Chromosomes as Barcodes: Discovery of a New Species of Black Fly (Diptera: Simuliidae) from California, USA †
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection of material
2.2. Chromosomal Procedures and Terminology
2.3. Morphological Procedures
2.4. Type Depositories
2.5. COI Barcoding Procedures and Sequence Relationship Analysis
3. Results
3.1. Diagnosis
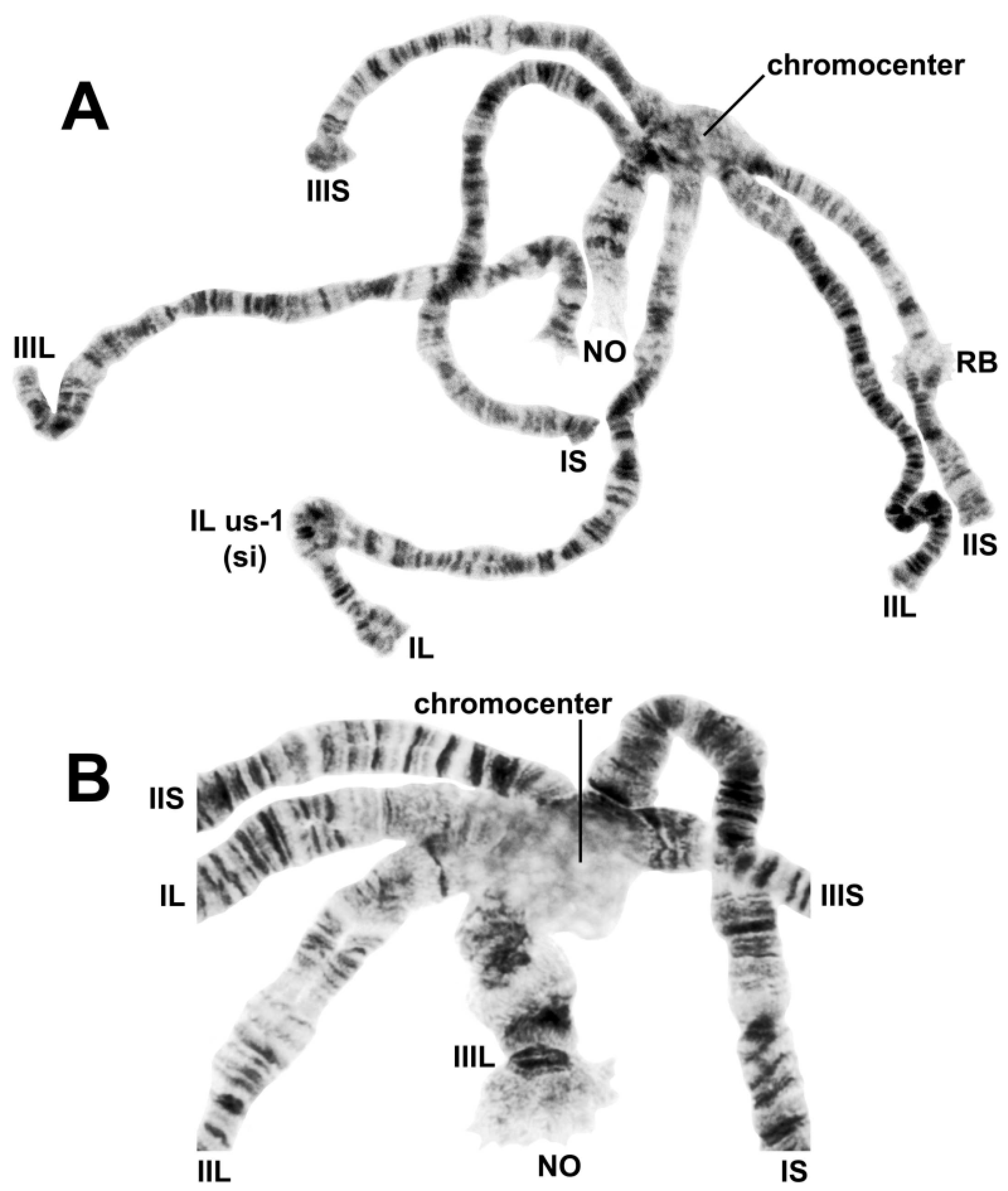
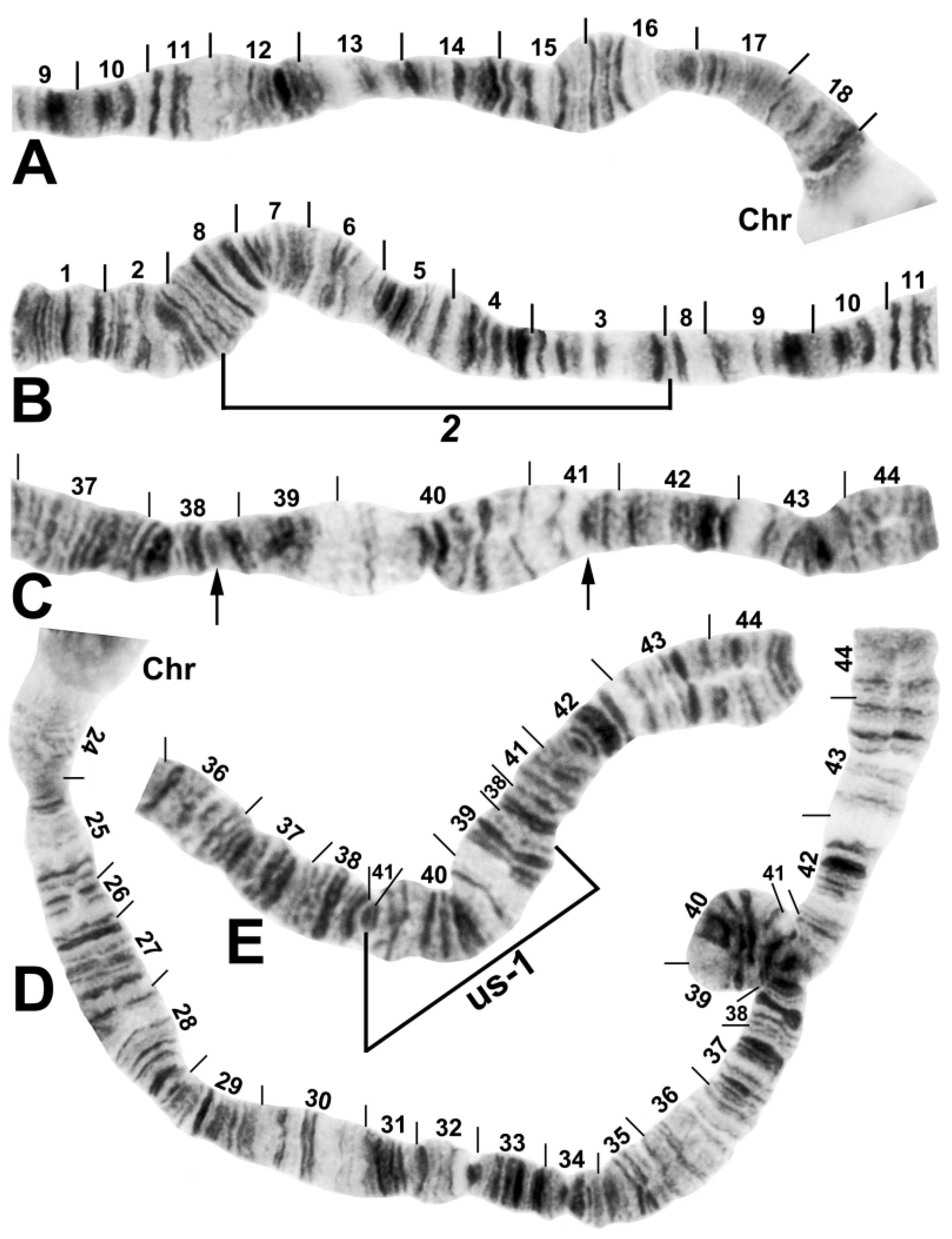
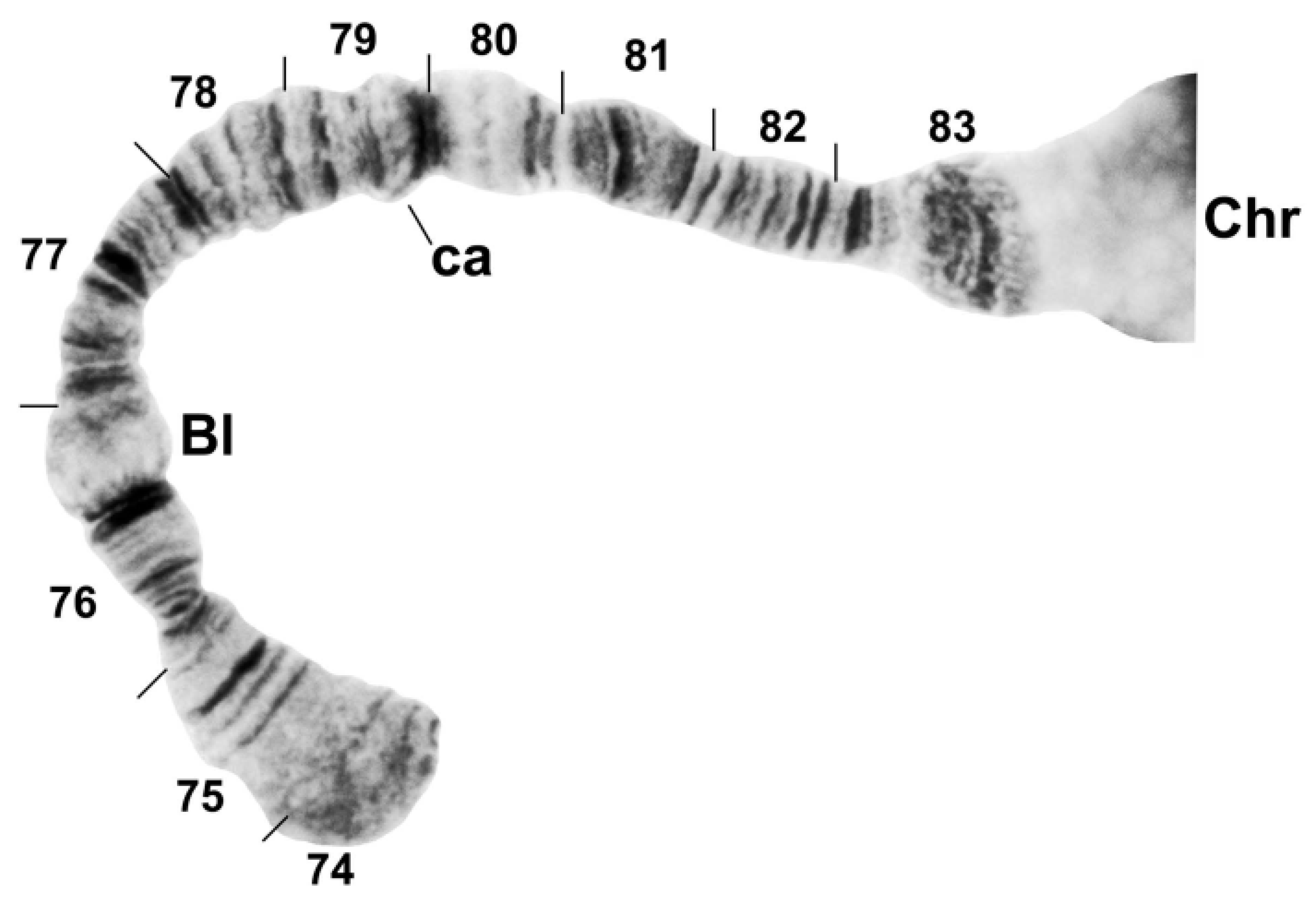
3.2. Chromosomal Description
- IIL-complex a: a[ed/hi/on/jk/bc]fg/lm/qp/r
- IIL-complex b: a[cb]kj/no/ih/defg/lm/qp/r
- IIL-complex c: abc[kj/no/ih/defg]lm/qp/r
- IIL-complex d: abc[gfed]hi/on/jklm/qp/r
- IIL-2: abcdefghi/on/jklm[qp]r
- IIL-3: abcdefghi/on[jklm]pqr
- IIL-1: abcdefghi[onmlkj]pqr
- Standard: abcdefghijklmnopqr
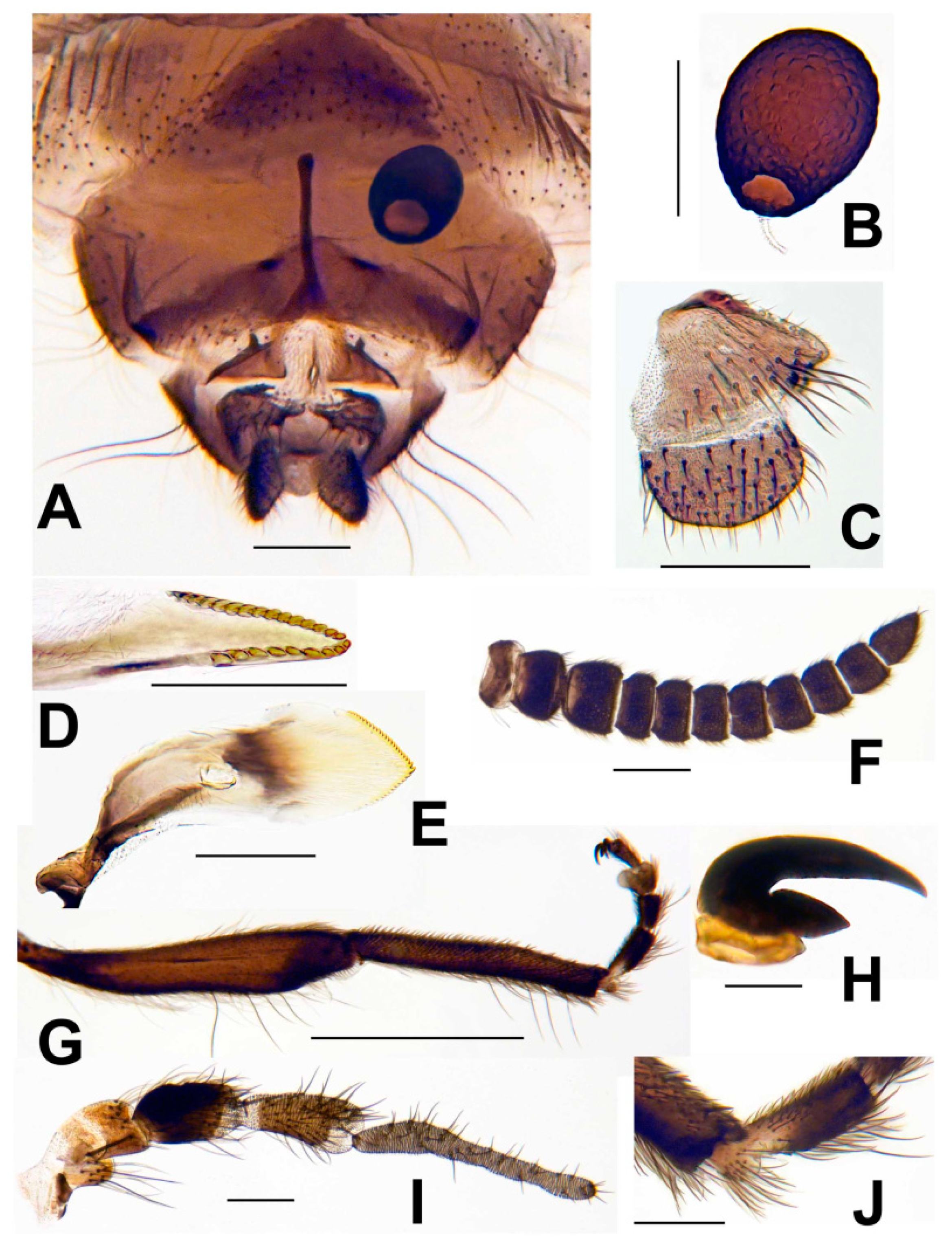
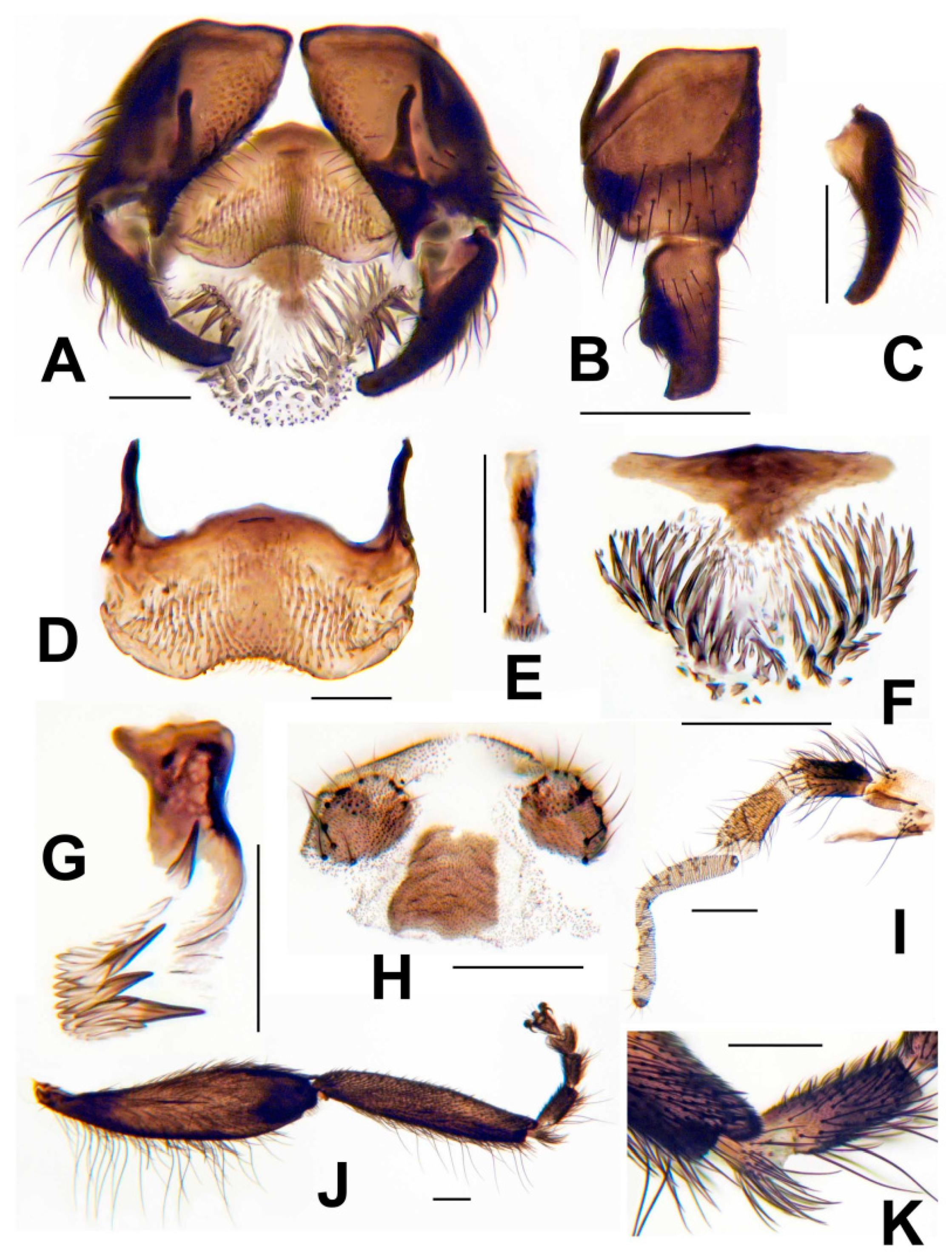
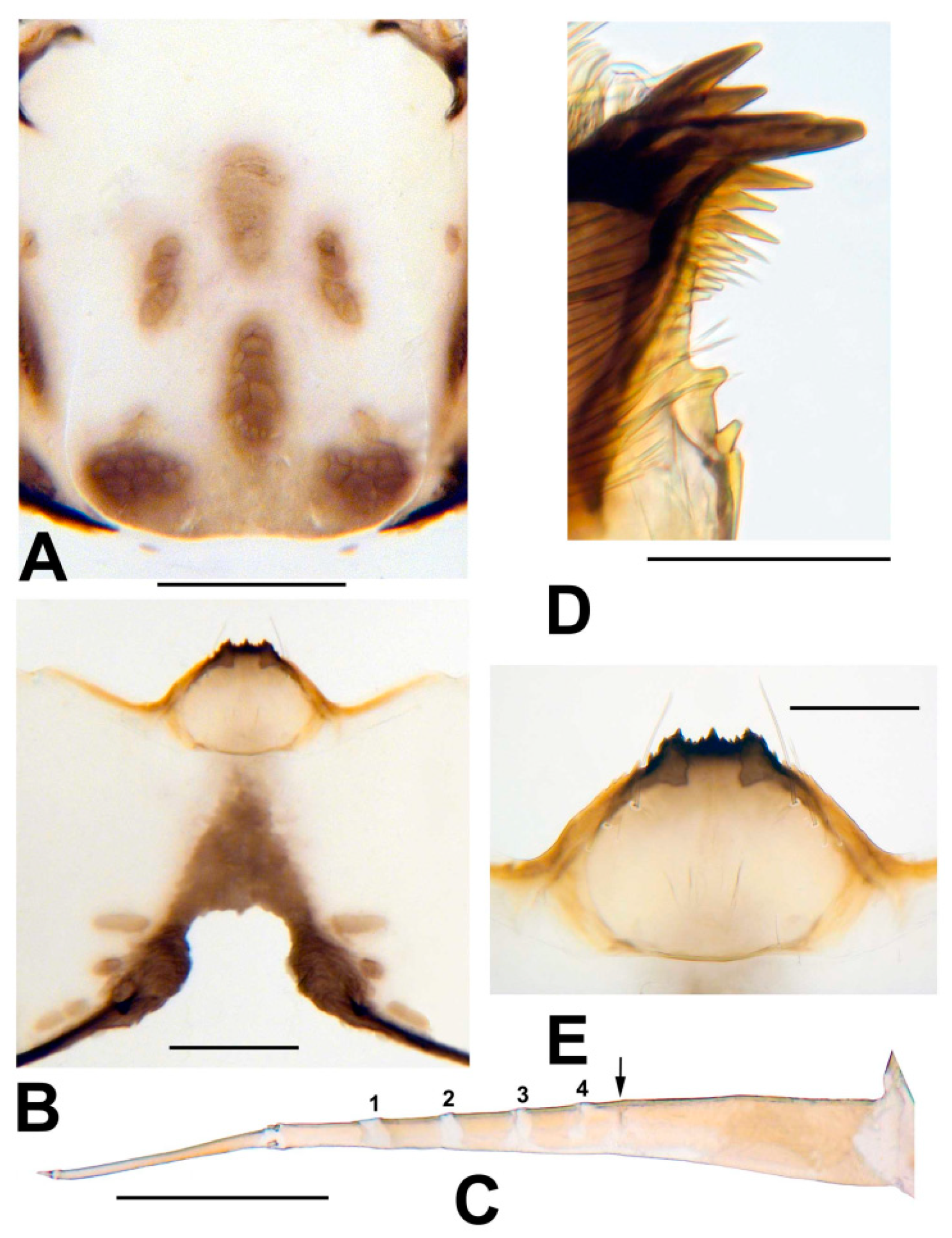
3.3. Morphological Description
3.4. Type material
3.5. Other Specimens Examined
3.6. Bionomics
3.7. Etymology
3.8. Morphological Comparisons with Other Species
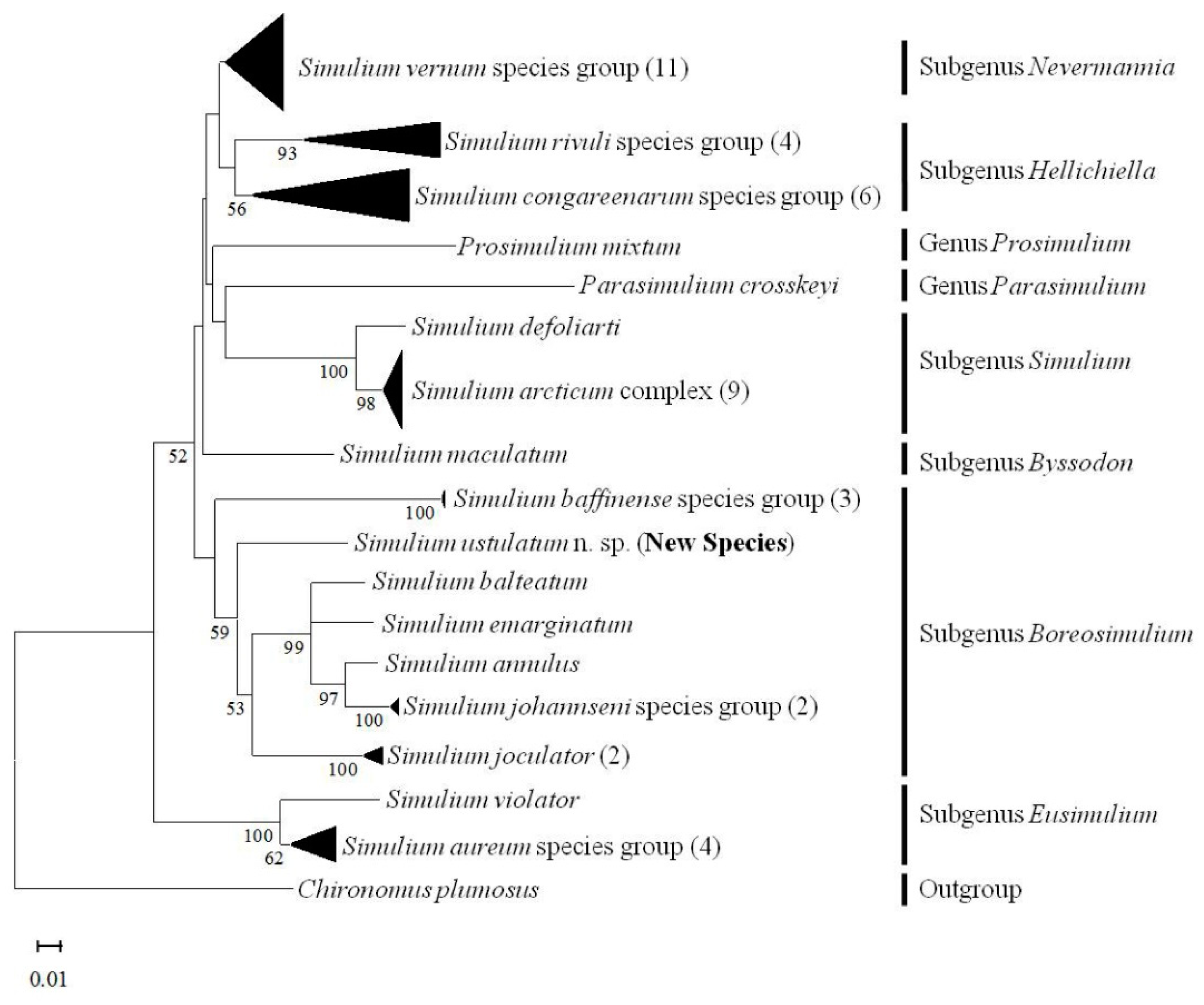
3.9. COI Barcoding
4. Discussion
4.1. Chromosomes as Barcodes
4.2. Chromosomal Similarities and Novelties
4.3. Implications of Discovery of the New Species
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L.; deWaard, J.R. Biological identifications through DNA barcodes. Proc. R. Soc. Lond. B 2003, 270, 313–321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhimulev, I.F.; Belyaeva, E.S.; Semeshin, V.F.; Koryakov, D.E.; Demakov, S.A.; Demakova, O.V.; Pokholkova, G.V.; Andreyeva, E.N. Polytene chromosomes: 70 years of genetic research. Int. Rev. Cytol. 2004, 241, 203–275. [Google Scholar] [CrossRef]
- Michailova, P.V. The polytene chromosomes and their significance to the systematics and phylogeny of the family Chironomidae, Diptera. Acta Zool. Fenn. 1989, 186, 1–107. [Google Scholar]
- Coluzzi, M.; Sabatini, A.; della Torre, A.; Di Deco, M.A.; Petrarca, V. A polytene chromosome analysis of the Anopheles gambiae species complex. Science 2002, 298, 1415–1418. [Google Scholar] [CrossRef]
- Tosi, D.; Pereira, M.A.Q.R.; Vilela, C.R. Polytene chromosomes and phylogenetic relationships in ten Drosophila species of the annulimana group (Diptera, Drosophilidae). Gen. Mol. Biol. 2007, 30, 1169–1180. [Google Scholar] [CrossRef]
- Chubareva, L.A.; Petrova, N.A. Tsitologicheskie Karty Politennykh Khromosom i Nekotorye Morfologicheskie Osobennosti Krovososushchikh Moshek Rossii i Sopredel’nykh Stran (Diptera: Simuliidae): Atlas [Cytological Maps of Polytene Chromosomes and Some Morphological Features of Bloodsucking Black Flies of Russia and Adjacent Countries (Diptera: Simuliidae): Atlas]; Tovarishchestvo Nauchnykh Izdanii KMK: St. Petersburg, Russia, 2008; p. 134. (In Russian) [Google Scholar]
- Adler, P.H.; Takaoka, H.; Sofian-Azirun, M.; Low, V.L.; Ya’cob, Z.; Chen, C.D.; Lau, K.W.; Pham, X.D. Vietnam, a hotspot for chromosomal diversity and cryptic species in black flies (Diptera: Simuliidae). PLoS ONE 2016, 11, e0163881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, J.; Cheng, B.; Zhu, G.; Wei, Y.; Tang, J.; Cao, J.; Ma, Y.; Sharakhova, M.V.; Xia, A.; Sharakhov, I.V. Structural divergence of chromosomes between malaria vectors Anopheles lesteri and Anopheles sinensis. Parasit. Vectors 2020, 9, 608. [Google Scholar] [CrossRef] [Green Version]
- Rothfels, K. Speciation in black flies. Genome 1989, 32, 500–509. [Google Scholar] [CrossRef]
- Green, C.A. Cladistic analysis of mosquito chromosome data (Anopheles (Cellia) Myzomyia). J. Hered. 1982, 73, 2–11. [Google Scholar] [CrossRef]
- Ayala, F.J.; Coluzzi, M. Chromosome speciation: Humans, Drosophila, and mosquitoes. Proc. Nat. Acad. Sci. USA 2005, 102, 6535–6542. [Google Scholar] [CrossRef] [Green Version]
- Kiknadze, I.I.; Gunderina, L.I.; Butler, M.G.; Wuelker, W.F.; Martin, J. Chromosomes and continents. In Biosphere Origin and Evolution; Dobretsov, N., Kolchanov, N., Rozanov, A., Zavarzin, G., Eds.; Springer Science + Business Media: New York, NY, USA, 2008; pp. 349–369. [Google Scholar] [CrossRef]
- Schaeffer, S.W.; Bhutkar, A.; McAllister, B.F.; Matsuda, M.; Matzkin, L.M.; O’Grady, P.M.; Rohde, C.; Valente, V.L.S.; Aguadé, M.; Anderson, W.W.; et al. Polytene chromosomal maps of 11 Drosophila species: The order of genomic scaffolds inferred from genetic and physical maps. Genetics 2008, 179, 1601–1655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coluzzi, M. Heterogeneities of the malaria vectorial system in tropical Africa and their significance in malaria epidemiology and control. Bull. World Health Org. 1984, 62, 107–113. [Google Scholar] [PubMed]
- Procunier, W.S. Cytological approaches to simuliid biosystematics in relation to the epidemiology and control of human onchocerciasis. Genome 1989, 32, 559–569. [Google Scholar] [CrossRef]
- Adler, P.H.; Cheke, R.A.; Post, R.J. Evolution, epidemiology, and population genetics of black flies (Diptera: Simuliidae). Infect. Genet. Evol. 2010, 10, 846–865. [Google Scholar] [CrossRef] [PubMed]
- Post, R.J.; Mustapha, M.; Krüger, A. Taxonomy and inventory of the cytospecies and cytotypes of the Simulium damnosum complex (Diptera: Simuliidae) in relation to onchocerciasis. Trop. Med. Int. Health 2007, 12, 1342–1353. [Google Scholar] [CrossRef] [PubMed]
- Adler, P.H. World Blackflies (Diptera: Simuliidae): A Comprehensive Revision of the Taxonomic and Geographical Inventory [2022]. 2022. Available online: http://biomia.sites.clemson.edu/pdfs/blackflyinventory.pdf (accessed on 29 May 2022).
- Adler, P.H.; Crosskey, R.W. Cytotaxonomy of the Simuliidae (Diptera): A systematic and bibliographic conspectus. Zootaxa 2015, 3975, 1–139. [Google Scholar] [CrossRef] [Green Version]
- Rothfels, K.H. Cytotaxonomy of black flies (Simuliidae). Annu. Rev. Entomol. 1979, 24, 507–539. [Google Scholar] [CrossRef]
- Adler, P.H.; Currie, D.C.; Wood, D.M. The Black Flies (Simuliidae) of North America; Cornell University Press: Ithaca, NY, USA, 2004; p. 941. [Google Scholar]
- Golini, V.I.; Rothfels, K. The polytene chromosomes of North American blackflies in the Eusimulium canonicolum group (Diptera: Simuliidae). Can. J. Zool. 1984, 62, 2097–2109. [Google Scholar] [CrossRef]
- Moulton, J.K.; Adler, P.H. Taxonomy and biology of Simulium clarkei Stone & Snoddy (Diptera: Simuliidae), a poorly known black fly of the southeastern United States. Zootaxa 2002, 31, 1–7. [Google Scholar] [CrossRef]
- Jinbo, U.; Kato, T.; Ito, M. Current progress in DNA barcoding and future implications for entomology. Entomol. Sci. 2011, 14, 107–124. [Google Scholar] [CrossRef]
- Craig, D.A. Mouthparts and feeding behaviour of Tahitian larval Simuliidae (Diptera: Nematocera). Quaest. Entomol. 1977, 13, 195–218. [Google Scholar]
- Yankovsky, A.V. Opredelitel’ Moshek (Diptera: Simuliidae) Rossii i Sopredel’nykh Territorii (Byvshego SSSR) [A Key for the Identification of Blackflies (Diptera: Simuliidae) of Russia and Adjacent Countries (Former USSR)]. In Handbook for the Identification of the Fauna of Russia 170; Zoological Institute: Saint Petersburg, Russia, 2003; p. 570. (In Russian) [Google Scholar]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, G. MEGA11: Molecular evolutionary genetics analysis version. 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Currie, D.C. Black flies (Diptera: Simuliidae) of the Yukon, with reference to the black-fly fauna of northwestern North America. In Insects of the Yukon; Danks, H.V., Downes, J.A., Eds.; Biological Survey of Canada (Terrestrial Arthropods): Ottawa, ON, Canada, 1997; pp. 563–614. [Google Scholar]
- Takaoka, H.; Otsuka, Y.; Fukuda, M. A new species of Simulium (Boreosimulium) from Hokkaido, Japan (Diptera: Simuliidae). Med. Entomol. Zool. 2006, 57, 35–43. [Google Scholar] [CrossRef] [Green Version]
- Sato, H.; Saito, K.; Fukuda, M.; Takahashi, M.; Takaoka, H. Taxonomic revision of Simulium konoi Takahasi (Diptera: Simuliidae) from Japan. Med. Entomol. Zool. 2004, 55, 313–324. [Google Scholar] [CrossRef] [Green Version]
- Rivera, J.; Currie, D.C. Identification of Nearctic black flies using DNA barcodes (Diptera: Simuliidae). Mol. Ecol. Resour. 2009, 9 (Suppl. 1), 224–236. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Bedo, D.G. Polytene chromosomes in pupal and adult blackflies (Diptera: Simuliidae). Chromosoma 1976, 57, 387–396. [Google Scholar] [CrossRef]
- Meier, R.; Blaimer, B.B.; Buenaventura, E.; Hartop, E.; vonRintelen, T.; Srivathsan, A.; Yeo, D. A re-analysis of the data in Sharkey et al.’s (2021) minimalist revision reveals that BINs do not deserve names, but BOLD Systems needs a stronger commitment to open science. Cladistics 2021, 38, 264–275. [Google Scholar] [CrossRef] [PubMed]
- Adler, P.H.; Srisuka, W.; Low, V.L.; Takaoka, H.; Saeung, A. High-elevation chromosomal diversity of black flies (Diptera: Simuliidae) in Thailand. Insect Syst. Divers. 2019, 3, 1. [Google Scholar] [CrossRef]
- Pramual, P.; Pangjanda, S. Effects of habitat specialization on population genetic structure of black fly Simulium weji Takaoka (Diptera: Simuliidae). J. Asia-Pac. Entomol. 2015, 18, 33–37. [Google Scholar] [CrossRef]
- Bedo, D.G. Cytogenetics and evolution of Simulium ornatipes Skuse (Diptera: Simuliidae). II. Temporal variation in chromosomal polymorphisms and homosequential sibling species. Evolution 1979, 33, 296–308. [Google Scholar] [CrossRef]
- Henderson, C.A.P. Homosequential species 2a and 2b within the Prosimulium onychodactylum complex (Diptera): Temporal heterogeneity, linkage disequilibrium, and Wahlund effect. Can. J. Zool. 1986, 64, 859–866. [Google Scholar] [CrossRef]
- Pangjanda, S.; Pramual, P. Tests of conspecificity for closely related black flies (Diptera: Simuliidae) species of the Simulium striatum group in Thailand. Zootaxa 2017, 4231, 421–430. [Google Scholar] [CrossRef]
- Brockhouse, C.; Bass, J.A.B.; Straus, N.A. Chromocentre polymorphism in polytene chromosomes of Simulium costatum (Diptera: Simuliidae). Genome 1989, 32, 510–515. [Google Scholar] [CrossRef]
- Thaijarern, J.; Pramual, P.; Adler, P.H. Limited differentiation among black flies in the Simulium multistriatum species group (Diptera: Simuliidae) in Thailand: Cryptic species, homosequential species, and homosequential cryptic species. Zool. J. Linn. Soc. 2018, 184, 1024–1054. [Google Scholar] [CrossRef]
- Rothfels, K.H.; Freeman, D.M. The salivary gland chromosomes of seven species of Prosimulium (Diptera, Simuliidae) in the mixtum (IIIL-1) group. Can. J. Zool. 1977, 55, 482–507. [Google Scholar] [CrossRef]
- Ferree, P.M.; Barbash, D.A. Species-specific heterochromatin prevents mitotic chromosome segregation to cause hybrid lethality in Drosophila. PLoS Biol. 2009, 7, e1000234. [Google Scholar] [CrossRef]





| Rearrangement | Frequency |
|---|---|
| IS-2 | 1.00 |
| IL us-1 | 0.58 1 |
| IIS-1 | 1.00 |
| IIL-1 | 1.00 |
| IIL-2 | 1.00 |
| IIL eu, us-1 | 1.00 |
| IIL-complex2 | 1.00 |
| IIL us-1 | 0.10 |
| IIL us-2 | 0.74 |
| IIIL-1 | 1.00 |
| IIIL-2 | 1.00 |
| IIIL ca, eu, em, Hi, us-1 | 0.71 |
| IIIL N.O. transposition | 1.00 |
| IIIL N.O. | * 3 |
| Species | Number of Hits * | Percent Identity |
|---|---|---|
| Simulium balteatum | 5 | 91.7–92.5 |
| Simulium emarginatum | 7 | 91.4–92.4 |
| Simulium sp. BIOUG24287-A03 | 1 | 92.20 |
| Simulium sp. BIOUG24289-E02 | 1 | 92.20 |
| Simulium annulus | 4 | 91.2–91.5 |
| Simulium johannseni | 4 | 91.1–91.4 |
| Simulium lundstromi | 9 | 90.1–90.7 |
| Simulium angustitarse | 3 | 90.3–90.6 |
| Simuliidae sp. BOLD:AAG9573 | 3 | 90.1–90.6 |
| Simulium maculatum | 1 | 90.30 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adler, P.H.; Huang, S. Chromosomes as Barcodes: Discovery of a New Species of Black Fly (Diptera: Simuliidae) from California, USA. Insects 2022, 13, 903. https://doi.org/10.3390/insects13100903
Adler PH, Huang S. Chromosomes as Barcodes: Discovery of a New Species of Black Fly (Diptera: Simuliidae) from California, USA. Insects. 2022; 13(10):903. https://doi.org/10.3390/insects13100903
Chicago/Turabian StyleAdler, Peter H., and Shaoming Huang. 2022. "Chromosomes as Barcodes: Discovery of a New Species of Black Fly (Diptera: Simuliidae) from California, USA" Insects 13, no. 10: 903. https://doi.org/10.3390/insects13100903
APA StyleAdler, P. H., & Huang, S. (2022). Chromosomes as Barcodes: Discovery of a New Species of Black Fly (Diptera: Simuliidae) from California, USA. Insects, 13(10), 903. https://doi.org/10.3390/insects13100903








