns under Hitchhiking or Direct Selection
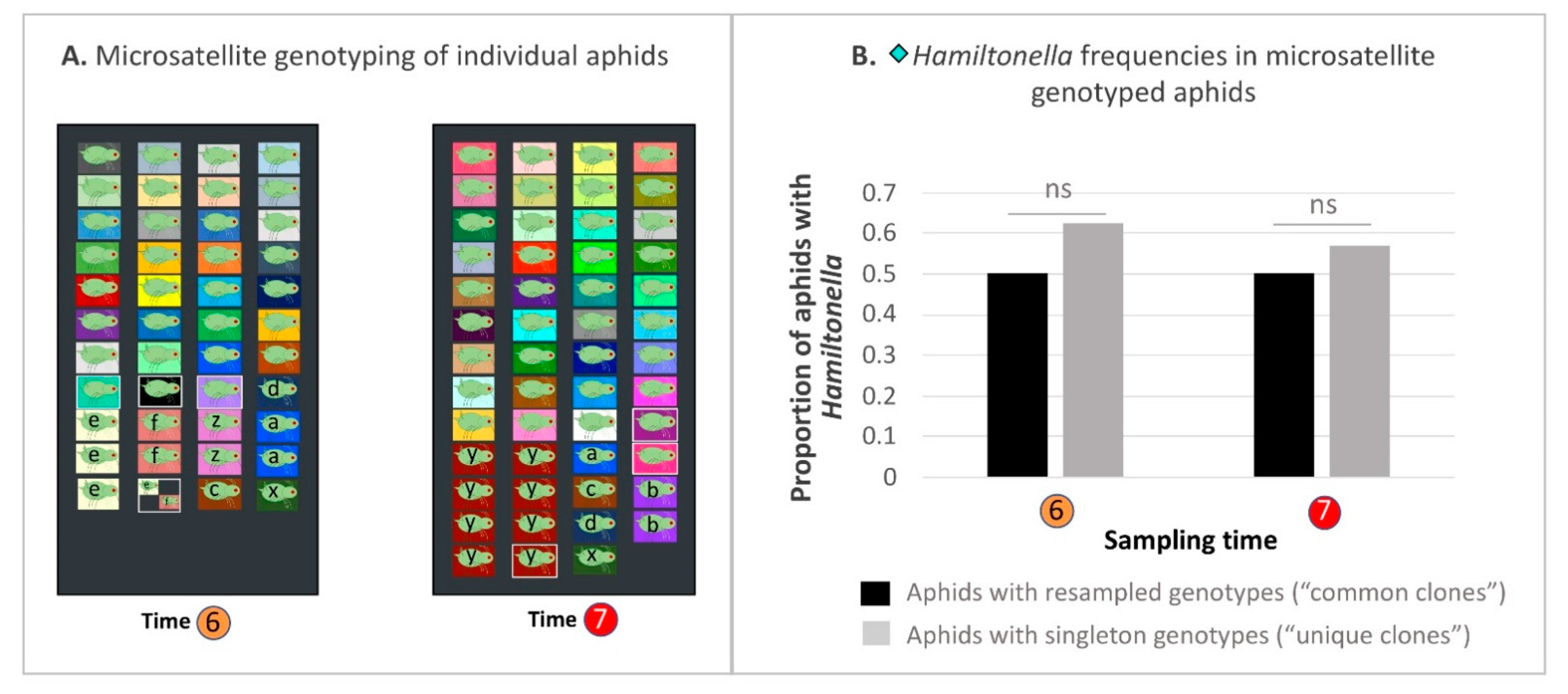
2.13. Hamiltonella-Focused Hitchhiking Effects—Microsatellite Genotyping to Ascertain Proliferation/Decline of Common Clones
2.14. Hamiltonella-Focused Hitchhiking Effects—Buchnera ibpA Gene Promoter Genotyping
2.15. Presentation of Statistical Results
3. Results
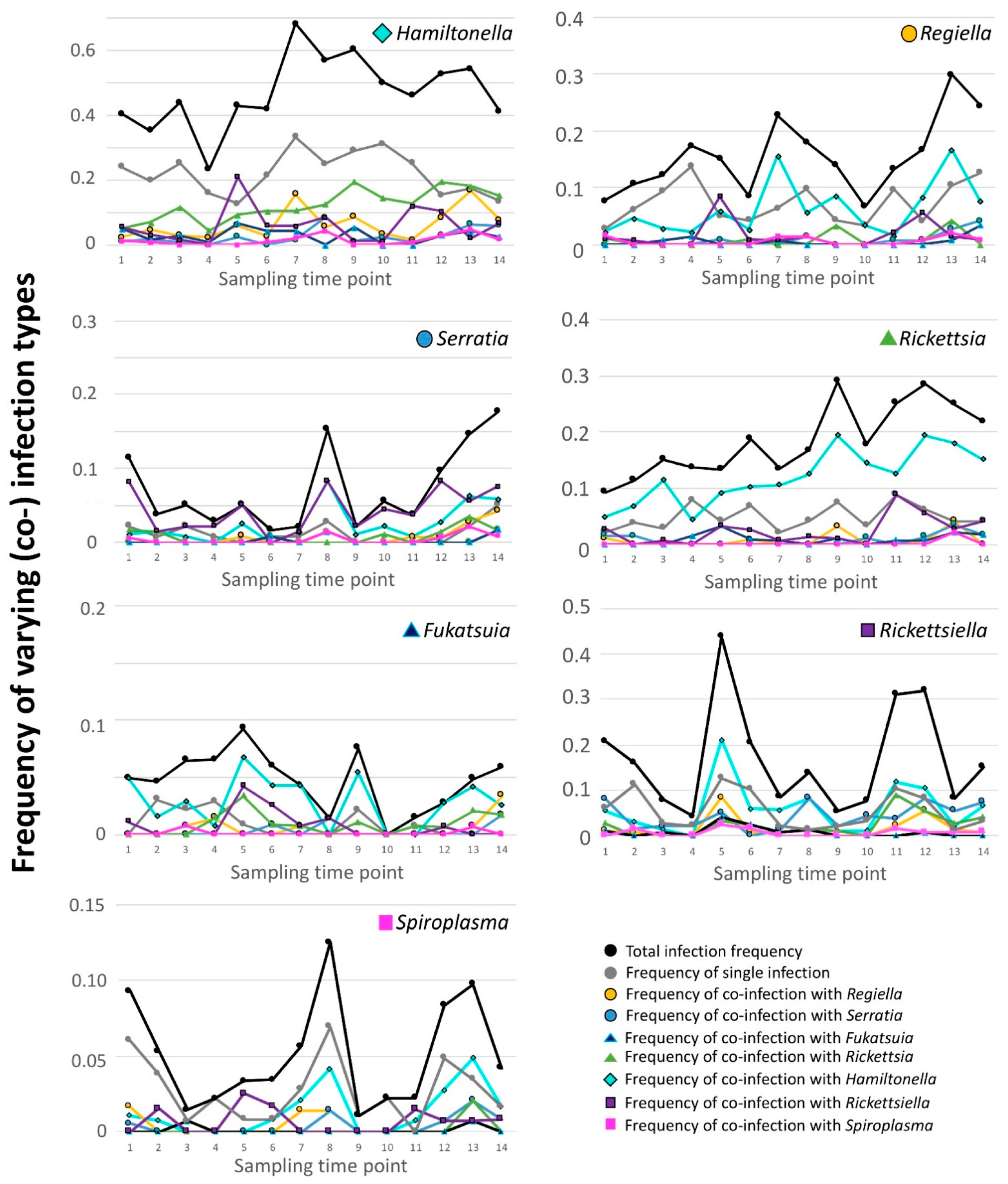
3.1. 2012 Longitudinal Field Study—Symbiont Frequency Variation over Time
3.2. Simultaneous Statistical Analyses of the 2012 Field Study—Symbiont Frequencies vs. Pandora Neoaphidis Pathogens
3.3. Simultaneous Statistical Analyses of the 2012 Field Study—Symbiont Frequencies vs. Parasitoids, Temperature, and Coccinellid Beetles
3.4. Time-Lagged Statistical Analyses of the 2012 Field Study—Comparing Symbiont Frequencies vs. a Priori Predicted Environmental Correlates Sampled 2-Weeks Prior
3.5. Common Symbiont Co-Infections in Our 2012 Study
3.6. Simultaneous Statistical Analyses of the 2012 Field Study—Do Originally Significant Variables Remain in Our Models after Accounting for Hitchhiking?
3.7. 2-Week Frequency Shifts in Our 2012 Field Study—Evidence for Hitchhiking?
3.8. Changes in Symbiont Frequency across Overwintering Periods
3.9. Hamiltonella-Focused Hitchhiking Effects—Aphid Microsatellite Genotyping and Buchnera ibpA Gene Promoter Genotyping
4. Discussion
4.1. Signals of Symbiont-Mediated, Seasonal Pea Aphid Adaptation
4.2. Past, Present, and Future Studies on Selection Targets and the Impacts of Co-Infection
4.3. Selection on Specific Co-Infections as Exceptions to the Rule
4.4. Hitchhiking among Maternally Inherited Elements
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tauber, C.A.; Tauber, M.J. Insect seasonal cycles: Genetics and evolution. Annu. Rev. Ecol. Syst. 1981, 12, 281–308. [Google Scholar] [CrossRef]
- Grainger, T.N.; Rudman, S.M.; Schmidt, P.; Levine, J.M. Competitive history shapes rapid evolution in a seasonal climate. Proc. Natl. Acad. Sci. USA 2021, 118, e2015772118. [Google Scholar] [CrossRef]
- Henter, H.J.; Via, S. The potential for coevolution in a host-parasitoid system. I. Genetic variation within an aphid population in susceptibility to a parasitic wasp. Evolution 1995, 49, 427–438. [Google Scholar] [PubMed]
- Altizer, S.; Dobson, A.; Hosseini, P.; Hudson, P.; Pascual, M.; Rohani, P. Seasonality and the dynamics of infectious diseases. Ecol. Lett. 2006, 9, 467–484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mappes, J.; Kokko, H.; Ojala, K.; Lindström, L. Seasonal changes in predator community switch the direction of selection for prey defenses. Nat. Commun. 2014, 5, 5016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Behrman, E.L.; Howick, V.M.; Kapun, M.; Staubach, F.; Bergland, A.O.; Petrov, D.A.; Lazzaro, B.P.; Schmidt, P.S. Rapid seasonal evolution in innate immunity of wild Drosophila melanogaster. Proc. R. Soc. B Biol. Sci. 2018, 285, 20172599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mateos, M.; Castrezana, S.J.; Nankivell, B.J.; Estes, A.M.; Markow, T.A.; Moran, N.A. Heritable endosymbionts of Drosophila. Genetics 2006, 174, 363–376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Douglas, A.E. Multiorganismal insects: Diversity and function of resident microorganisms. Annu. Rev. Entomol. 2015, 60, 17–34. [Google Scholar] [CrossRef] [Green Version]
- Zchori-Fein, E.; Gottlieb, Y.; Kelly, S.E.; Brown, J.K.; Wilson, J.M.; Karr, T.L.; Hunter, M.S. A newly discovered bacterium associated with parthenogenesis and a change in host selection behavior in parasitoid wasps. Proc. Natl. Acad. Sci. USA 2001, 98, 12555–12560. [Google Scholar] [CrossRef] [Green Version]
- Feldhaar, H.; Straka, J.; Krischke, M.; Berthold, K.; Stoll, S.; Mueller, M.J.; Gross, R. Nutritional upgrading for omnivorous carpenter ants by the endosymbiont Blochmannia. BMC Biol. 2007, 5, 48. [Google Scholar] [CrossRef] [Green Version]
- Teixeira, L.; Ferreira, Á.; Ashburner, M. The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster. PLoS Biol. 2008, 6, e1000002. [Google Scholar] [CrossRef] [Green Version]
- Douglas, A.E. The microbial dimension in insect nutritional ecology. Funct. Ecol. 2009, 23, 38–47. [Google Scholar] [CrossRef]
- Feldhaar, H. Bacterial symbionts as mediators of ecologically important traits of insect hosts. Ecol. Entomol. 2011, 36, 533–543. [Google Scholar] [CrossRef]
- Anbutsu, H.; Moriyama, M.; Nikoh, N.; Hosokawa, T.; Futahashi, R.; Tanahashi, M.; Meng, X.-Y.; Kuriwada, T.; Mori, N.; Oshima, K.; et al. Small genome symbiont underlies cuticle hardness in beetles. Proc. Natl. Acad. Sci. USA 2017, 114, E8382–E8391. [Google Scholar] [CrossRef] [Green Version]
- Tsuchida, T.; Koga, R.; Fukatsu, T. Host plant specialization governed by facultative symbiont. Science 2004, 303, 1989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hansen, A.K.; Moran, N.A. The impact of microbial symbionts on host plant utilization by herbivorous insects. Mol. Ecol. 2014, 23, 1473–1496. [Google Scholar] [CrossRef]
- McLean, A.H.C.; van Asch, M.; Ferrari, J.; Godfray, H.C.J. Effects of bacterial secondary symbionts on host plant use in pea aphids. Proc. R. Soc. B 2011, 278, 278760–278766. [Google Scholar] [CrossRef]
- Scarborough, C.L.; Ferrari, J.; Godfray, H.C.J. Aphid protected from pathogen by endosymbiont. Science 2005, 310, 1781. [Google Scholar] [CrossRef] [PubMed]
- Kriesner, P.; Conner, W.R.; Weeks, A.R.; Turelli, M.; Hoffmann, A.A. Persistence of a Wolbachia infection frequency cline in Drosophila melanogaster and the possible role of reproductive dormancy. Evolution 2016, 70, 979–997. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corbin, C.; Heyworth, E.R.; Ferrari, J.; Hurst, G.D. Heritable symbionts in a world of varying temperature. Heredity 2017, 118, 10–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, B.; Leonard, S.P.; Li, Y.; Moran, N.A. Obligate bacterial endosymbionts limit thermal tolerance of insect host species. Proc. Natl. Acad. Sci. USA 2019, 116, 24712–24718. [Google Scholar] [CrossRef]
- Oliver, K.M.; Perlman, S.J. Chapter Eight—Toxin-mediated protection against natural enemies by insect defensive symbionts. In Advances in Insect Physiology; Oliver, K.M., Russell, J.A., Eds.; Academic Press: Cambridge, UK, 2020; Volume 58, pp. 277–316. [Google Scholar]
- Jaenike, J.; Unckless, R.; Cockburn, S.N.; Boelio, L.M.; Perlman, S.J. Adaptation via symbiosis: Recent spread of a Drosophila defensive symbiont. Science 2010, 329, 212–215. [Google Scholar] [CrossRef]
- Himler, A.G.; Adachi-Hagimori, T.; Bergen, J.E.; Kozuch, A.; Kelly, S.E.; Tabashnik, B.E.; Chiel, E.; Duckworth, V.E.; Dennehy, T.J.; Zchori-Fein, E.; et al. Rapid spread of a bacterial symbiont in an invasive whitefly is driven by fitness benefits and female bias. Science 2011, 332, 254–256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliver, K.M.; Smith, A.H.; Russell, J.A. Defensive symbiosis in the real world–advancing ecological studies of heritable, protective bacteria in aphids and beyond. Funct. Ecol. 2014, 28, 341–355. [Google Scholar] [CrossRef] [Green Version]
- Werren, J.H.; Baldo, L.; Clark, M.E. Wolbachia: Master manipulators of invertebrate biology. Nat. Rev. Microbiol. 2008, 6, 741–751. [Google Scholar] [CrossRef]
- Zug, R.; Hammerstein, P. Still a host of hosts for Wolbachia: Analysis of recent data suggests that 40% of terrestrial arthropod species are infected. PLoS ONE 2012, 7, e38544. [Google Scholar] [CrossRef] [Green Version]
- Gerth, M.; Gansauge, M.T.; Weigert, A.; Bleidorn, C. Phylogenomic analyses uncover origin and spread of the Wolbachia pandemic. Nat. Commun. 2014, 5, 5117. [Google Scholar] [CrossRef] [Green Version]
- Ju, J.-F.; Bing, X.-L.; Zhao, D.-S.; Guo, Y.; Xi, Z.; Hoffmann, A.A.; Zhang, K.-J.; Huang, H.-J.; Gong, J.-T.; Zhang, X.; et al. Wolbachia supplement biotin and riboflavin to enhance reproduction in planthoppers. ISME J. 2020, 14, 676–687. [Google Scholar] [CrossRef] [PubMed]
- Newton, I.L.; Rice, D.W. The Jekyll and Hyde symbiont: Could Wolbachia be a nutritional mutualist? J. Bacteriol. 2020, 202, e00589-19. [Google Scholar] [CrossRef] [PubMed]
- Russell, J.A.; Funaro, C.F.; Giraldo, Y.M.; Goldman-Huertas, B.; Suh, D.; Kronauer, D.J.; Moreau, C.S.; Pierce, N.E. A veritable menagerie of heritable bacteria from ants, butterflies, and beyond: Broad molecular surveys and a systematic review. PLoS ONE 2012, 7, e51027. [Google Scholar] [CrossRef] [PubMed]
- Zchori-Fein, E.; Perlman, S.J. Distribution of the bacterial symbiont Cardinium in arthropods. Mol. Ecol. 2004, 13, 2009–2016. [Google Scholar] [CrossRef] [PubMed]
- Duron, O.; Bouchon, D.; Boutin, S.; Bellamy, L.; Zhou, L.; Engelstädter, J.; Hurst, G.D. The diversity of reproductive parasites among arthropods: Wolbachia do not walk alone. BMC Biol. 2008, 6, 27. [Google Scholar] [CrossRef] [Green Version]
- Weinert, L.A.; Werren, J.H.; Aebi, A.; Stone, G.N.; Jiggins, F.M. Evolution and diversity of Rickettsia bacteria. BMC Biol. 2009, 7, 6. [Google Scholar] [CrossRef] [PubMed]
- Jousselin, E.; Cœur d’Acier, A.; Vanlerberghe-Masutti, F.; Duron, O. Evolution and diversity of Arsenophonus endosymbionts in aphids. Mol. Ecol. 2013, 22, 260–270. [Google Scholar] [CrossRef]
- Hosokawa, T.; Kaiwa, N.; Matsuura, Y.; Kikuchi, Y.; Fukatsu, T. Infection prevalence of Sodalis symbionts among stinkbugs. Zool. Lett. 2015, 1, 5. [Google Scholar] [CrossRef] [Green Version]
- Zchori-Fein, E.; Lahav, T.; Freilich, S. Variations in the identity and complexity of endosymbiont combinations in whitefly hosts. Front. Microbiol. 2014, 5, 310. [Google Scholar] [CrossRef] [Green Version]
- Zytynska, S.E.; Weisser, W.W. The natural occurrence of secondary bacterial symbionts in aphids. Ecol. Entomol. 2016, 41, 13–26. [Google Scholar] [CrossRef] [Green Version]
- Sudakaran, S.; Kost, C.; Kaltenpoth, M. Symbiont acquisition and replacement as a source of ecological innovation. Trends Microbiol. 2017, 25, 375–390. [Google Scholar] [CrossRef]
- Moran, N.A.; McCutcheon, J.P.; Nakabachi, A. Genomics and evolution of heritable bacterial symbionts. Annu. Rev. Genet. 2008, 42, 165–190. [Google Scholar] [CrossRef] [Green Version]
- Fisher, R.M.; Henry, L.M.; Cornwallis, C.K.; Kiers, E.T.; West, S.A. The evolution of host-symbiont dependence. Nat. Commun. 2017, 8, 15973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliver, K.M.; Degnan, P.H.; Burke, G.R.; Moran, N.A. Facultative symbionts in aphids and the horizontal transfer of ecologically important traits. Annu. Rev. Entomol. 2010, 55, 247–266. [Google Scholar] [CrossRef] [Green Version]
- Zytynska, S.E.; Tighiouart, K.; Frago, E. Benefits and costs of hosting facultative symbionts in plant-sucking insects: A meta-analysis. Mol. Ecol. 2021, 30, 2483–2494. [Google Scholar] [CrossRef] [PubMed]
- Hosokawa, T.; Koga, R.; Kikuchi, Y.; Meng, X.Y.; Fukatsu, T. Wolbachia as a bacteriocyte-associated nutritional mutualist. Proc. Natl. Acad. Sci. USA 2010, 107, 769–774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wernegreen, J.J.; Kauppinen, S.N.; Brady, S.G.; Ward, P.S. One nutritional symbiosis begat another: Phylogenetic evidence that the ant tribe Camponotini acquired Blochmannia by tending sap-feeding insects. BMC Evol. Biol. 2009, 9, 292. [Google Scholar] [CrossRef] [Green Version]
- Meseguer, A.S.; Manzano-Marín, A.; Coeur D’Acier, A.; Clamens, A.L.; Godefroid, M.; Jousselin, E. Buchnera has changed flatmate but the repeated replacement of co-obligate symbionts is not associated with the ecological expansions of their aphid hosts. Mol. Ecol. 2017, 26, 2363–2378. [Google Scholar] [CrossRef] [PubMed]
- Morrow, J.L.; Hall, A.A.; Riegler, M. Symbionts in waiting: The dynamics of incipient endosymbiont complementation and replacement in minimal bacterial communities of psyllids. Microbiome 2017, 5, 58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Říhová, J.; Batani, G.; Rodríguez-Ruano, S.M.; Martinů, J.; Vácha, F.; Nováková, E.; Hypša, V. A new symbiotic lineage related to Neisseria and Snodgrassella arises from the dynamic and diverse microbiomes in sucking lice. Mol. Ecol. 2021, 30, 2178–2196. [Google Scholar] [CrossRef] [PubMed]
- Simon, J.C.; Carre, S.; Boutin, M.; Prunier-Leterme, N.; Sabater-Muñoz, B.; Latorre, A.; Bournoville, R. Host-based divergence in populations of the pea aphid: Insights from nuclear markers and the prevalence of facultative symbionts. Proc. R. Soc. B-Biol. Sci. 2003, 270, 1703–1712. [Google Scholar] [CrossRef] [Green Version]
- Toju, H.; Fukatsu, T. Diversity and infection prevalence of endosymbionts in natural populations of the chestnut weevil: Relevance of local climate and host plants. Mol. Ecol. 2011, 20, 853–868. [Google Scholar] [CrossRef]
- Ferrari, J.; West, J.A.; Via, S.; Godfray, H.C.J. Population genetic structure and secondary symbionts in host-associated populations of the pea aphid complex. Evolution 2012, 66, 375–390. [Google Scholar] [CrossRef]
- Perlman, S.J.; Kelly, S.E.; Hunter, M.S. Population biology of cytoplasmic incompatibility: Maintenance and spread of Cardinium symbionts in a parasitic wasp. Genetics 2008, 178, 1003–1011. [Google Scholar] [CrossRef] [Green Version]
- Osaka, R.; Nomura, M.; Watada, M.; Kageyama, D. Negative effects of low temperatures on the vertical transmission and infection density of a Spiroplasma endosymbiont in Drosophila hydei. Curr. Microbiol. 2008, 57, 335–339. [Google Scholar] [CrossRef]
- Hague, M.T.; Mavengere, H.; Matute, D.R.; Cooper, B.S. Environmental and genetic contributions to imperfect wMel-like Wolbachia transmission and frequency variation. Genetics 2020, 215, 1117–1132. [Google Scholar] [CrossRef]
- Turelli, M.; Hoffmann, A.A. Rapid spread of an inherited incompatibility factor in California Drosophila. Nature 1991, 353, 440–442. [Google Scholar] [CrossRef] [PubMed]
- Schuler, H.; Bertheau, C.; Egan, S.P.; Feder, J.L.; Riegler, M.; Schlick-Steiner, B.C.; Steiner, F.M.; Johannesen, J.; Kern, P.; Tuba, K.; et al. Evidence for a recent horizontal transmission and spatial spread of Wolbachia from endemic Rhagoletis cerasi (Diptera: Tephritidae) to invasive Rhagoletis cingulata in Europe. Mol. Ecol. 2013, 22, 4101–4111. [Google Scholar] [CrossRef] [PubMed]
- Kriesner, P.; Hoffmann, A.A.; Lee, S.F.; Turelli, M.; Weeks, A.R. Rapid sequential spread of two Wolbachia variants in Drosophila simulans. PLoS Pathog. 2013, 9, e1003607. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, A.A.; Hercus, M.; Dagher, H. Population dynamics of the Wolbachia infection causing cytoplasmic incompatibility in Drosophila melanogaster. Genetics 1998, 148, 221–231. [Google Scholar] [CrossRef]
- Osaka, R.; Watada, M.; Kageyama, D.; Nomura, M. Population dynamics of a maternally-transmitted Spiroplasma infection in Drosophila hydei. Symbiosis 2010, 52, 41–45. [Google Scholar] [CrossRef]
- Smith, A.H.; O’Connor, M.P.; Deal, B.; Kotzer, C.; Lee, A.; Wagner, B.; Joffe, J.; Woloszynek, S.; Oliver, K.M.; Russell, J.A. Does getting defensive get you anywhere?—Seasonal balancing selection, temperature, and parasitoids shape real-world, protective endosymbiont dynamics in the pea aphid. Mol. Ecol. 2021, 30, 2449–2472. [Google Scholar] [CrossRef]
- Dyer, K.A.; Jaenike, J. Evolutionarily stable infection by a male-killing endosymbiont in Drosophila innubila. Genetics 2004, 168, 1443–1455. [Google Scholar] [CrossRef] [Green Version]
- Cockburn, S.N.; Haselkorn, T.S.; Hamilton, P.T.; Landzberg, E.; Jaenike, J.; Perlman, S.J. Dynamics of the continent-wide spread of a Drosophila defensive symbiont. Ecol. Lett. 2013, 16, 609–616. [Google Scholar] [CrossRef]
- Russell, J.A.; Weldon, S.; Smith, A.H.; Kim, K.L.; Hu, Y.; Łukasik, P.; Doll, S.; Anastopoulos, I.; Novin, M.; Oliver, K.M. Uncovering symbiont-driven genetic diversity across North American pea aphids. Mol. Ecol. 2013, 22, 2045–2059. [Google Scholar] [CrossRef]
- Smith, A.H.; Łukasik, P.; O’Connor, M.P.; Lee, A.; Mayo, G.; Drott, M.T.; Doll, S.; Tuttle, R.; Disciullo, R.A.; Messina, A.; et al. Patterns, causes and consequences of defensive microbiome dynamics across multiple scales. Mol. Ecol. 2015, 24, 1135–1149. [Google Scholar] [CrossRef]
- Campbell, A.; Frazer, B.D.; Gilbert, N.; Gutierrez, A.P.; Mackauer, M. Temperature requirements of some aphids and their para- sites. J. Appl. Ecol. 1974, 11, 431–438. [Google Scholar] [CrossRef]
- Wyatt, I.J.; White, P.F. Simple estimation of intrinsic increase rates for aphids and tetranychid mites. J. Appl. Ecol. 1977, 14, 757–766. [Google Scholar] [CrossRef]
- Rock, D.I.; Smith, A.H.; Joffe, J.; Albertus, A.; Wong, N.; O’Connor, M.; Oliver, K.M.; Russell, J.A. Context-dependent vertical transmission shapes strong endosymbiont community structure in the pea aphid, Acyrthosiphon pisum. Mol. Ecol. 2018, 27, 2039–2056. [Google Scholar] [CrossRef] [PubMed]
- Oliver, K.M.; Higashi, C.H. Variations on a protective theme: Hamiltonella defensa infections in aphids variably impact parasitoid success. Curr. Opin. Insect Sci. 2019, 32, 1–7. [Google Scholar] [CrossRef]
- Ives, A.R.; Barton, B.T.; Penczykowski, R.M.; Harmon, J.P.; Kim, K.L.; Oliver, K.; Radeloff, V.C. Self-perpetuating ecological–evolutionary dynamics in an agricultural host–parasite system. Nat. Ecol. Evol. 2020, 4, 702–711. [Google Scholar] [CrossRef] [PubMed]
- Oliver, K.M.; Campos, J.; Moran, N.A.; Hunter, M.S. Population dynamics of defensive symbionts in aphids. Proc. R. Soc. B-Biol. Sci. 2008, 275, 293–299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doremus, M.R.; Smith, A.H.; Kim, K.L.; Holder, A.J.; Russell, J.A.; Oliver, K.M. Breakdown of a defensive symbiosis, but not endogenous defenses, at elevated temperatures. Mol. Ecol. 2018, 27, 2138–2151. [Google Scholar] [CrossRef]
- Higashi, C.H.; Barton, B.T.; Oliver, K.M. Warmer nights offer no respite for a defensive mutualism. J. Anim. Ecol. 2020, 89, 1895–1905. [Google Scholar] [CrossRef]
- Russell, J.A.; Moran, N.A. Horizontal transfer of bacterial symbionts: Heritability and fitness effects in a novel aphid host. Appl. Environ. Microbiol. 2005, 71, 7987–7994. [Google Scholar] [CrossRef] [Green Version]
- Russell, J.A.; Moran, N.A. Costs and benefits of symbiont infection in aphids: Variation among symbionts and across temperatures. Proc. R. Soc. B-Biol. Sci. 2006, 273, 603–610. [Google Scholar] [CrossRef] [Green Version]
- Simon, J.-C.; Boutin, S.; Tsuchida, T.; Koga, R.; Le Gallic, J.-F.; Frantz, A.; Outreman, Y.; Fukatsu, T. Facultative symbiont infections affect aphid reproduction. PLoS ONE 2011, 6, e21831. [Google Scholar] [CrossRef] [Green Version]
- Heyworth, E.R.; Smee, M.R.; Ferrari, J. Aphid facultative symbionts aid recovery of their obligate symbiont and their host after heat stress. Front. Ecol. Evol. 2020, 8, 56. [Google Scholar] [CrossRef] [Green Version]
- Charlat, S.; Duplouy, A.; Hornett, E.A.; Dyson, E.A.; Davies, N.; Roderick, G.K.; Wedell, N.; Hurst, G.D. The joint evolutionary histories of Wolbachia and mitochondria in Hypolimnas bolina. BMC Evol. Biol. 2009, 9, 64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schuler, H.; Köppler, K.; Daxböck-Horvath, S.; Rasool, B.; Krumböck, S.; Schwarz, D.; Hoffmeister, T.S.; Schlick-Steiner, B.; Steiner, F.M.; Telschow, A.; et al. The hitchhiker’s guide to Europe: The infection dynamics of an ongoing Wolbachia invasion and mitochondrial selective sweep in Rhagoletis cerasi. Mol. Ecol. 2016, 25, 1595–1609. [Google Scholar] [CrossRef]
- Hrček, J.; McLean, A.H.; Godfray, H.C.J. Symbionts modify interactions between insects and natural enemies in the field. J. Anim. Ecol. 2016, 85, 1605–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rothacher, L.; Ferrer-Suay, M.; Vorburger, C. Bacterial endosymbionts protect aphids in the field and alter parasitoid community composition. Ecology 2016, 97, 1712–1723. [Google Scholar] [CrossRef]
- Frago, E.; Mala, M.; Weldegergis, B.T.; Yang, C.; McLean, A.; Godfray, H.C.J.; Gols, R.; Dicke, M. Symbionts protect aphids from parasitic wasps by attenuating herbivore-induced plant volatiles. Nat. Commun. 2017, 8, 1860. [Google Scholar] [CrossRef] [Green Version]
- Russell, J.A.; Latorre, A.; Sabater-Munoz, B.; Moya, A.; Moran, N.A. Side-stepping secondary symbionts: Widespread horizon- tal transfer across and beyond the Aphidoidea. Mol. Ecol. 2003, 12, 1061–1075. [Google Scholar] [CrossRef] [PubMed]
- Fukatsu, T.; Tsuchida, T.; Nikoh, N.; Koga, R. Spiroplasma symbiont of the pea aphid, Acyrthosiphon pisum (Insecta: Homoptera). Appl. Environ. Microbiol. 2001, 67, 1284–1291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pons, I.; Scieur, N.; Dhondt, L.; Renard, M.E.; Renoz, F.; Hance, T. Ubiquity of the symbiont Serratia symbiotica in the aphid natural environment: Distribution, diversity and evolution at a multitrophic level. bioRxiv 2021. [Google Scholar] [CrossRef]
- Manzano-Marín, A.; Simon, J.C.; Latorre, A. Reinventing the wheel and making it round again: Evolutionary convergence in Buchnera–Serratia symbiotic consortia between the distantly related Lachninae aphids Tuberolachnus salignus and Cinara cedri. Genome Biol. Evol. 2016, 8, 1440–1458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Łukasik, P.; van Asch, M.; Guo, H.; Ferrari, J.; Charles, J.; Godfray, H. Unrelated facultative endosymbionts protect aphids against a fungal pathogen. Ecol. Lett. 2013, 16, 214–218. [Google Scholar] [CrossRef] [PubMed]
- Parker, B.J.; Spragg, C.J.; Altincicek, B.; Gerardo, N.M. Symbiont-mediated protection against fungal pathogens in pea aphids: A role for pathogen specificity? Appl. Environ. Microbiol. 2013, 79, 2455–2458. [Google Scholar] [CrossRef] [Green Version]
- Chen, D.Q.; Montllor, C.B.; Purcell, A.H. Fitness effects of two facultative endosymbiotic bacteria on the pea aphid, Acyrthosiphon pisum, and the blue alfalfa aphid, A. kondoi. Entomol. Exp. Appl. 2000, 95, 315–323. [Google Scholar] [CrossRef]
- Montllor, C.B.; Maxmen, A.; Purcell, A.H. Facultative bacterial endosymbionts benefit pea aphids Acyrthosiphon pisum under heat stress. Ecol. Entomol. 2002, 27, 189–195. [Google Scholar] [CrossRef]
- Mathé-Hubert, H.; Kaech, H.; Ganesanandamoorthy, P.; Vorburger, C. Evolutionary costs and benefits of infection with diverse strains of Spiroplasma in pea aphids. Evolution 2019, 73, 1466–1481. [Google Scholar] [CrossRef]
- McLean, A.H.C.; Hrček, J.; Parker, B.J.; Mathé-Hubert, H.; Kaech, H.; Paine, C.; Godfray, H.C.J. Multiple phenotypes conferred by a single insect symbiont are independent. Proc. R. Soc. B-Biol. Sci. 2020, 287, 20200562. [Google Scholar] [CrossRef]
- Heyworth, E.R.; Ferrari, J. A facultative endosymbiont in aphids can provide diverse ecological benefits. J. Evol. Biol. 2015, 28, 1753–1760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smee, M.R.; Raines, S.A.; Ferrari, J. Genetic identity and genotype × genotype interactions between symbionts outweigh species level effects in an insect microbiome. ISME J. 2021, 15, 2537–2546. [Google Scholar] [CrossRef] [PubMed]
- Doremus, M.R.; Oliver, K.M. Aphid heritable symbiont exploits defensive mutualism. Appl. Environ. Microbiol. 2017, 83, e03276-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinez, A.J.; Doremus, M.R.; Kraft, L.J.; Kim, K.L.; Oliver, K.M. Multi-modal defenses in aphids offer redundant protection and increased costs likely impeding a protective mutualism. J. Anim. Ecol. 2018, 87, 464–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dunbar, H.E.; Wilson, A.C.C.; Ferguson, N.R.; Moran, N.A. Aphid thermal tolerance is governed by a point mutation in bacterial symbionts. PLoS Biol. 2007, 5, e96. [Google Scholar] [CrossRef]
- Burke, G.R.; McLaughlin, H.J.; Simon, J.C.; Moran, N.A. Dynamics of a recurrent Buchnera mutation that affects thermal tolerance of pea aphid hosts. Genetics 2010, 186, 367–372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliver, K.M.; Russell, J.A.; Moran, N.A.; Hunter, M.S. Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. Proc. Natl. Acad. Sci. USA 2003, 100, 1803–1807. [Google Scholar] [CrossRef] [Green Version]
- Costopoulos, K.; Kovacs, J.L.; Kamins, A.; Gerardo, N.M. Aphid facultative symbionts reduce survival of the predatory lady beetle Hippodamia convergens. BMC Ecol. 2014, 14, 5. [Google Scholar] [CrossRef] [Green Version]
- Straub, C.S.; Faselt, J.A.; Keyser, E.S.; Traugott, M. Host plant resistance promotes a secondary pest population. Ecosphere 2020, 11, e03073. [Google Scholar] [CrossRef]
- Rubin, D.B. Multiple imputation after 18+ years. J. Am. Stat. Assoc. 1996, 91, 473–489. [Google Scholar] [CrossRef]
- Nakagawa, S. Missing data: Mechanisms, methods, and messages. In Ecological Statistics: Contemporary Theory and Application; Fox, G.A., Negrete-Yankelevich, S., Sosa, V.J., Eds.; Oxford University Press: Oxford, UK, 2015; pp. 81–105. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Weldon, S.R.; Russell, J.A.; Oliver, K.M. More is not always better: Coinfections with defensive symbionts generate highly variable outcomes. Appl. Environ. Microbiol. 2019, 86, e02537-19. [Google Scholar] [CrossRef]
- McLean, A.H.; Parker, B.J.; Hrček, J.; Kavanagh, J.C.; Wellham, P.A.; Godfray, H.C.J. Consequences of symbiont co-infections for insect host phenotypes. J. Anim. Ecol. 2018, 87, 478–488. [Google Scholar] [CrossRef]
- Moran, N.A. The evolution of aphid life cycles. Annu. Rev. Entomol. 1992, 37, 321–348. [Google Scholar] [CrossRef]
- Wilson, A.C.; Sunnucks, P.; Hales, D.F. Heritable genetic variation and potential for adaptive evolution in asexual aphids (Aphidoidea). Biol. J. Linn. Soc. 2003, 79, 115–135. [Google Scholar] [CrossRef] [Green Version]
- Lenhart, P.A.; White, J.A. Endosymbionts facilitate rapid evolution in a polyphagous herbivore. J. Evol. Biol. 2020, 33, 1507–1511. [Google Scholar] [CrossRef]
- Martel, S.I.; Ossa, C.G.; Simon, J.C.; Figueroa, C.C.; Bozinovic, F. Latitudinal trend in the reproductive mode of the pea aphid Acyrthosiphon pisum invading a wide climatic range. Ecol. Evol. 2020, 10, 8289–8298. [Google Scholar] [CrossRef] [PubMed]
- Caillaud, M.C.; Mondor-Genson, G.; Levine-Wilkinson, S.; Mieuzet, L.; Frantz, A.; Simon, J.C.; Coeur d’Acier, A. Microsatellite DNA markers for the pea aphid Acyrthosiphon pisum. Mol. Ecol. Notes 2004, 4, 446–448. [Google Scholar] [CrossRef]
- Wilson, A.C.; Massonnet, B.; Simon, J.C.; Prunier-Leterme, N.; Dolatti, L.; Llewellyn, K.S.; Figueroa, C.C.; Ramirez, C.C.; Blackman, R.L.; Estoup, A.; et al. Cross-species amplification of microsatellite loci in aphids: Assessment and application. Mol. Ecol. Notes 2004, 4, 104–109. [Google Scholar] [CrossRef]
- Kovacs, J.L.; Wolf, C.; Voisin, D.; Wolf, S. Evidence of indirect symbiont conferred protection against the predatory lady beetle Harmonia axyridis in the pea aphid. BMC Ecol. 2017, 17, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harmon, J.P.; Moran, N.A.; Ives, A.R. Species response to environmental change: Impacts of food web interactions and evolution. Science 2009, 323, 1347–1350. [Google Scholar] [CrossRef] [Green Version]
- Tsuchida, T.; Koga, R.; Shibao, H.; Matsumoto, T.; Fukatsu, T. Diversity and geographic distribution of secondary endosymbiotic bacteria in natural populations of the pea aphid, Acyrthosiphon pisum. Mol. Ecol. 2002, 11, 2123–2135. [Google Scholar] [CrossRef]
- Henry, L.M.; Peccoud, J.; Simon, J.C.; Hadfield, J.D.; Maiden, M.J.; Ferrari, J.; Godfray, H.C.J. Horizontally transmitted symbionts and host colonization of ecological niches. Curr. Biol. 2013, 23, 1713–1717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leonardo, T.E.; Mondor, E.B. Symbiont modifies host life-history traits that affect gene flow. Proc. R. Soc. B Biol. Sci. 2006, 273, 1079–1084. [Google Scholar] [CrossRef] [Green Version]
- Mathé-Hubert, H.; Kaech, H.; Hertaeg, C.; Jaenike, J.; Vorburger, C. Nonrandom associations of maternally transmitted symbionts in insects: The roles of drift versus biased cotransmission and selection. Mol. Ecol. 2019, 28, 53300–55346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gehrer, L.; Vorburger, C. Parasitoids as vectors of facultative bacterial endosymbionts in aphids. Biol. Lett. 2012, 8, 613–615. [Google Scholar] [CrossRef] [PubMed]
- Moran, N.A.; Jarvik, T. Lateral transfer of genes from fungi underlies carotenoid production in aphids. Science 2010, 328, 624–627. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Bickel, R.D.; Parker, B.J.; Ziabari, S.S.; Liu, F.; Vellichirammal, N.N.; Simon, J.-C.; DLBrisson, J.A.S. A large genomic insertion containing a duplicated follistatin gene is linked to the pea aphid male wing dimorphism. eLife 2020, 9, e50608. [Google Scholar] [CrossRef] [PubMed]
- Nichols, H.L.; Goldstein, E.B.; Saleh Ziabari, O.; Parker, B.J. Intraspecific variation in immune gene expression and heritable symbiont density. PLoS Pathog. 2021, 17, e1009552. [Google Scholar] [CrossRef]
- Via, S.; Shaw, A.J. Short-term evolution in the size and shape of pea aphids. Evolution 1996, 50, 163–173. [Google Scholar]
- Losey, J.E.; Ives, A.R.; Harmon, J.; Ballentyne, F.; Brown, C. A polymorphism maintained by opposite patterns of parasitism and predation. Nature 1997, 388, 269–272. [Google Scholar] [CrossRef]
- Siddiqui, W.H.; Barlow, C.A.; Randolph, P.A. Effects of some constant and alternating temperatures on population growth of the pea aphid, Acyrthosiphon pisum (Homoptera: Aphididae). Can. Entomol. 1973, 105, 145–156. [Google Scholar] [CrossRef]
- Bergland, A.O.; Behrman, E.L.; O’Brien, K.R.; Schmidt, P.S.; Petrov, D.A. Genomic evidence of rapid and stable adaptive oscillations over seasonal time scales in Drosophila. PLoS Genet. 2014, 10, e1004775. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clay, K.; Holan, J.; Rudgers, J.A. Herbivores cause a rapid increase in hereditary symbiosis and alter plant community composition. Proc. Natl. Acad. Sci. USA 2005, 102, 12465–12470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barton, B.T.; Schmitz, O.J. Opposite effects of daytime and nighttime warming on top-down control of plant diversity. Ecology 2018, 99, 13–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morag, N.; Klement, E.; Saroya, Y.; Lensky, I.; Gottlieb, Y. Prevalence of the symbiont Cardinium in Culicoides (Diptera: Ceratopogonidae) vector species is associated with land surface temperature. FASEB J. 2012, 26, 4025–4034. [Google Scholar] [CrossRef]
- Chen, D.Q.; Purcell, A.H. Occurrence and transmission of facultative endosymbionts in aphids. Curr. Microbiol. 1997, 34, 220–225. [Google Scholar] [CrossRef]
- Darby, A.C.; Douglas, A.E. Elucidation of the transmission patterns of an insect-borne bacterium. Appl. Environ. Microbiol. 2003, 69, 4403–4407. [Google Scholar] [CrossRef] [Green Version]
- Skaljac, M.; Vogel, H.; Wielsch, N.; Mihajlovic, S.; Vilcinskas, A. Transmission of a protease-secreting bacterial symbiont among pea aphids via host plants. Front. Physiol. 2019, 10, 438. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.D.; Lei, H.X.; Chen, F.F. Infection pattern and negative effects of a facultative endosymbiont on its insect host are environment-dependent. Sci. Rep. 2019, 9, 4013. [Google Scholar] [CrossRef]
- Chiel, E.; Gottlieb, Y.; Zchori-Fein, E.; Mozes-Daube, N.; Katzir, N.; Inbar, M.; Ghanim, M. Biotype-dependent secondary symbiont communities in sympatric populations of Bemisia tabaci. Bull. Entomol. Res. 2007, 97, 407–413. [Google Scholar] [CrossRef] [Green Version]
- Ferrari, J.; Vavre, F. Bacterial symbionts in insects or the story of communities affecting communities. Philos. Trans. R. Soc. B-Biol. Sci. 2011, 366, 1389–1400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaenike, J. Population genetics of beneficial heritable symbionts. Trends Ecol. Evol. 2012, 27, 226–232. [Google Scholar] [CrossRef]
- Łukasik, P.; Guo, H.; van Asch, M.; Ferrari, J.; Godfray, H.C.J. Protection against a fungal pathogen conferred by the aphid facultative endosymbionts Rickettsia and Spiroplasma is expressed in multiple host genotypes and species and is not influenced by co-infection with another symbiont. J. Evol. Biol. 2013, 26, 2654–2661. [Google Scholar] [CrossRef]
- Oliver, K.M.; Moran, N.A.; Hunter, M.S. Costs and benefits of a superinfection of facultative symbionts in aphids. Proc. R. Soc. B-Biol. Sci. 2006, 273, 1273–1280. [Google Scholar] [CrossRef]
- Cayetano, L.; Rothacher, L.; Simon, J.C.; Vorburger, C. Cheaper is not always worse: Strongly protective isolates of a defensive symbiont are less costly to the aphid host. Proc. R. Soc. B Biol. Sci. 2015, 282, 20142333. [Google Scholar] [CrossRef]
- Parker, B.J.; Hrček, J.; McLean, A.H.C.; Godfray, H.C.J. Genotype specificity among hosts, pathogens, and beneficial microbes influences the strength of symbiont-mediated protection. Evolution 2017, 71, 1222–1231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, J.; Butler, S.; Sanchez, G.; Mateos, M. Male killing Spiroplasma protects Drosophila melanogaster against two parasitoid wasps. Heredity 2014, 112, 399–408. [Google Scholar] [CrossRef]
- Watanabe, M.; Miura, K.; Hunter, M.S.; Wajnberg, E. Superinfection of cytoplasmic incompatibility-inducing Wolbachia is not additive in Orius strigicollis (Hemiptera: Anthocoridae). Heredity 2011, 106, 642–648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goto, S.; Anbutsu, H.; Fukatsu, T. Asymmetrical interactions between Wolbachia and Spiroplasma endosymbionts coexisting in the same insect host. Appl. Environ. Microbiol. 2006, 72, 4805–4810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Audsley, M.; Seleznev, A.; Albert Joubert, D.; Woolfit, M.; O’Neill, S.L.; McGraw, E.A. Wolbachia infection alters the relative abundance of resident bacteria in adult Aedes aegypti mosquitoes, but not larvae. Mol. Ecol. 2018, 27, 297–309. [Google Scholar] [CrossRef]
- Dittmer, J.; Bouchon, D. Feminizing Wolbachia influence microbiota composition in the terrestrial isopod Armadillidium vulgare. Sci. Rep. 2018, 8, 6998. [Google Scholar] [CrossRef]
- Guay, J.-F.; Boudreault, S.; Michaud, D.; Cloutier, C. Impact of environmental stress on aphid clonal resistance to parasitoids: Role of Hamiltonella defensa bacterial symbiosis in association with a new facultative symbiont of the pea aphid. J. Insect. Physiol. 2009, 55, 919–926. [Google Scholar] [CrossRef]
- Hurst, G.D.D.; Jiggins, F.M. Problems with mitochondrial DNA as a marker in population, phylogeographic and phylogenetic studies: The effects of inherited symbionts. Proc. R. Soc. B Biol. 2006, 272, 1525–1534. [Google Scholar] [CrossRef] [PubMed]
- Johnstone, R.A.; Hurst, G.D.D. Maternally inherited male-killing microorganisms may confound interpretation of mitochondrial DNA variability. Biol. J. Linn. Soc. 1996, 58, 453–470. [Google Scholar] [CrossRef]
- Hinrich, J.; Schulenburg, G.V.; Hurst, G.D.D.; Tetzlaff, D.; Booth, G.E.; Zakharov, I.A.; Majerus, M.E.N. History of infection with different male-killing bacteria in the two-spot ladybird beetle Adalia bipunctata revealed through mitochondrial DNA sequence analysis. Genetics 2002, 160, 1075–1086. [Google Scholar] [CrossRef]
- Jiggins, F.M. Male-killing Wolbachia and mitochondrial DNA: Selective sweeps, hybrid introgression and parasite population dynamics. Genetics 2003, 164, 5–12. [Google Scholar] [CrossRef] [PubMed]
- Shoemaker, D.D.; Dyer, K.A.; Ahrens, M.; McAbee, K.; Jaenike, J. Decreased diversity but increased substitution rate in host mtDNA as a consequence of Wolbachia endosymbiont infection. Genetics 2004, 168, 2049–2058. [Google Scholar] [CrossRef] [Green Version]




| Symbiont Tested Across 2012 | AIC in Model with vs. without Period † | p-Value Representing the Difference between the 2 Models †† |
|---|---|---|
| Fukatsuia | 231.59 vs. 230.16 | 0.0264 ** |
| Hamiltonella * | 408.2 vs. 471.4 | 1.99 × 10−13 |
| Regiella | 328.87 vs. 359.89 | 1.78 × 10−7 |
| Rickettsia | 352.20 vs. 370.78 | 2.46 × 10−5 |
| Rickettsiella | 351.05 vs. 485.35 | 2.20 × 10−16 |
| Serratia | 263.67 vs. 300.53 | 1.61 × 10−8 |
| Spiroplasma | 244.46 vs. 255.79 | 3.68 × 10−4 |
| Symbiont | Final Model † | Parameter Estimates and Associated Statistics †† | ||||
|---|---|---|---|---|---|---|
| Estimate | Std. Error | z Value | Pr(>|z|) | |||
| Regiella | P(Reg)~lgAphDen + lgLeafHop + Ppandora + (1|fField) | (Intercept) | −2.6827 | 0.26804 | −10.009 | <2.0 × 10−16 |
| lgAphDen | 0.2366 | 0.11165 | 2.119 | 0.0341 | ||
| lgLeafHop | 0.32962 | 0.08362 | 3.942 | 8.08 × 10−5 | ||
| Ppandora | 1.76821 | 0.32237 | 5.485 | 4.13 × 10−8 | ||
| Rickettsia | P(Rick)~lgAervi + lgLeafHop + Paervi + (1|fField) | (Intercept) | −1.18695 | 0.10071 | −11.786 | <2.0 × 10−16 |
| lgAervi | −0.22329 | 0.11341 | −1.969 | 0.04897 | ||
| lgLeafHop | −0.20100 | 0.07654 | −2.626 | 0.00863 | ||
| Paervi | −1.12356 | 0.43613 | −2.576 | 0.00999 | ||
| Rickettsiella | P(Rkla)~stdmaVap + lgAphDen + Psurv + (1|fField) | (Intercept) | −2.79286 | 0.27472 | −10.166 | <2.0 × 10−16 |
| stdmaVap | −0.33461 | 0.07735 | −4.326 | 1.52 × 10−5 | ||
| lgAphDen | 0.40121 | 0.10271 | 3.906 | 9.37 × 10−5 | ||
| Psurv | 1.01093 | 0.24990 | 4.045 | 5.23 × 10−5 | ||
| Serratia | P(Ser)~lgLeafHop + lgCoccin + Ppandora + (1|fField) | (Intercept) | −2.6756 | 0.1743 | −15.351 | <2.0 × 10−16 |
| lgLeafHop | −0.3763 | 0.1052 | −3.577 | 3.48 × 10−4 | ||
| lgCoccin | −0.3640 | 0.1870 | −1.947 | 0.05158 | ||
| Ppandora | 1.4043 | 0.3770 | 3.725 | 1.96 × 10−4 | ||
| Spiroplasma | P(Spi)~lgAphDen + lgAervi + lgCoccin + Paervi + Ppandora + (1|fField) | (Intercept) | −5.0718 | 0.4726 | −10.733 | <2.0 × 10−16 |
| lgAphDen | 0.7999 | 0.2033 | 3.934 | 8.34 × 10−5 | ||
| lgAervi | −0.4758 | 0.2173 | −2.190 | 0.02852 | ||
| lgCoccin | 1.0285 | 0.2573 | 3.997 | 6.41 × 10−5 | ||
| Paervi | 1.8833 | 0.8617 | 2.186 | 0.02884 | ||
| Ppandora | 2.0677 | 0.5807 | 3.561 | 3.69 × 10−4 | ||
| Focal Symbiont | Time (2-Week Time Interval or Month Overwintering pd.) | Change in Focal Symbiont Prevalence | p-Value for Prevalence Change † | Most Common Partner | p-Value for Prevalence Change after Removing Co-Infections w/Common Partner |
|---|---|---|---|---|---|
| Hamiltonella | 3–4 | −0.207 | 2.32 × 10−4 | Rickettsia | 2.61 × 10−3 |
| 4–5 | 0.197 | 7.50 × 10−4 | Rickettsiella | 0.3368 | |
| 6–7 | 0.262 | 2.62 × 10−5 | Regiella | 1.12 × 10−3 | |
| Overwintering: 2012–2013 | −0.108 | 0.0673 †† | Rickettsia | 0.9433 | |
| Overwintering: 2013–2014 | −0.234 | 3.81 × 10−5 †† | Rickettsiella | 4.73 × 10−3 | |
| Rickettsiella | 4–5 | 0.393 | 7.04 × 10−13 | Hamiltonella | 2.42 × 10−6 |
| 5–6 | −0.232 | 8.35 × 10−4 | Hamiltonella | 0.0154 | |
| 10–11 | 0.301 | 3.92 × 10−4 | Hamiltonella | 0.0433 | |
| 12–13 | −0.236 | 2.91 × 10−7 | Serratia | 3.18 × 10−5 | |
| Overwintering: 2012–2013 | 0.187 | 4.82 × 10−4 | Serratia | 4.13 × 10−5 | |
| Overwintering: 2013–2014 | −0.094 | 0.0183 | Serratia | 9.93 × 10−8 | |
| Hamiltonella | 0.892 | ||||
| Serratia + Hamiltonella | 6.20 × 10−5 | ||||
| Regiella | Overwintering: 2012–2013 | −0.115 | 0.0286 | Hamiltonella | 4.27 × 10−3 |
| Rickettsia | Overwintering: 2012–2013 | −0.144 | 8.96 × 10−3 | Hamiltonella | 0.3869 |
| Overwintering: 2013–2014 | 0.048 | 0.0987 | Hamiltonella | 0.0862 | |
| Serratia | Overwintering: 2013–2014 | 0.203 | 2.82 × 10−8 | Rickettsiella | 0.0113 |
| Spiroplasma | Overwintering: 2013–2014 | 0.167 | 6.80 × 10−6 | Hamiltonella | 2.96 × 10−6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carpenter, M.; Peng, L.; Smith, A.H.; Joffe, J.; O’Connor, M.; Oliver, K.M.; Russell, J.A. Frequent Drivers, Occasional Passengers: Signals of Symbiont-Driven Seasonal Adaptation and Hitchhiking in the Pea Aphid, Acyrthosiphon pisum. Insects 2021, 12, 805. https://doi.org/10.3390/insects12090805
Carpenter M, Peng L, Smith AH, Joffe J, O’Connor M, Oliver KM, Russell JA. Frequent Drivers, Occasional Passengers: Signals of Symbiont-Driven Seasonal Adaptation and Hitchhiking in the Pea Aphid, Acyrthosiphon pisum. Insects. 2021; 12(9):805. https://doi.org/10.3390/insects12090805
Chicago/Turabian StyleCarpenter, Melissa, Linyao Peng, Andrew H. Smith, Jonah Joffe, Michael O’Connor, Kerry M. Oliver, and Jacob A. Russell. 2021. "Frequent Drivers, Occasional Passengers: Signals of Symbiont-Driven Seasonal Adaptation and Hitchhiking in the Pea Aphid, Acyrthosiphon pisum" Insects 12, no. 9: 805. https://doi.org/10.3390/insects12090805
APA StyleCarpenter, M., Peng, L., Smith, A. H., Joffe, J., O’Connor, M., Oliver, K. M., & Russell, J. A. (2021). Frequent Drivers, Occasional Passengers: Signals of Symbiont-Driven Seasonal Adaptation and Hitchhiking in the Pea Aphid, Acyrthosiphon pisum. Insects, 12(9), 805. https://doi.org/10.3390/insects12090805