Simple Summary
In this study, we conducted a transcriptome analysis of adult antennae in Plagiodera versicolora (Coleoptera: Chrysomelidae) and identified a total of 98 candidate chemosensory genes, encoding 40 odorant receptors (ORs), 7 ionotropic receptors (IRs), 13 gustatory receptors (GRs), 10 chemosensory proteins (CSPs), 24 odorant binding proteins (OBPs), and 4 sensory neuron membrane proteins (SNMPs). The tissue expression profiles showed that almost all PverOBPs and PverORs were highly expressed in the antennae. In addition, the results revealed that PverOBP10, PverOBP12, PverOBP18, PverOR24, and PverOR35 showed female-biased expression profiles, indicating that these receptors may be involved in some female-specific behaviors such as oviposition site seeking. This work greatly promotes the understanding of the olfactory system and will help provide insight for functional studies of the chemoreception mechanism in P. versicolora.
Abstract
Insects can sense surrounding chemical signals by their accurate chemosensory systems. This system plays a vital role in the life history of insects. Several gene families participate in chemosensory processes, including odorant receptors (ORs), ionotropic receptors (IRs), gustatory receptors (GRs), chemosensory proteins (CSPs), odorant binding proteins (OBPs), and sensory neuron membrane proteins (SNMPs). Plagiodera versicolora (Coleoptera: Chrysomelidae), is a leaf-eating forest pest found in salicaceous trees worldwide. In this study, a transcriptome analysis of male and female adult antennae in P. versicolora individuals was conducted, which identified a total of 98 candidate chemosensory genes including 40 ORs, 7 IRs, 13 GRs, 10 CSPs, 24 OBPs, and 4 SNMPs. Subsequently, the tissue expression profiles of 15 P. versicolora OBPs (PverOBPs) and 39 ORs (PverORs) were conducted by quantitative real-time PCR. The data showed that almost all PverOBPs and PverORs were highly expressed in the male and female antennae. In addition, several OBPs and ORs (PverOBP10, PverOBP12, PverOBP18, PverOR24, and PverOR35) had higher expression levels in female antennae than those in the male antennae, indicating that these genes may be taking part in some female-specific behaviors, such as find mates, oviposition site, etc. This study deeply promotes further understanding of the chemosensory system and functional studies of the chemoreception genes in P. versicolora.
1. Introduction
Insects depend on a complex chemosensory system to find mates, oviposition sites, foods, and to evade predators or toxic compounds [1]. Generally, the antennae, proboscis, legs, and labial palps are the main chemosensory organs in insects [2]. The chemosensory system involves several different types of genes, including odorant receptors (ORs), odorant binding proteins (OBPs), chemosensory proteins (CSPs), gustatory receptors (GRs), ionotropic receptors (IRs), and sensory neuron membrane proteins (SNMPs) [3,4,5,6]. The peripheral level of the insect chemosensory system general includes several major steps; after entering into the sensilla lymph through pores on the sensilla wall, the hydrophobic odorant molecules are bound by soluble olfactory proteins (e.g., OBPs and CSPs) in the sensilla lymph. This complex is subsequently transported to corresponding chemosensory receptors (ORs, GRs, and IRs proteins), which finally induces an action potential and guides insects’ behavior [4,7,8,9,10]. For example, ApisOBP3 and ApisOBP7 are known to bind, and transport (E)-β-farnesene (the alarm pheromone) to ApisOR5 in Acyrthosiphon pisum, which made aphids escape by their walking away and falling down from the host plant.
Most chemosensory OBPs contain 120–140 amino acids and have a common folding style of six α-helical domains, among which six conserved cysteines form three interlocked disulphide bridges to stabilize the compact structure [11,12]. OBPs are water soluble and are extracellular proteins that are expressed in non-neuronal support cells of the chemosensory sensilla [13,14]. Additionally, OBPs could be categorized into pheromone binding proteins (PBPs) and general odorant binding proteins (GOBPs) in Lepidoptera [15]. OBPs were first identified and studied in Antheraea polyphemus [3].
Following this research, more and more OBPs have been identified and studied among insects [16,17,18]. Insect ORs encode seven transmembrane domain proteins with an inverted membrane topology (intracellular N terminus and an extracellular C terminus) when compared with vertebrate ORs [19,20]. ORs are located in the dendrite membrane of olfactory sensory neurons (OSNs), and are housed within the olfactory sensilla (mainly on the antenna) [2,21]. ORs are considered to play a central role in identifying distinct odorants and activating the OSNs [22,23]. Insect ORs form a novel heteromeric ligand-gated ion channel. These heteromers contain two subunits: a divergent conventional ligand-binding OR subunit [24,25,26,27] and a highly conserved co-receptor (Orco) subunit [28,29,30]. The insect OR protein family was first described in Drosophila [22].
The willow leaf beetle Plagiodera versicolora (Coleoptera: Chrysomelidae), is a leaf-eating forest pest, which mainly damages salicaceous trees, including willows (Salix) and poplars (Populus) [31,32]. However, there is a gap in the research regarding the identified chemosensory gene families in P. versicolora. In this study, we conducted a transcriptome analysis of male and female adults’ antenna in P. versicolora, using second-generation Illumina RNA sequencing, and then, candidate chemosensory genes including OBPs, CSPs, ORs, GRs, IRs and SNMPs were identified. The phylogenetic relationships of these genes to other insect species were analyzed. We also examined the temporal expression profiles of OBP and OR genes by performing quantitative real time PCR (RT-qPCR).
2. Materials and Methods
2.1. Insect and Tissue Collection
The adults and larvae of P. versicolora were collected from Sha Lake Park in Wuhan, China. The rearing conditions were 28 ± 1 °C, with 70 ± 5% RH (relative humidity) and a photoperiod of 12:12 h (light: dark). Larvae and adults were fed with the fresh leaf of willows. Around 300 male and female antennae tissues were excised from 3 day-old adults. After collection, all samples were immediately frozen in liquid nitrogen and stored at −80 °C for RNA extraction.
2.2. RNA Extraction, cDNA Library Construction, and Illumina Sequencing
Total RNA was extracted from collected samples using Trizol Reagent (Invitrogen, Carlsbad, CA, USA) according to protocol. The quality of RNA was checked with a NanoDrop-2000 (Thermo Scientific, Waltham, MA, USA). The Illumina sequencing of the samples was performed by Berry Genomics (Beijing, China). The cDNA library was synthetized with NEBNext® Ultra mRNA Library Prep Kit for Illumina (NEB, Ipswich, MA, USA) following manufacturer’s instructions. The mRNAs were enriched from total RNA using Oligo(dT)-attached magnetic beads and mRNAs were fragmented into short sequences within an RNA fragmentation buffer.
Next, first-strand cDNA was generated with random hexamers by using mRNAs as the template. The buffer, dNTPs, RNase H, and DNA polymerase I were used to synthesize the second strand cDNA. Then, end repair was performed on these double strands of cDNA with a dA-tail was added. After the end repair and ligation of adaptors, the PCR was performed to enrich the cDNA. Finally, the cDNA library was sequenced using the Illumina Novaseq platform.
2.3. De Novo Assembly and Gene Annotation
After removing the raw reads containing the adaptor sequences, low-quality reads, and repeated reads, the clean reads were obtained. The transcriptome was assembled according to these clean reads by using Trinity 2.8.5 to generate a set of transcripts. These transcripts were annotated according to the following databases: NR, Swissprot, KEGG, KOG/COG, and a search conducted in the National Center for Biotechnology Information (NCBI). The open reading frame (ORFs) of each gene were predicted with an ORF finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html (accessed on 10 May 2021). Signal peptides in these sequences were predicted following the SignalP 2.0 server (http://www.cbs.dtu.dk/services/SignalP-2.0/#submission (accessed on 10 May 2021).
2.4. Sequence and Phylogenetic Analyses
The ORFs and amino acid sequences of chemosensory genes (File S1) were obtained and identified, after removing redundant sequences. Phylogenetic trees were constructed with amino acid sequences from P. versicolora and other insect species, including Colaphellus bowringi [33], Ips typographus [34], Dendroctonus ponderosae [34], Dendroctonus valens [35], Monochamus alternatus [36], Dastarcus helophoroides [36], Anoplophora chinensis [37] Basilepta melanopus [38], Tribolium castaneum [39], Megacyllene caryae [40], Plutella xylostella [41], Helicoverpa armigera [42,43,44], Bombyx mori [20,45,46], Drosophila melanogaster [47,48,49,50], Aedes aegypti [51], and Anopheles gambiae [52]. Then, we used ClustalX 1.83 to align amino acid sequences and the MEGA6 neighbor-joining method was used to construct the phylogenetic trees. Lastly, the different trees were viewed and edited with FigTree 1.4.2 software (http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 15 July 2021).
2.5. Quantitative Real-Time PCR (RT-qPCR) Analysis
RT-qPCR was conducted to determine the expression profiles of male and female insects. The cDNAs were synthesized with HiScript® III RT SuperMix for RT-qPCR (+gDNA wiper) (Vazyme, Nanjing, China), based on the manufacturer’s instructions. The specific primers were designed and are listed in Table S1. RT-qPCR was conducted on the CFX Connect Real-Time System (Bio-Rad, Hercules, CA, USA) with a ChamQTM Universal SYBR® RT-qPCR Master Mix (Vazyme, Nanjing, China), following the manufacturer’s instructions. Reaction programs were set at 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s and 60 °C for 34 s. The RPS18 gene [53] was used as a reference to normalize the relative expression levels of OR and OBP genes. For each gene, three biological replicates were conducted. Gene expression levels were analyzed using the 2−ΔΔCT method [54]. The one-way analysis of variance (ANOVA) followed by the Tukey’s HSD test was used to test gene expression using SPSS 26.0 software (SPSS Inc., Chicago, IL, USA).
3. Results
3.1. Overview of the Sequence Assembly
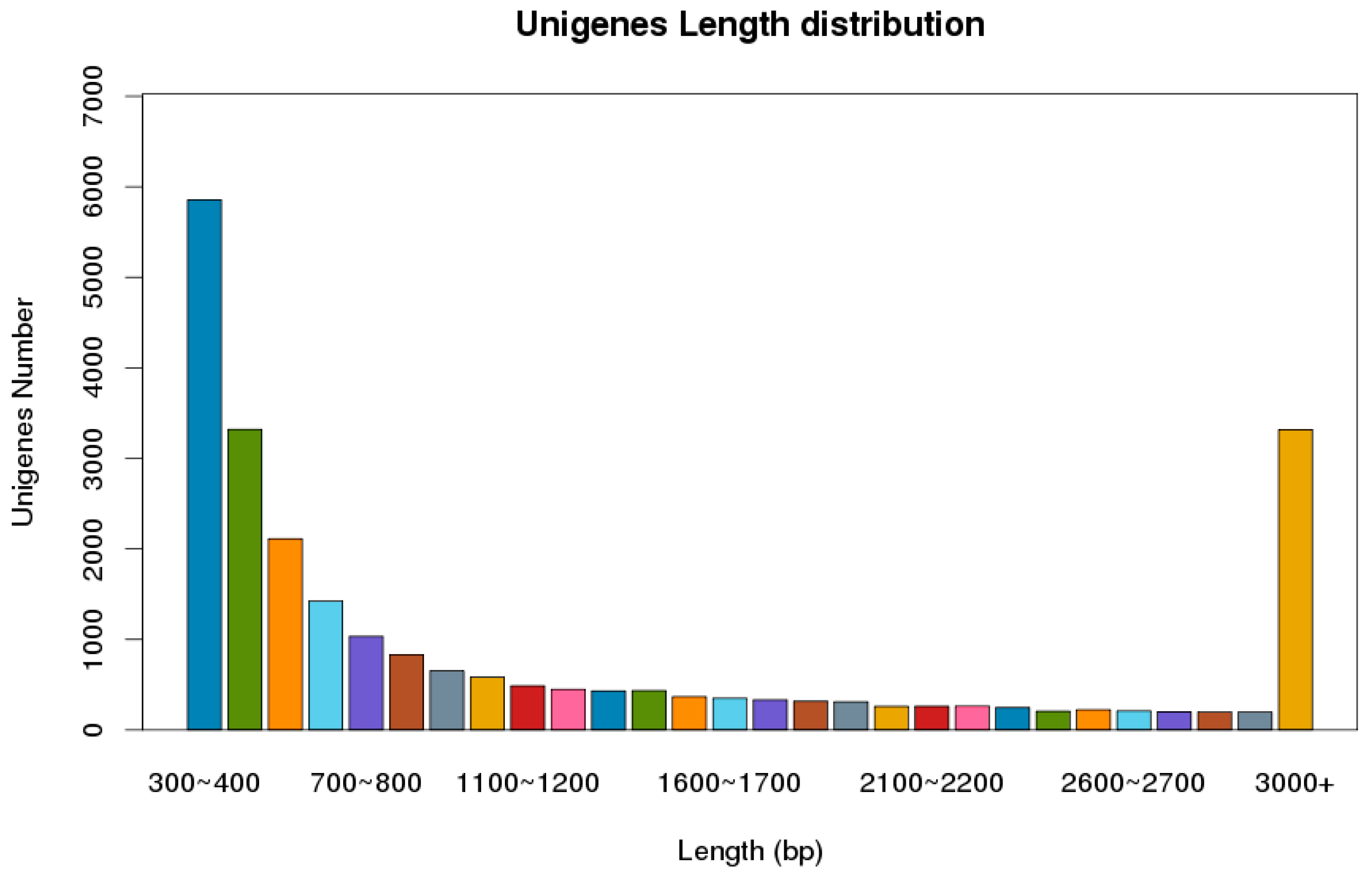
The next-generation sequencing of the cDNA library, using the Illumina Novaseq platform, was constructed from the male and female adult antennae of P. versicolora. In total, 59,893,741 clean reads were obtained with a Q20 percentage of 97.83%. About 24,862 unigenes, with a total length of 34,555,981 and an N50 length of 2675 bp, were identified. Statistics showed that 63.1% of the 15,687 unigenes were greater than 500 bp in length (Figure 1). In total, 11,925 unigenes were matched to entries in the NCBI non-redundant (NR) protein database (http://www.ncbi.nlm.nih.gov/protein (accessed on 10 April 2021) by a BLASTX search.
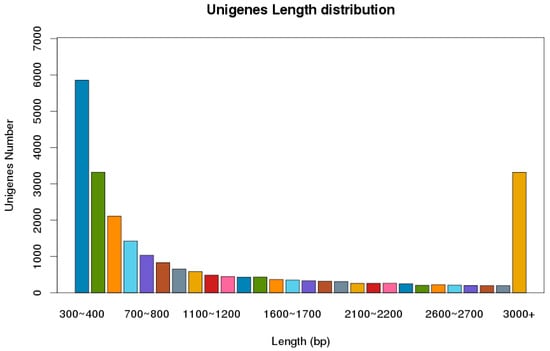
Figure 1.
Distribution of unigene size in the P. versicolora transcriptome assembly.
3.2. Overview of Gene Ontology (GO) Annotation
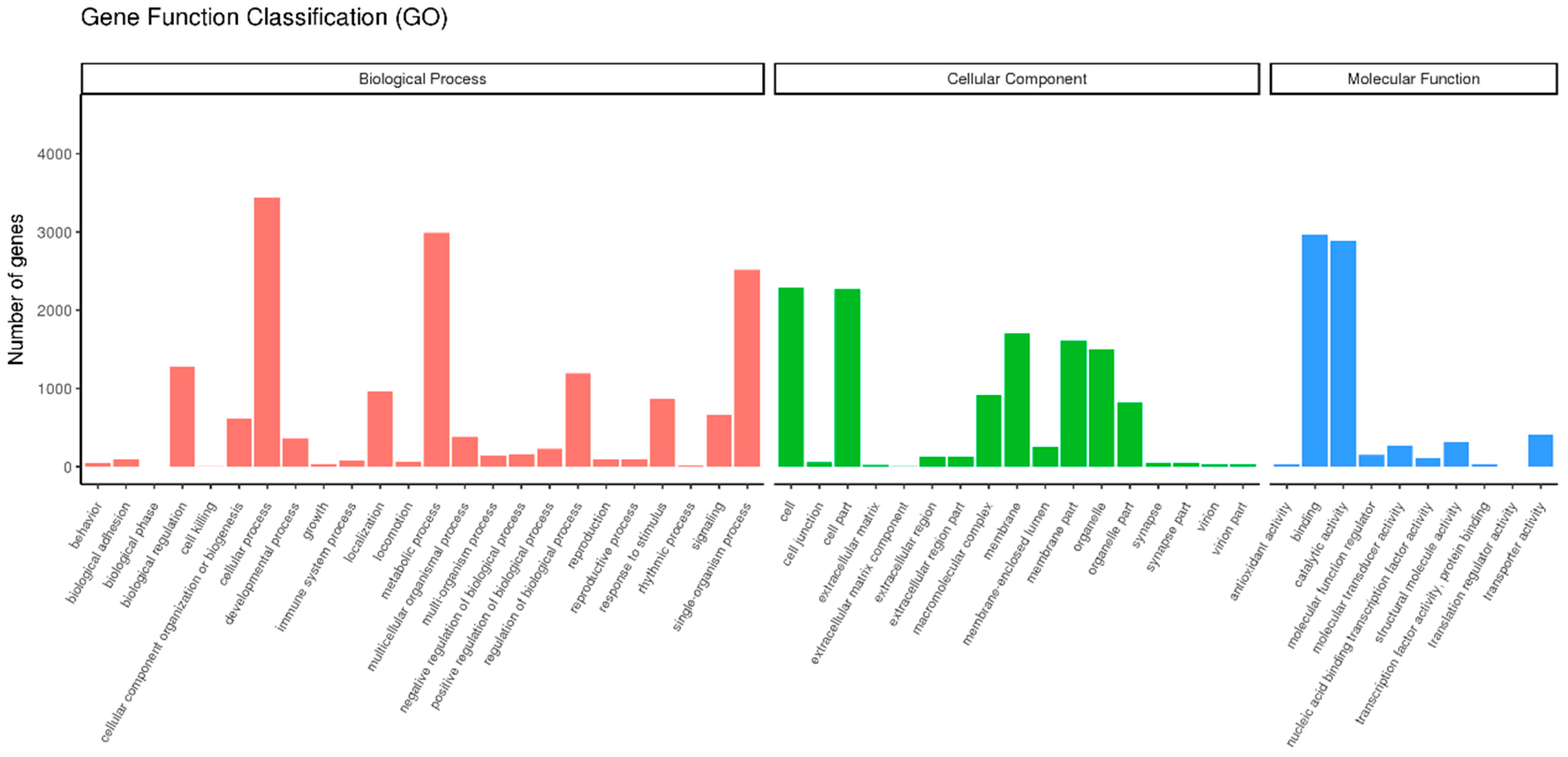
The transcripts were classified into different functional categories based on their GO annotation. Overall, these unigenes could be placed into three functional categories: cellular components (11,847), biological processes (16,293), and molecular function (7150) (Figure 2). In the class of molecular function, the genes expressed in the antennae were mostly related to binding (2966), catalytic (2887), and transporter activities (406), indicating that some unigenes in these sub-categories might have a connection with chemosensory behavior in insects.
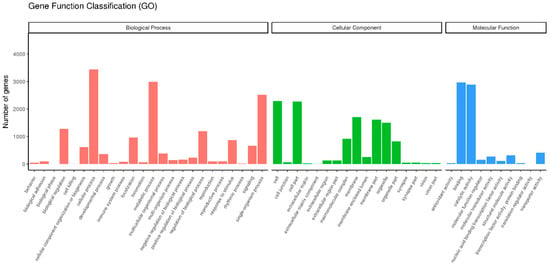
Figure 2.
Gene ontology (GO) classification of P. versicolora unigenes.
3.3. Identification of the Candidate Chemosensory Genes
Based on similarity analyses of the sequences tested, a total of 98 candidate chemosensory genes from the male and female antennae transcriptomes of P. versicolora were identified. These included 40 ORs, 7 IRs, 13 GRs, 10 CSPs, 24 OBPs, and 4 SNMPs (Tables S2 and S3). When compared with insects of Coleoptera, where the chemosensory genes had been identified by transcriptome tests, the number of chemosensory genes identified in this study was similar to those found in C. bowringi (104 chemosensory genes), D. ponderosae (111 chemosensory genes), and was higher than that of M. alternatus (52 chemosensory genes).
3.4. OBPs
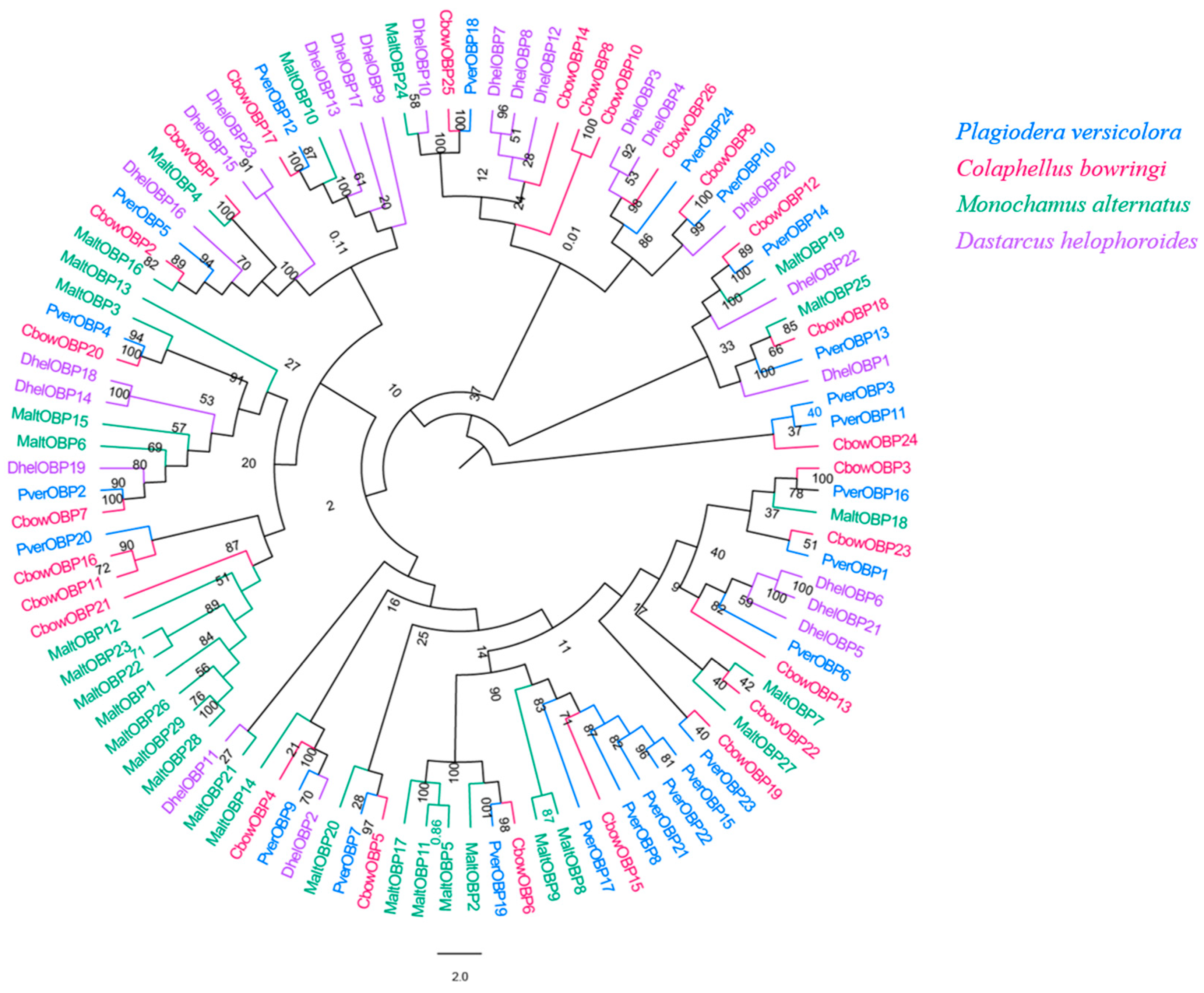
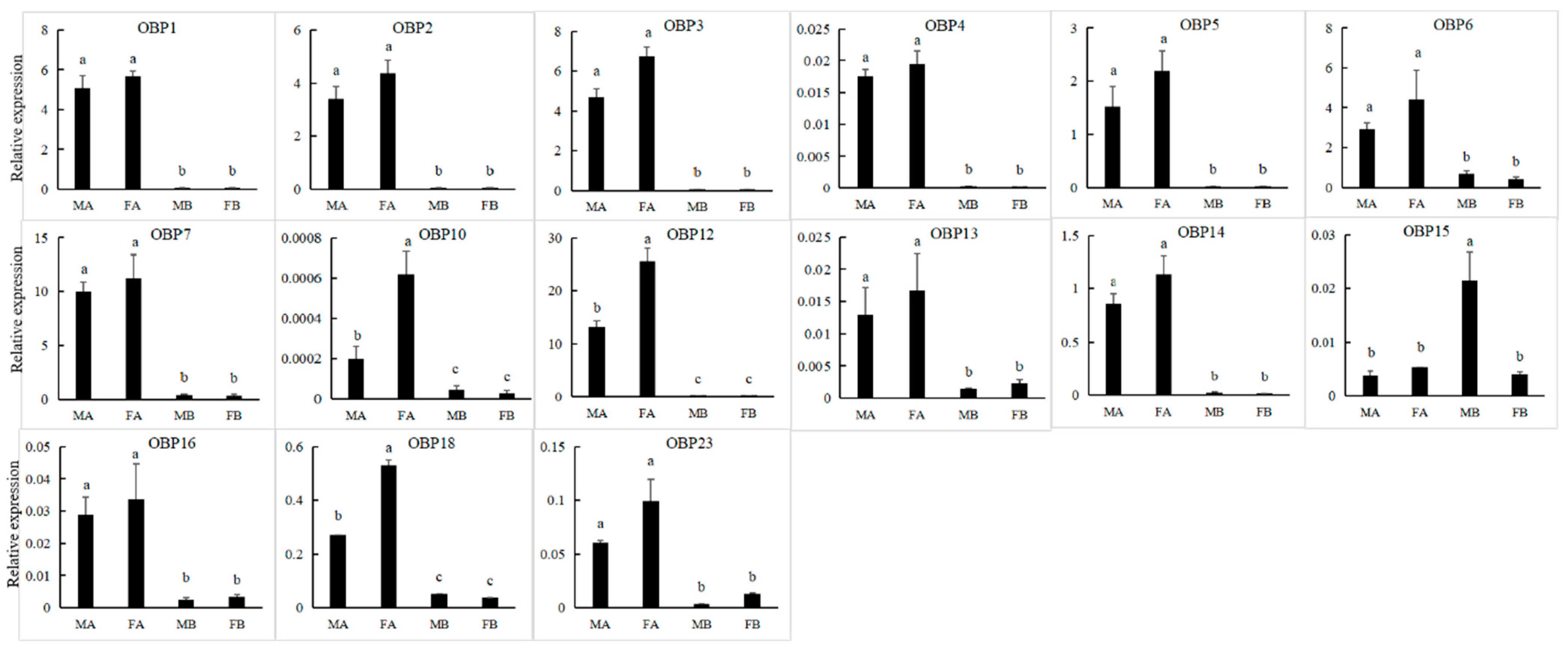
We obtained 24 unigenes encoding candidate OBPs in P. versicolora (PverOBPs), which is less than that observed in M. alternatus (29) and C. bowringi (26), but more than that observed in D. helophoroides (23). Sequence analysis showed that 23 OBPs have complete ORFs and encoded 125 to 226 amino acids, but only three OBPs have no signal peptide sequences (Table S2). The result of the phylogenetic tree showed that PverOBP4 and PverOBP12 were clustered with the functionally characterized MaltOBP13 and MaltOBP10, respectively. In addition, several PverOBPs (OBP18, 10, 14, 16, 19, 7, 2, and 4) were clustered with CbowOBPs (OBP25, 26, 12, 3, 6, 5, 7, and 20, respectively) (Figure 3). The tissue expression profiles revealed that three PverOBPs (PverOBP10, 12 and 18) had a higher expression level in female antennae than male antennae. Among these PverOBPs, except for PverOBP15, the remaining 14 candidate genes were specifically expressed in the antennae with low or no expression level in the body (Figure 4).
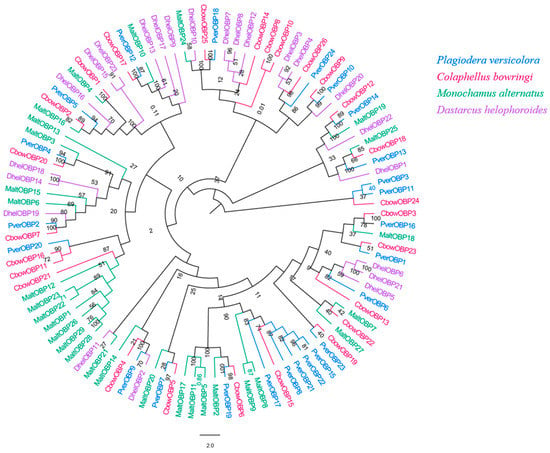
Figure 3.
Phylogenetic tree of insect OBP. The P. versicolora genes are shown in blue. The tree was constructed using MEGA6 with the Neighbor-joining method.
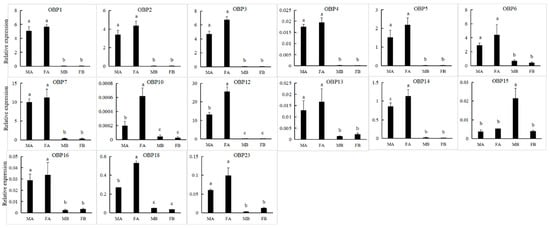
Figure 4.
Expression levels of PverOBP genes in different tissues assessed by RT-qPCR. MA, male antennae; FA, female antennae; MB, male body (without antennae); FB, female body (without antennae). Error bars, r, are represented by the standard error of three biological replicates. Different letters (a–c) indicate significant differences (p < 0.05) of male or female based on one-way ANOVA.
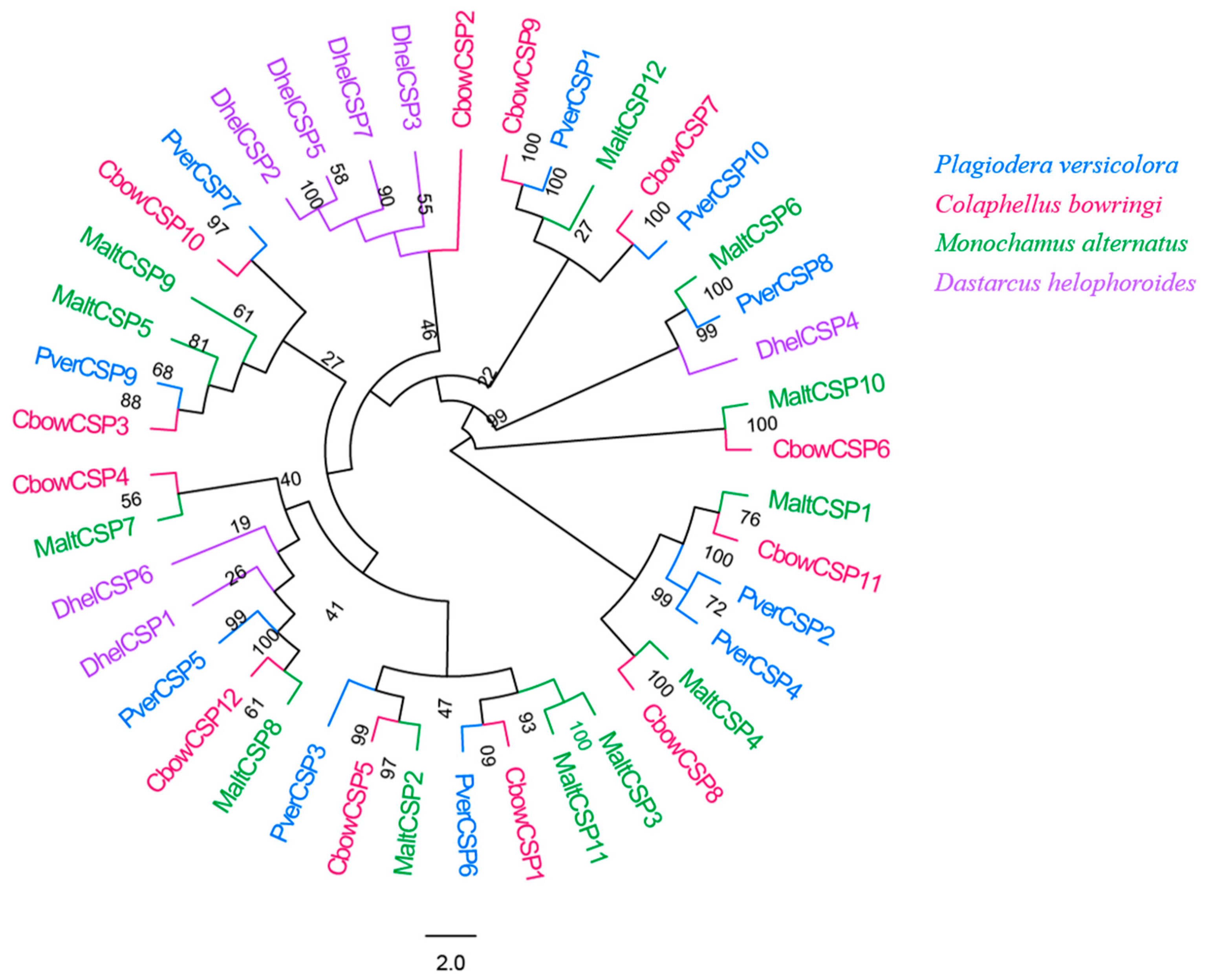
3.5. CSPs
In total, 10 different candidate unigenes encoding for CSPs were obtained in P. versicolora (PverCSPs), based on the transcriptomes of the antennae (Table S2). Among these CSPs, all had full-length ORFs with a predicted signal peptide. The phylogenetic trees were divided into several branches (Figure 5). The results showed that five PverCSPs (PverCSP1, 6, 7, 9 and 10) were orthologs of known CbowCSPs (CSP9, 1, 10, 3 and 7, respectively) from other insects (Figure 5).
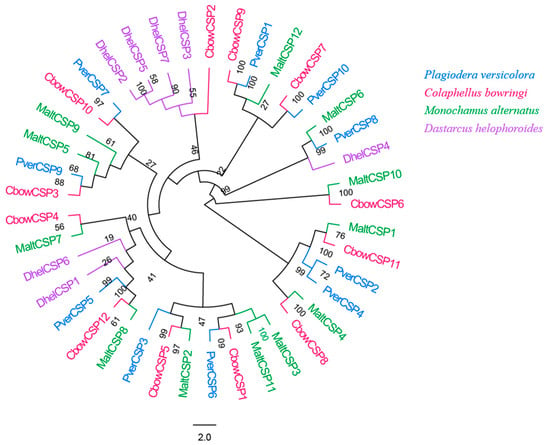
Figure 5.
Phylogenetic tree of insect CSP. The P. versicolora genes are shown in blue. The tree was constructed using MEGA6 with the Neighbor-joining method.
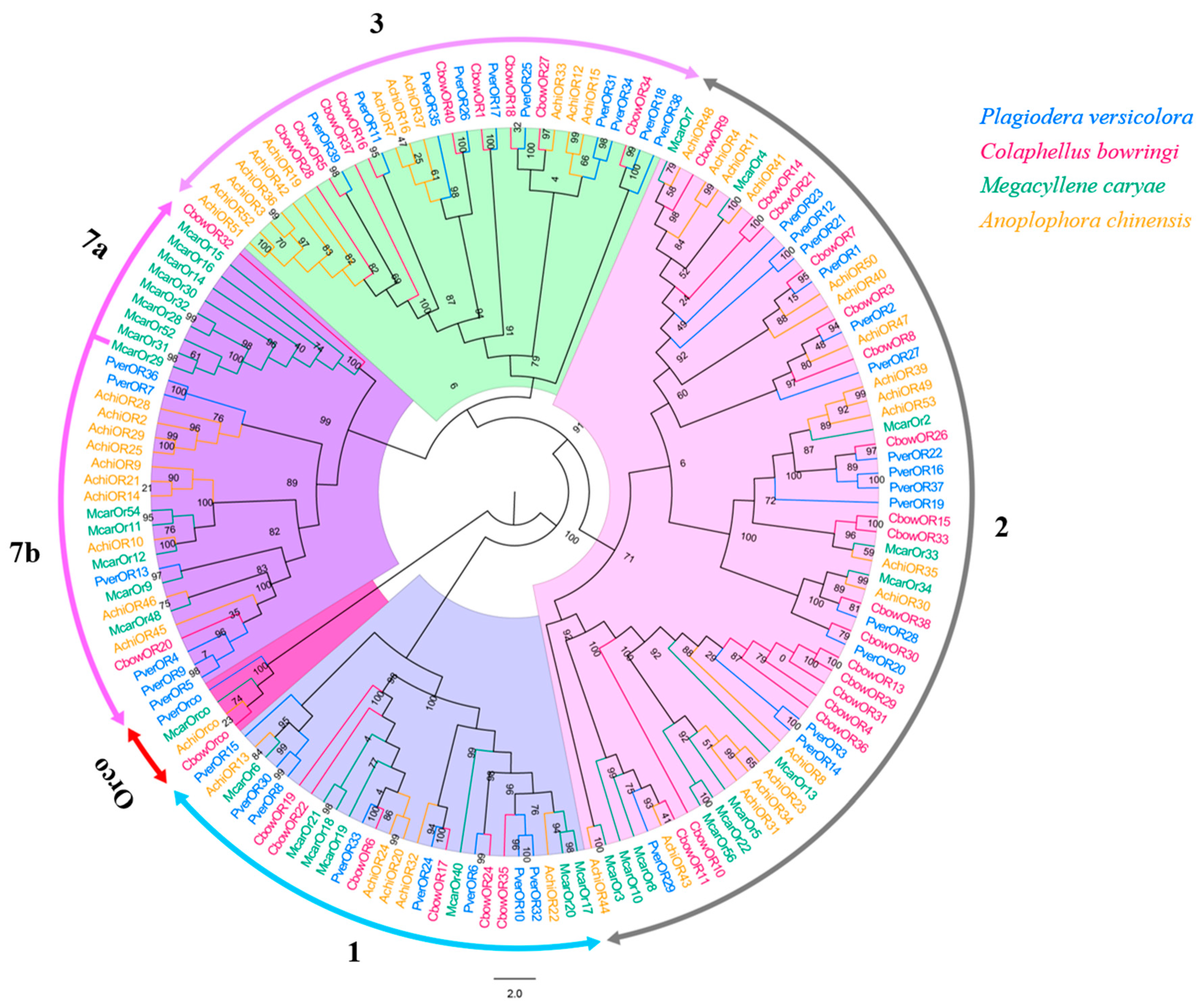
3.6. ORs
Forty different unigenes for candidate ORs were identified in P. versicolora (PverOR), among which 35 ORs contained a complete ORFs that encoded 372 to 479 amino acids (Table S3). The phylogenetic analysis showed that a PverOR gene displayed a high homology with the conserved Orco gene family in other three insects (C. bowringi, M. alternatus, and A. chinensis), which was designated as PverOrco. The results show that ORs were separated into five subfamilies, those being 1–3, 7a and 7b. We found that three PverORs (PverOR6, 10 and 32) and an McarOR20 that have been functionally characterized were clustered within a subgroup. Additionally, PverOR24 was clustered with CbowOR17 and AchiOR32 in the tree (Figure 6). Among these PverORs, except for PverOR27 (which had a similar expression level between the antennae and bodies), the remaining candidate genes were specifically expressed at higher levels in the antennae than in the bodies. The results of RT-qPCR showed that PverOR24 and PverOR35 were highly expressed in female antennae (Figure 7).
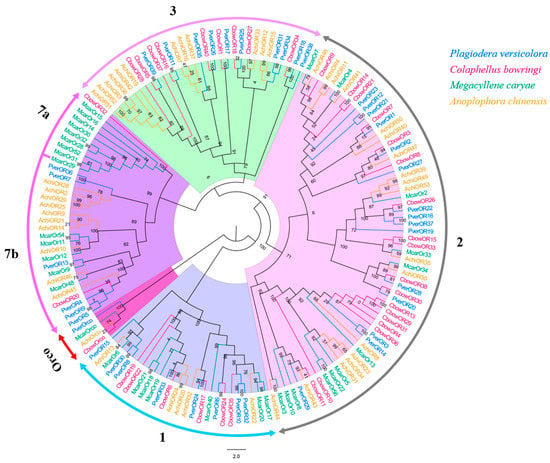
Figure 6.
Phylogenetic tree of insect OR. The P. versicolora genes are shown in blue. The tree was constructed using MEGA6 with the Neighbor-joining method.

Figure 7.
Expression levels of PverOR genes in different tissues assessed by RT-qPCR. MA, male antennae; FA, female antennae; MB, male body (without antennae); FB, female body (without antennae). Error bars, r, are represented by standard error of three biological replicates. Different letters (a–c) indicate significant differences (p < 0.05) of male or female based on one-way ANOVA.
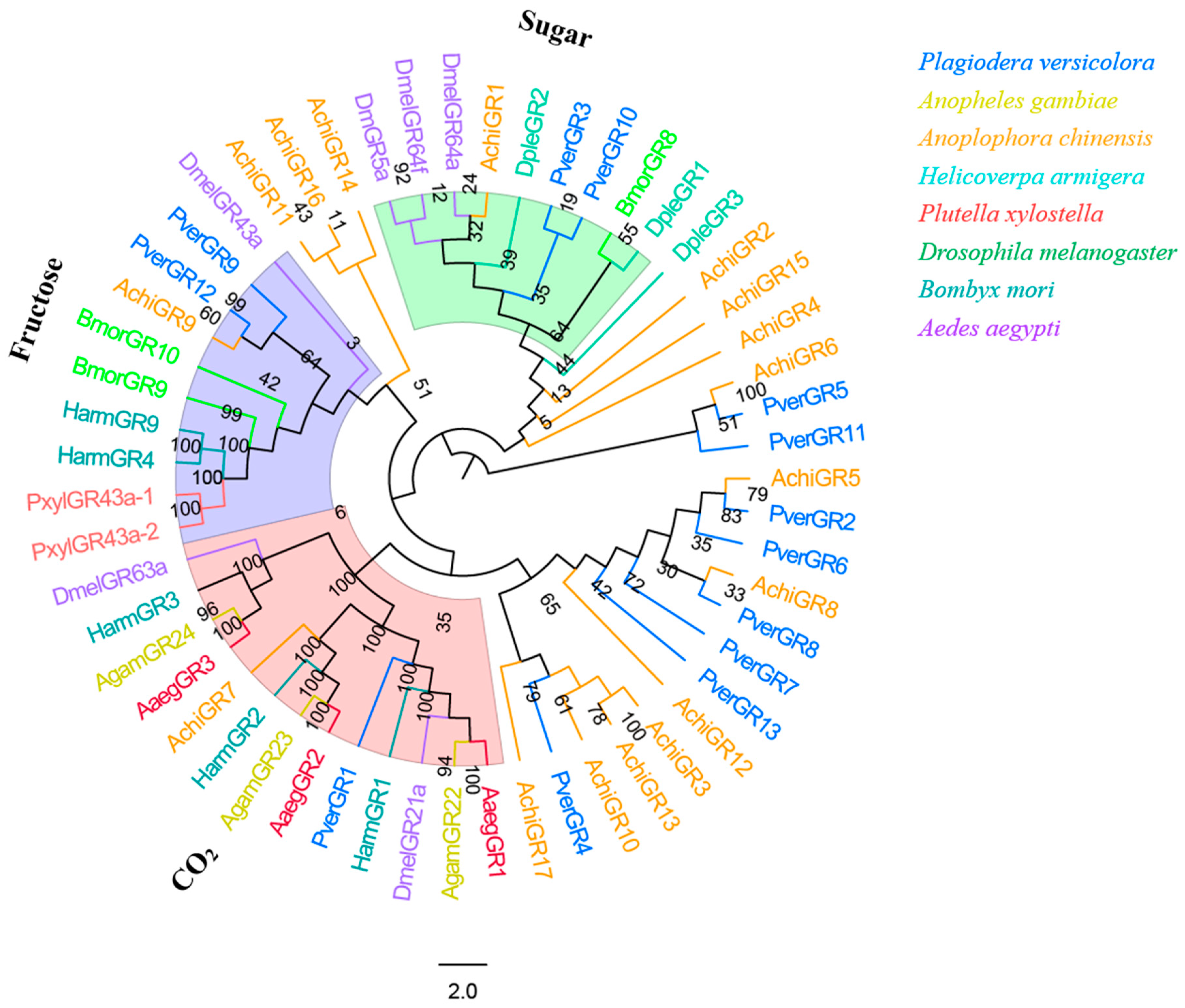
3.7. GRs
Bioinformatic analysis identified 13 candidate GRs in the P. versicolora (PverGRs) antennal transcriptome, seven of which have full-length ORFs (Table S3). GR sequences in P. versicolora and other insects were used for the phylogenetic analysis. The tree showed that PverGR1 was clustered in the CO2 receptors subfamily, two PverGRs (GR3 and GR10) were clustered together with the sugar receptor (including trehalose, glucose, sucrose, etc., expect for fructose) subfamily, and PverGR9 and PverGR12 were clustered together with the fructose receptor subgroup (Figure 8).
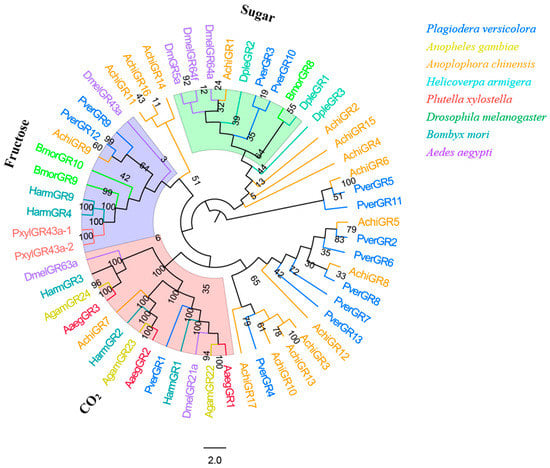
Figure 8.
Phylogenetic tree of insect GR. The P. versicolora genes are shown in blue. The tree was constructed using MEGA6 with the Neighbor-joining method. The sugar subfamily does not include the fructose receptors.
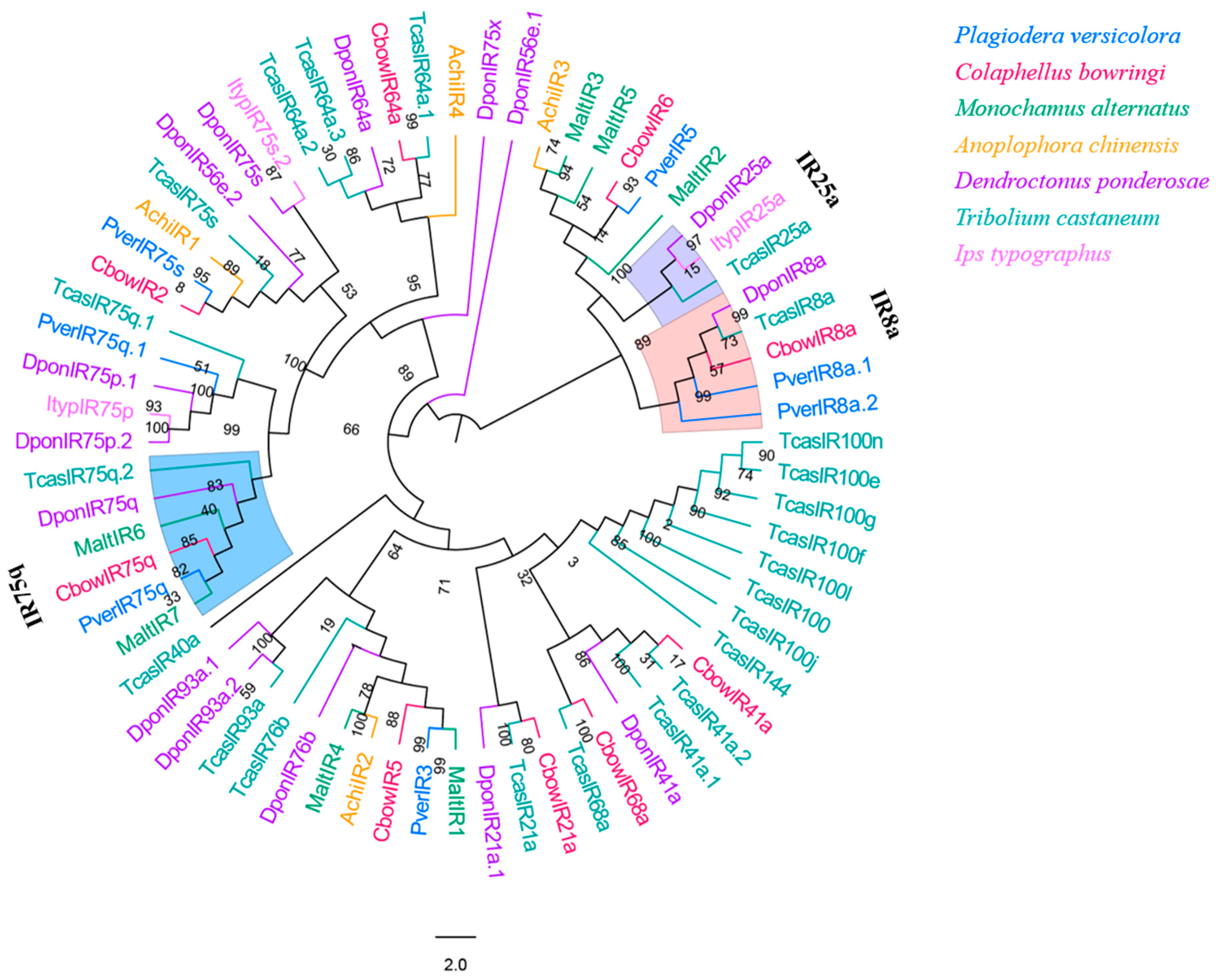
3.8. IRs
Seven IR genes were identified in P. versicolora from the male and female antennal transcriptomes. Only four of these IRs had a full-length ORF (PverIR2, PverIR4, PverIR5, and PverIR6) that encoded 639 to 877 amino acids (Table S3). The phylogenetic analysis of IRs from six species of Coleopterans showed that (Figure 9) these IRs can be divided into several different subfamilies. PverIR1 (named PverIR75q) clustered with CbowIR75q, DponIR75q, and TcasIR75q, suggesting it is part of the IR75q group. The results show that PverIR4 (named PverIR8a.1) and PverIR7 (named PverIR8a.2) were classified into IR8a coreceptor subgroup (Figure 9).
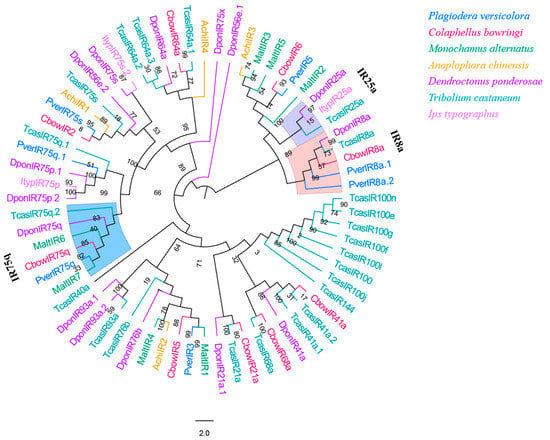
Figure 9.
Phylogenetic tree of insect IR. The P. versicolora genes are shown in blue. The tree was constructed using MEGA6 with the Neighbor-joining method.
3.9. SNMPs
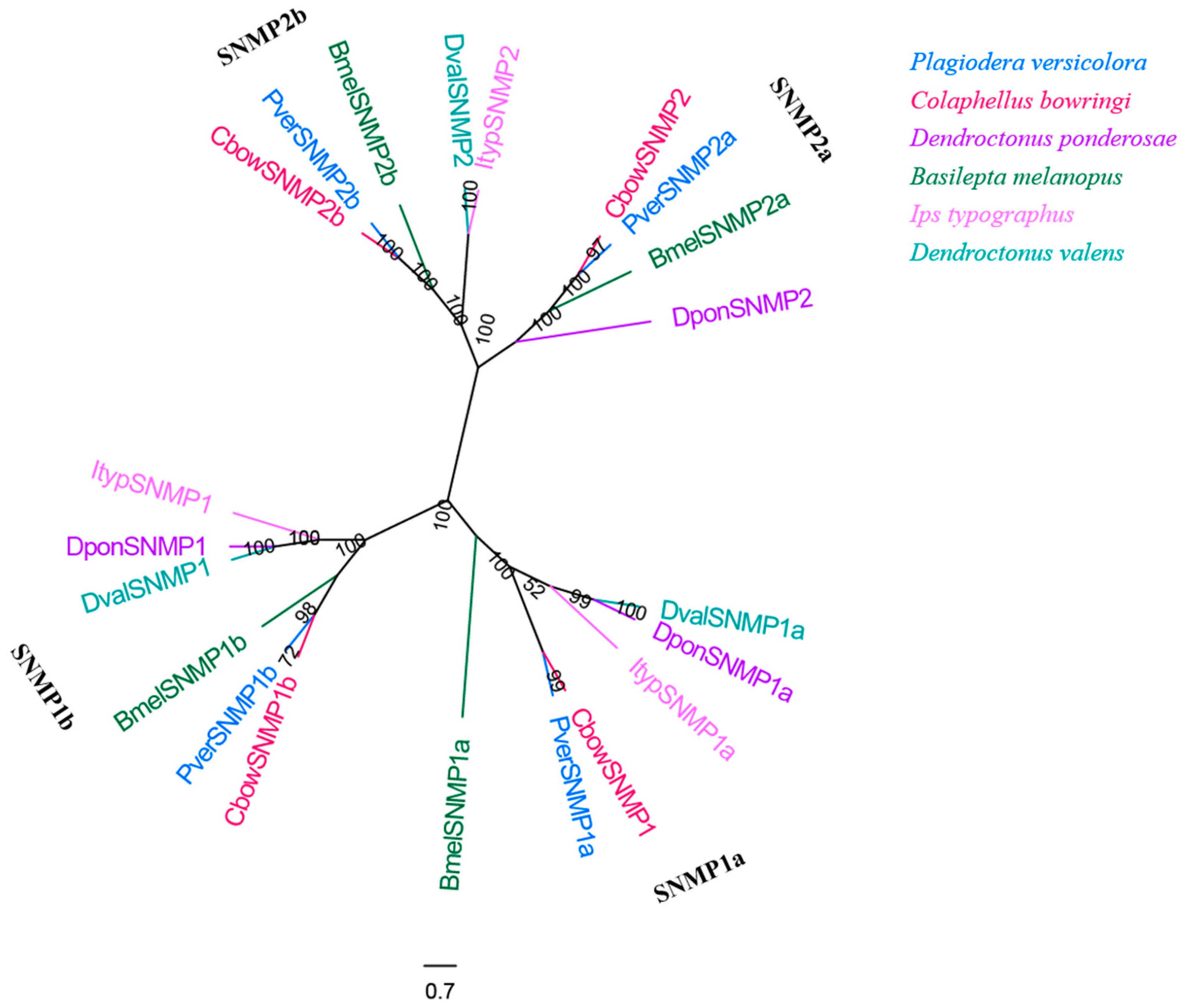
Four SNMP genes with complete ORFs were obtained from the male and female antennal transcriptomes of P. versicolora (Table S3). This number is similar to that observed in C. bowringi but is higher than in other insects used in the phylogenetic tree. The results also show that four PverSNMP genes were clustered into the Coleoptera SNMP1 group (SNMP1a and SNMP1b subgroup) and SNMP2 group (SNMP2a and SNMP2b subgroup) (Figure 10). Insects generally have two representatives of SNMPs (SNMP1 and SNMP2), although the copy numbers of each lineal orthologue seems to differ between species [55].
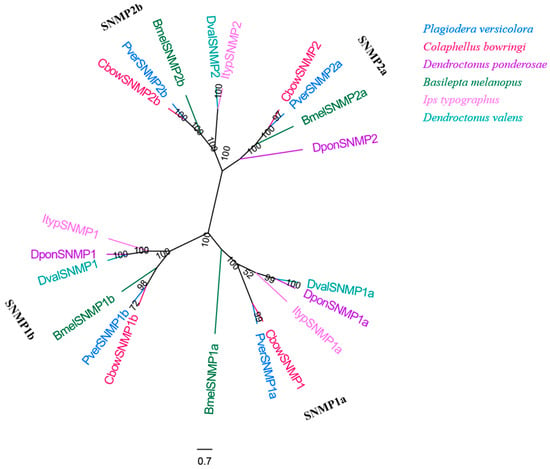
Figure 10.
Phylogenetic tree of insect SNMP. The P. versicolora genes are shown in blue. The tree was constructed using MEGA6 with the Neighbor-joining method.
4. Discussion
Although Coleoptera is the largest insect order when compared to Dipterans and Lepidopterans, there has been little research into the molecular mechanism of chemoreception. In the present study, the antennal transcriptome of a Coleoptera beetle, P. versicolora, was sequenced and analyzed. A total of 24,862 unigenes were identified and 76.5% of them were over 300 bp in length, suggesting the high depth and quality of the transcriptome. Busco analysis was conducted to evaluate the completeness of the transcriptome (File S2). Based on the transcriptome analysis, we identified 98 chemosensory genes in P. versicolora (Tables S2 and S3), and the phylogenetic trees were constructed with other insect chemosensory sequences (Figure 3, Figure 5, Figure 6, Figure 8, Figure 9 and Figure 10). In addition, the spatial expression patterns of OBPs and ORs have been assessed through RT-qPCR analysis (Figure 4 and Figure 7).
OBP-binding odorant molecules consist of the first step in the olfactory process. The number of PverOBPs (24) observed is similar to those found in C. bowringi (26) and D. helophoroides (23), and less than that of M. alternatus (29). The phylogenetic analysis showed that PverOBP4, 6 and 12 were clustered together with functionally characterized DhelOBP13, 21 and 10, respectively. The results indicated that the three PverOBPs may have same function as in D. helophoroides. In the present study, the majority of PverOBPs (14 out of 15 OBPs) were highly expressed in antennae, as shown by the RT-qPCR test, which is consistent with the expression profile of genes in other insects, such as Dioryctria abietella [56] and Galleria mellonella [57]. In addition, PverOBP10, 12 and 18 exhibited highly abundant expression levels in female antennae (Figure 4), suggesting that these OBPs may play an important role in antennal recognition processes, though further verification is needed. PverOBP15 was highly expressed in the bodies when compared to other OBPs (Figure 4), which may be involved in delivering and detecting some specific semiochemicals. In line with our results, several other studies showed that OBPs are differentially expressed in the body; for example, AtumOBP5, AtumOBP17, and AtumOBP21 have a higher expression in the forelegs [58].
PverGR1 was clustered in the CO2 receptors subfamily, PverGR9 and PverGR12 in the fructose receptors subgroup, and PverGR3 and PverGR10 in the sugar receptors subfamily (Figure 8), indicating that these GRs might be taking part in the detection of CO2, sugar, and fructose [45,47,59]. Other GRs that do not belong to these three categories might be involved in other taste perception processes. Previous studies showed that an insect usually has three CO2 receptors (these are allocated into three different groups: one, two, and three), e.g., AaegGr1-3 were reported in A. aegypti [51], AgamGr22-24 in A. gambiae [52], and HarmGr1-3 in H. armigera [42]. However, only one CO2 receptor gene (PverGR1) was identified in P. versicolora, which belongs to the GR1 subfamily. Considering that gustatory sensilla are mainly distributed in the mouthparts (proboscises, labial palps), antennae, wings, legs, and ovipositor [60,61], we may identify more PverGR genes from the transcriptome of other tissues in the future.
The number of PverORs (40) is greater than that of harmonia axyridis (26) [62] and less than that of Rhynchophorus palmarum (63) [63]. In the OR phylogenetic tree, PverOR6, PverOR10, and PverOR32 were clustered into the same subgroup, with a functionally characterized pheromone receptor, McarOR20, the receptor of (2S,3R)-2,3-hexanediol and 3-hydroxyhexan-2-one in M. caryae [40]. The results indicate that these PverORs may be associated with the detection of the above pheromones or other active compounds. The discovery of new attractive substances would be helpful for the identification of sex pheromone compounds in P. versicolora. PverOR24 and PverOR35 genes were identified with significantly higher expression levels in female antennae than male antennae. Considering previous studies of the insect OR functions [64,65], female-biased PverORs may be involved in the detection of odors that play a critical role in female behavior, such as mating or oviposition. In addition, PverOR24 clusters with CbowOR17 and AchiOR32, as this might be relevant in the light of discovering pheromone receptors. The specific functions of these PverORs need to be explored in the future.
Collectively, a total of 98 chemosensory gene families, including 40 ORs, 7 IRs, 13 GRs, 10 CSPs, 24 OBPs, and 4 SNMPs, were identified through the transcriptomic analysis of P. versicolora. The results regarding the expression profiles of these chemosensory genes demonstrated that most PverOBPs and PverORs were highly expressed in the antennae. Besides, PverOBP10, PverOBP12, PverOBP18, PverOR24, and PverOR35 have a higher expression level in female antennae than male, indicating the genes might be crucial for regulating female-specific behaviors. In this study, we provide a comprehensive sequence resource of chemosensory receptors and insight into the P. versicolora chemical ecology, which lays a basis for further functional studies of the olfactory system in this pest, and sheds light on a new perspective of pheromone-based pest management in the future.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/insects13010036/s1, Table S1: Primers for RT-qPCR of PverOBP and PverOR genes in P. versicolora. Table S2: The Blastx match of P. versicolora candidate CSP and OBP genes. Table S3: The Blastx match of P. versicolora candidate GR, IR, OR and SNMP genes. File S1: The amino acid sequences of Plagiodera versicolora putative chemosensory receptor genes. File S2: The busco analysis of P. versicolora transcriptome.
Author Contributions
Conceptualization, X.L. and M.L.; methodology, X.L., N.T., Y.L. and Z.W.; software, X.L., P.L. and M.M.; formal analysis, X.L., N.T., P.L. and M.M.; investigation, X.L., N.T., Z.W. and Y.L.; resources, X.L.; data curation, X.L., N.T., Z.W., Y.L. and P.L.; writing—original draft preparation, X.L.; writing—review and editing, X.L. and M.L.; supervision, M.L.; funding acquisition, M.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National talent project (1070017364).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We thank Wang Liao (Hubei University, Wuhan, China) for help with drawing the graphical abstract. We thank Jinmeng Guo (Nanjing Agricultural University, Nanjing, China) for help in the phylogenetic analyses.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Field, L.M.; Pickett, J.A.; Wadhams, L.J. Molecular studies in insect olfaction. Insect Mol. Biol. 2000, 9, 545–551. [Google Scholar] [CrossRef] [PubMed]
- Zacharuk, R.; Shields, V. Sensilla of Immature Insects. Annu. Rev. Entomol. 1991, 36, 331–354. [Google Scholar] [CrossRef]
- Vogt, R.G.; Riddiford, L.M. Pheromone binding and inactivation by moth antennae. Nature 1981, 293, 161–163. [Google Scholar] [CrossRef]
- Leal, W.S. Odorant reception in insects: Roles of receptors, binding proteins, and degrading enzymes. Annu. Rev. Entomol. 2013, 58, 373–391. [Google Scholar] [CrossRef]
- Andersson, M.N.; Videvall, E.; Walden, K.K.O.; Harris, M.O.; Robertson, H.M.; Löfstedt, C. Sex- and tissue-specific profiles of chemosensory gene expression in a herbivorous gall-inducing fly (Diptera: Cecidomyiidae). BMC Genom. 2014, 15, 501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pelosi, P.; Iovinella, I.; Felicioli, A.; Dani, F.R. Soluble proteins of chemical communication: An overview across arthropods. Front. Physiol. 2014, 5, 320. [Google Scholar] [CrossRef] [Green Version]
- Rybczynski, R.; Reagan, J.; Lerner, M.R. A pheromone-degrading aldehyde oxidase in the antennae of the moth Manduca sexta. J. Neurosci. Off. J. Soc. Neurosci. 1989, 9, 1341–1353. [Google Scholar] [CrossRef]
- Rützler, M.; Zwiebel, L.J. Molecular biology of insect olfaction: Recent progress and conceptual models. J. Comp. Physiol. A Neuroethol. Sens. Neural. Behav. Physiol. 2005, 191, 777–790. [Google Scholar] [CrossRef]
- Sánchez-Gracia, A.; Vieira, F.G.; Rozas, J. Molecular evolution of the major chemosensory gene families in insects. Heredity 2009, 103, 208–216. [Google Scholar] [CrossRef]
- Jacquin-Joly, E.; Legeai, F.; Montagné, N.; Monsempes, C.; François, M.-C.; Poulain, J.; Gavory, F.; Walker, W.B.; Hansson, B.S.; Larsson, M.C. Candidate chemosensory genes in female antennae of the noctuid moth Spodoptera littoralis. Int. J. Biol. Sci. 2012, 8, 1036–1050. [Google Scholar] [CrossRef] [Green Version]
- Leal, W.S.; Nikonova, L.; Peng, G. Disulfide structure of the pheromone binding protein from the silkworm moth, Bombyx mori. FEBS Lett. 1999, 464, 85–90. [Google Scholar] [CrossRef] [Green Version]
- Scaloni, A.; Monti, M.; Angeli, S.; Pelosi, P. Structural analysis and disulfide-bridge pairing of two odorant-binding proteins from Bombyx mori. Biochem. Biophys. Res. Commun. 1999, 266, 386–391. [Google Scholar] [CrossRef]
- Vogt, R.G.; Riddiford, L.M.; Prestwich, G.D. Kinetic properties of a sex pheromone-degrading enzyme: The sensillar esterase of Antheraea polyphemus. Proc. Natl. Acad. Sci. USA 1985, 82, 8827–8831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, P.; Hooper, A.M.; Pickett, J.A.; Leal, W.S. Specificity determinants of the silkworm moth sex pheromone. PLoS ONE 2012, 7, e44190. [Google Scholar] [CrossRef]
- Vogt, R.G.; Große-Wilde, E.; Zhou, J.-J. The Lepidoptera Odorant Binding Protein gene family: Gene gain and loss within the GOBP/PBP complex of moths and butterflies. Insect Biochem. Mol. Biol. 2015, 62, 142–153. [Google Scholar] [CrossRef]
- Zhang, S.; Zhang, Z.; Wang, H.; Kong, X. Antennal transcriptome analysis and comparison of olfactory genes in two sympatric defoliators, Dendrolimus houi and Dendrolimus kikuchii (Lepidoptera: Lasiocampidae). Insect Biochem. Mol. Biol. 2014, 52, 69–81. [Google Scholar] [CrossRef]
- Zhu, J.; Ban, L.; Song, L.-M.; Liu, Y.; Pelosi, P.; Wang, G. General odorant-binding proteins and sex pheromone guide larvae of Plutella xylostella to better food. Insect Biochem. Mol. Biol. 2016, 72, 10–19. [Google Scholar] [CrossRef]
- He, P.; Engsontia, P.; Chen, G.-L.; Yin, Q.; Wang, J.; Lu, X.; Zhang, Y.-N.; Li, Z.-Q.; He, M. Molecular characterization and evolution of a chemosensory receptor gene family in three notorious rice planthoppers, Nilaparvata lugens, Sogatella furcifera and Laodelphax striatellus, based on genome and transcriptome analyses. Pest Manag. Sci. 2018, 74, 2156–2167. [Google Scholar] [CrossRef] [PubMed]
- Benton, R.; Vannice, K.S.; Vosshall, L.B. An essential role for a CD36-related receptor in pheromone detection in Drosophila. Nature 2007, 450, 289–293. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.-J.; Anderson, A.R.; Trowell, S.C.; Luo, A.-R.; Xiang, Z.-H.; Xia, Q.-Y. Topological and functional characterization of an insect gustatory receptor. PLoS ONE 2011, 6, e24111. [Google Scholar] [CrossRef] [Green Version]
- Schneider, D. Insect Antennae. Annu. Rev. Entomol. 1964, 9, 103–122. [Google Scholar] [CrossRef]
- Clyne, P.J.; Warr, C.G.; Freeman, M.R.; Lessing, D.; Kim, J.; Carlson, J.R. A novel family of divergent seven-transmembrane proteins: Candidate odorant receptors in Drosophila. Neuron 1999, 22, 327–338. [Google Scholar] [CrossRef] [Green Version]
- Hallem, E.A.; Ho, M.G.; Carlson, J.R. The molecular basis of odor coding in the Drosophila antenna. Cell 2004, 117, 965–979. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larsson, M.C.; Domingos, A.I.; Jones, W.D.; Chiappe, M.E.; Amrein, H.; Vosshall, L.B. Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron 2004, 43, 703–714. [Google Scholar] [CrossRef] [Green Version]
- Benton, R.; Sachse, S.; Michnick, S.W.; Vosshall, L.B. Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol. 2006, 4, e20. [Google Scholar] [CrossRef] [Green Version]
- Hansson, B.S.; Stensmyr, M.C. Evolution of insect olfaction. Neuron 2011, 72, 698–711. [Google Scholar] [CrossRef] [Green Version]
- Andersson, M.N.; Lofstedt, C.; Newcomb, R.D. Insect olfaction and the evolution of receptor tuning. Front. Ecol. Evol. 2015, 3, 53. [Google Scholar] [CrossRef] [Green Version]
- Sato, K.; Pellegrino, M.; Nakagawa, T.; Nakagawa, T.; Vosshall, L.B.; Touhara, K. Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature 2008, 452, 1002–1006. [Google Scholar] [CrossRef] [PubMed]
- Wicher, D.; Schäfer, R.; Bauernfeind, R.; Stensmyr, M.C.; Heller, R.; Heinemann, S.H.; Hansson, B.S. Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels. Nature 2008, 452, 1007–1011. [Google Scholar] [CrossRef]
- Butterwick, J.A.; Del Mármol, J.; Kim, K.H.; Kahlson, M.A.; Rogow, J.A.; Walz, T.; Ruta, V. Cryo-EM structure of the insect olfactory receptor Orco. Nature 2018, 560, 447–452. [Google Scholar] [CrossRef]
- Utsumi, S.; Ando, Y.; Ohgushi, T. Evolution of feeding preference in a leaf beetle: The importance of phenotypic plasticity of a host plant. Ecol. Lett. 2009, 12, 920–929. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Xu, S.; Sun, L.; Zhang, Y.; Luo, J.; Bock, R.; Zhang, J. Synergistic action of the gut microbiota in environmental RNA interference in a leaf beetle. Microbiome 2021, 9, 98. [Google Scholar] [CrossRef]
- Li, X.-M.; Zhu, X.-Y.; Wang, Z.-Q.; Wang, Y.; He, P.; Chen, G.; Sun, L.; Deng, D.-G.; Zhang, Y.-N. Candidate chemosensory genes identified in Colaphellus bowringi by antennal transcriptome analysis. BMC Genom. 2015, 16, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Andersson, M.N.; Grosse-Wilde, E.; Keeling, C.I.; Bengtsson, J.M.; Yuen, M.M.S.; Li, M.; Hillbur, Y.; Bohlmann, J.; Hansson, B.S.; Schlyter, F. Antennal transcriptome analysis of the chemosensory gene families in the tree killing bark beetles, Ips typographus and Dendroctonus ponderosae (Coleoptera: Curculionidae: Scolytinae). BMC Genom. 2013, 14, 198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, X.-C.; Zhang, Y.-N.; Kang, K.; Dong, S.-L.; Zhang, L.-W. Antennal Transcriptome Analysis of Odorant Reception Genes in the Red Turpentine Beetle (RTB), Dendroctonus valens. PLoS ONE 2015, 10, e0125159. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Li, D.-Z.; Min, S.-F.; Mi, F.; Zhou, S.-S.; Wang, M.-Q. Analysis of chemosensory gene families in the beetle Monochamus alternatus and its parasitoid Dastarcus helophoroides. Comp. Biochem. Physiol. Part D Genom. Proteom. 2014, 11, 1–8. [Google Scholar] [CrossRef]
- Sun, L.; Zhang, Y.-N.; Qian, J.-L.; Kang, K.; Zhang, X.-Q.; Deng, J.-D.; Tang, Y.-P.; Chen, C.; Hansen, L.; Xu, T.; et al. Identification and Expression Patterns of Anoplophora chinensis (Forster) Chemosensory Receptor Genes from the Antennal Transcriptome. Front. Physiol. 2018, 9, 90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, L.-Y.; Li, W.; Liu, H.-Y.; Xiang, F.; Kang, Y.-K.; Yin, X.; Huang, A.-P.; Wang, Y.-J. Systemic identification and analyses of genes potentially involved in chemosensory in the devastating tea pest Basilepta melanopus. Comp. Biochem. Physiol. Part D Genom. Proteom. 2019, 31, 100586. [Google Scholar] [CrossRef]
- Engsontia, P.; Sanderson, A.P.; Cobb, M.; Walden, K.K.O.; Robertson, H.M.; Brown, S. The red flour beetle’s large nose: An expanded odorant receptor gene family in Tribolium castaneum. Insect Biochem. Mol. Biol. 2008, 38, 387–397. [Google Scholar] [CrossRef]
- Mitchell, R.F.; Hughes, D.T.; Luetje, C.W.; Millar, J.G.; Soriano-Agatón, F.; Hanks, L.M.; Robertson, H.M. Sequencing and characterizing odorant receptors of the cerambycid beetle Megacyllene caryae. Insect Biochem. Mol. Biol. 2012, 42, 499–505. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.-L.; Sun, S.-J.; Hou, W.; Zhang, J.; Yan, Q.; Dong, S.-L. Functional characterization of two spliced variants of fructose gustatory receptor in the diamondback moth, Plutella xylostella. Pestic. Biochem. Physiol. 2020, 164, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Zhang, H.-J.; Anderson, A. A sugar gustatory receptor identified from the foregut of cotton bollworm Helicoverpa armigera. J. Chem. Ecol. 2012, 38, 1513–1520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, X.-J.; Ning, C.; Guo, H.; Jia, Y.-Y.; Huang, L.-Q.; Qu, M.-J.; Wang, C.-Z. A gustatory receptor tuned to D-fructose in antennal sensilla chaetica of Helicoverpa armigera. Insect Biochem. Mol. Biol. 2015, 60, 39–46. [Google Scholar] [CrossRef]
- Xu, W.; Anderson, A. Carbon dioxide receptor genes in cotton bollworm Helicoverpa armigera. Sci. Nat. 2015, 102, 11. [Google Scholar] [CrossRef]
- Sato, K.; Tanaka, K.; Touhara, K. Sugar-regulated cation channel formed by an insect gustatory receptor. Proc. Natl. Acad. Sci. USA 2011, 108, 11680–11685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kikuta, S.; Endo, H.; Tomita, N.; Takada, T.; Morita, C.; Asaoka, K.; Sato, R. Characterization of a ligand-gated cation channel based on an inositol receptor in the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 2016, 74, 12–20. [Google Scholar] [CrossRef]
- Jones, W.D.; Cayirlioglu, P.; Kadow, I.G.; Vosshall, L.B. Two chemosensory receptors together mediate carbon dioxide detection in Drosophila. Nature 2007, 445, 86–90. [Google Scholar] [CrossRef]
- Jiao, Y.; Moon, S.J.; Montell, C. A Drosophila gustatory receptor required for the responses to sucrose, glucose, and maltose identified by mRNA tagging. Proc. Natl. Acad. Sci. USA 2007, 104, 14110–14115. [Google Scholar] [CrossRef] [Green Version]
- Jiao, Y.; Moon, S.J.; Wang, X.; Ren, Q.; Montell, C. Gr64f is required in combination with other gustatory receptors for sugar detection in Drosophila. Curr. Biol. 2008, 18, 1797–1801. [Google Scholar] [CrossRef] [Green Version]
- Miyamoto, T.; Slone, J.; Song, X.; Amrein, H. A fructose receptor functions as a nutrient sensor in the Drosophila brain. Cell 2012, 151, 1113–1125. [Google Scholar] [CrossRef] [Green Version]
- Erdelyan, C.N.G.; Mahood, T.H.; Bader, T.S.Y.; Whyard, S. Functional validation of the carbon dioxide receptor genes in Aedes aegypti mosquitoes using RNA interference. Insect Mol. Biol. 2012, 21, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.; Qiu, Y.T.; Wang, G.; Kwon, J.Y.; Rutzler, M.; Kwon, H.-W.; Pitts, R.J.; van Loon, J.J.A.; Takken, W.; Carlson, J.R.; et al. Odor coding in the maxillary palp of the malaria vector mosquito Anopheles gambiae. Curr. Biol. 2007, 17, 1533–1544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, M.; Tu, C.; Luo, J.; Lu, M.; Zhang, S.; Xu, L. Metabolic and immunological effects of gut microbiota in leaf beetles at the local and systemic levels. Integr. Zool. 2021, 16, 313–323. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Nichols, Z.; Vogt, R.G. The SNMP/CD36 gene family in Diptera, Hymenoptera and Coleoptera: Drosophila melanogaster, D. pseudoobscura, Anopheles gambiae, Aedes aegypti, Apis mellifera, and Tribolium castaneum. Insect Biochem. Mol. Biol. 2008, 38, 398–415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xing, Y.; Thanasirungkul, W.; Adeel, M.M.; Yu, J.; Aslam, A.; Chi, D.-F. Identification and analysis of olfactory genes in Dioryctria abietella based on the antennal transcriptome. Comp. Biochem. Physiol. Part D Genom. Proteom. 2021, 38, 100814. [Google Scholar] [CrossRef]
- Zhao, H.-X.; Xiao, W.-Y.; Ji, C.-H.; Ren, Q.; Xia, X.-S.; Zhang, X.-F.; Huang, W.-Z. Candidate chemosensory genes identified from the greater wax moth, Galleria mellonella, through a transcriptomic analysis. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef]
- Wu, L.; Zhai, X.; Li, L.; Li, Q.; Liu, F.; Zhao, H. Identification and Expression Profile of Chemosensory Genes in the Small Hive Beetle Aethina tumida. Insects 2021, 12, 661. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.-L.; Yan, Q.; Yang, Y.-L.; Hou, W.; Miao, C.-L.; Peng, Y.-C.; Dong, S.-L. A Gustatory Receptor GR8 Tunes Specifically to D-Fructose in the Common Cutworm Spodoptera litura. Insects 2019, 10, 272. [Google Scholar] [CrossRef] [Green Version]
- Stocker, R.F. The organization of the chemosensory system in Drosophila melanogaster: A rewiew. Cell Tissue Res. 1994, 275, 3–26. [Google Scholar] [CrossRef]
- Zhang, Y.-F.; van Loon, J.J.A.; Wang, C.-Z. Tarsal taste neuron activity and proboscis extension reflex in response to sugars and amino acids in Helicoverpa armigera (Hubner). J. Exp. Biol. 2010, 213, 2889–2895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rondoni, G.; Roman, A.; Meslin, C.; Montagné, N.; Jacquin-Joly, E. Antennal transcriptome analysis and identification of candidate chemosensory genes of the harlequin ladybird beetle, harmonia axyridis (pallas) (cole-optera: Coccinellidae). Insects 2021, 12, 209. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, F.; Johny, J.; Walker, W.B.; Guan, Q.; Antony, B. Antennal transcriptome sequencing and identification of candidate chemoreceptor proteins from an invasive pest, the American palm weevil, Rhynchophorus palmarum. Sci. Rep. 2021, 11, 8334. [Google Scholar] [CrossRef] [PubMed]
- Anderson, A.R.; Wanner, K.W.; Trowell, S.C.; Warr, C.G.; Jaquin-Joly, E.; Zagatti, P.; Robertson, H.; Newcomb, R.D. Molecular basis of female-specific odorant responses in Bombyx Mori. Insect Biochem. Mol. Biol. 2009, 39, 189–197. [Google Scholar] [CrossRef]
- Pelletier, J.; Hughes, D.T.; Luetje, C.W.; Leal, W.S. An odorant receptor from the southern house mosquito Culex pipiens quinquefasciatus sensitive to oviposition attractants. PLoS ONE 2010, 5, e10090. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).