New Cell Lines Derived from Laboratory Colony Triatoma infestans and Rhodnius prolixus, Vectors of Trypanosoma cruzi, Do Not Harbour Triatoma Virus
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Insects
2.2. Preparation of Primary Cell Cultures
2.3. Subculture and Cell Line Generation
2.4. Cryopreservation and Resuscitation of Cultured Cells
2.5. Species Confirmation and Screening for Contaminating Bacteria
2.6. SISPA Screening for TrV and Other RNA Viruses
2.7. Deposition of Sequences in Public Databases
3. Results
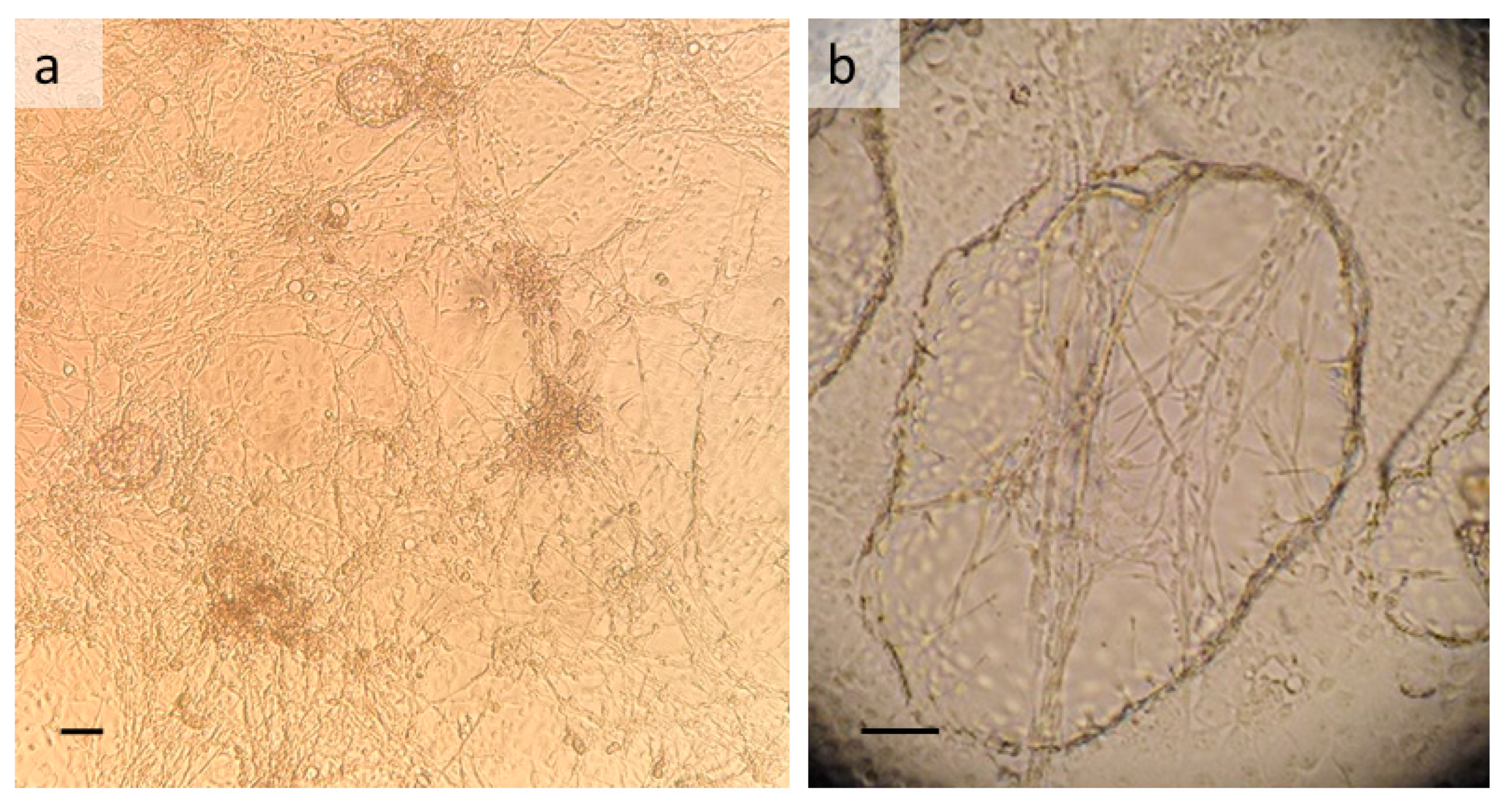
3.1. T. infestans Cell Line TIE/LULS54
3.2. R. prolixus Cell Lines RPE/LULS53 and RPE/LULS57
3.3. Confirmation of Species Origin and Screening for Contaminating Microorganisms
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Perez-Molina, J.A.; Molina, I. Chagas disease. Lancet 2018, 391, 82–94. [Google Scholar] [CrossRef]
- Pessoa, G.C.D.; Vinas, P.A.; Rosa, A.C.L.; Diotaiuti, L. History of insecticide resistance of Triatominae vectors. Rev. Soc. Bras. Med. Trop. 2015, 48, 380–389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muscio, O.A.; La Torre, J.L.; Scodeller, E.A. Characterization of Triatoma virus, a Picorna-like virus isolated from the triatomine bug Triatoma infestans. J. Gen. Virol. 2018, 69, 2929–2934. [Google Scholar] [CrossRef] [PubMed]
- Squires, G.; Pous, J.; Agirre, J.; Rozas-Dennis, G.S.; Costabel, M.D.; Marti, G.A.; Navaza, J.; Bressanelli, S.; Guerin, D.M.A.; Rey, F.A. Structure of the Triatoma virus capsid. Acta Cryst. 2013, D69, 1026–1037. [Google Scholar]
- Marti, G.A.; Gonzalez, E.T.; Garcia, J.J.; Viguera, A.R.; Guerin, D.M.A.; Echeverria, M.G. AC-ELISA and RT-PCR assays for the diagnosis of Triatoma virus (TrV) in triatomines (Hemiptera: Reduviidae) species. Arch. Virol. 2008, 153, 1427–1432. [Google Scholar] [CrossRef] [PubMed]
- Muscio, O.A.; La Torre, J.L.; Bonder, M.A.; Scodeller, E.A. Triatoma virus pathogenicity in laboratory colonies of Triatoma infestans (Hemiptera: Reduviidae). J. Med. Entomol. 1997, 34, 253–256. [Google Scholar] [CrossRef]
- Marti, G.A.; Ragone, P.; Balsalobre, A.; Ceccarelli, S.; Susevich, M.L.; Diosque, P.; Echeverria, M.G.; Rabinovich, J.E. Can Triatoma virus inhibit infection of Trypanosoma cruzi (Chagas, 1909) in Triatoma infestans (Klug)? A cross infection and co-infection study. J. Invertebr. Pathol. 2017, 150, 101–105. [Google Scholar] [CrossRef] [PubMed]
- Marti, G.A.; Echeverria, M.G.; Susevich, M.L.; Ceccarelli, S.; Balsalobre, A.; Rabinovich, J.E.; Diotaiuti, L.; Guerin, D.M.A. Exploration for Triatoma virus (TrV) infection in laboratory-reared triatomines of Latin America: A collaborative study. Int. J. Trop. Insect Sci. 2013, 33, 294–304. [Google Scholar] [CrossRef]
- Sanchez-Eugenia, R.; Mendez, F.; Querido, J.F.B.; Silva, M.S.; Guerin, D.M.A.; Rodriguez, J.F. Triatoma virus structural polyprotein expression, processing and assembly into virus-like particles. J. Gen. Virol. 2015, 96, 64–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Susevic, M.L.; Marti, G.A.; Metz, G.E.; Echeverria, M.G. First study of different insect cells to Triatoma virus infection. Curr. Microbiol. 2015, 70, 470–475. [Google Scholar] [CrossRef] [PubMed]
- Marti, G.A.; Bonica, M.B.; Susevich, M.L.; Reynaldi, F.; Micieli, M.V.; Echeverria, M.G. Host range of Triatoma virus does not extend to Aedes aegypti and Apis mellifera. J. Invertebr. Pathol. 2020, 173, 107383. [Google Scholar] [CrossRef]
- Varma, M.G.R.; Pudney, M. The culture of embryonic cells from the bug Triatoma maculata (Erichson) (Hemiptera: Reduviidae). Exp. Cell Res. 1967, 45, 671–675. [Google Scholar] [CrossRef]
- Pudney, M.; Lanar, D. Establishment and characterization of a cell line (BTC-32) from the triatomine bug, Triatoma infestans (Klug) (Hemiptera: Reduviidae). Ann. Trop. Med. Parasitol. 1977, 71, 109–118. [Google Scholar] [CrossRef]
- Lanar, D.E. Growth and differentiation of Trypanosoma cruzi cultivated with a Triatoma infestans embryo cell line. J. Protozool. 1979, 26, 457–462. [Google Scholar] [CrossRef]
- Kurtti, T.J.; Tsang, K.R.; Brooks, M.A. The spread of infection by the microsporidan, Nosema disstriae, in insect cell lines. J. Protozool. 1983, 30, 652–657. [Google Scholar] [CrossRef]
- Chrzastek, K.; Lee, D.; Smith, D.; Sharma, P.; Suarez, D.L.; Pantin-Jackwood, M.; Kapczynski, D.R. Use of Sequence-Independent, Single-Primer-Amplification (SISPA) for rapid detection, identification, and characterization of avian RNA viruses. Virology 2017, 509, 159–166. [Google Scholar] [CrossRef]
- Brito, T.F.; Coelho, V.L.; Cardoso, M.A.; Brito, I.A.A.; Berni, M.A.; Zenk, F.L.; Iovino, N.; Pane, A. Transovarial transmission of a core virome in the Chagas disease vector Rhodnius prolixus. PLoS Pathog. 2021, 17, e1009780. [Google Scholar] [CrossRef]
- Whitten, M.M.A.; Mello, C.B.; Gomes, S.A.O.; Nigam, Y.; Azambuja, P.; Garcia, E.S.; Ratcliffe, N.A. Role of superoxide and reactive nitrogen intermediates in Rhodnius prolixus (Reduviidae)/Trypanosoma rangeli interactions. Exp. Parasitol. 2001, 98, 44–57. [Google Scholar] [CrossRef]
- Buxton, P.A. The biology of a blood-sucking bug, Rhodnius prolixus. Trans. Entomol. Soc. Lond. 1930, 78, 227–236. [Google Scholar] [CrossRef]
- Alberdi, M.P.; Nijhof, A.M.; Jongejan, F.; Bell-Sakyi, L. Tick cell culture isolation and growth of Rickettsia raoultii from Dutch Dermacentor reticulatus ticks. Ticks Tick Borne Dis. 2012, 3, 349–354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bell-Sakyi, L. Continuous cell lines from the tick Hyalomma anatolicum anatolicum. J. Parasitol. 1991, 77, 1006–1008. [Google Scholar] [CrossRef] [PubMed]
- Medlin, L.; Elwood, H.J.; Stickel, S.; Sogin, M.L. The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene 1988, 71, 491–499. [Google Scholar] [CrossRef] [Green Version]
- Marcilla, A.; Bragues, M.D.; Ramsey, J.M.; Magallon-Gastelum, E.; Salazar-Schettino, P.M.; Abad-Franch, F.; Dujardin, J.-P.; Schofield, C.J.; Mas-Coma, S. The ITS-2 of the nuclear rDNA as a molecular marker for populations, species, and phylogenetic relationships in Triatominae (Hemiptera: Reduviidae), vectors of Chagas disease. Mol. Phylogenet. Evol. 2001, 18, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, F.A.; Peretolchina, T.; Lazoski, C.; Harris, K.; Dotson, E.M.; Abad-Franch, F.; Tamayo, E.; Pennington, P.M.; Monroy, C.; Cordon-Rosales, C.; et al. Phylogeographic pattern and extensive mitochondrial DNA divergence disclose a species complex within the Chagas disease vector Triatoma dimidiata. PLoS ONE 2013, 8, e70974. [Google Scholar] [CrossRef]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef] [Green Version]
- Mutunga, M.; Preston, P.M.; Sumption, K.J. Nitric oxide is produced by Cowdria ruminantium-infected bovine pulmonary endothelial cells in vitro and is stimulated by gamma interferon. Infect. Immun. 1998, 66, 2115–2121. [Google Scholar] [CrossRef] [Green Version]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- De Coster, W.; D’Hert, S.; Schultz, D.T.; Cruts, M.; Van Broeckhoven, C. NanoPack: Visualizing and processing long-read sequencing data. Bioinformatics 2018, 34, 2666–2669. [Google Scholar] [CrossRef] [Green Version]
- Ondov, B.D.; Bergman, N.H.; Phillippy, A.M. Interactive metagenomic visualization in a Web browser. Bioinformatics 2011, 12, 385. [Google Scholar] [CrossRef] [Green Version]
- Marti, J.M. Recentrifuge: Robust comparative analysis and contamination removal for metagenomics. PLoS Comput. Biol. 2019, 15, e1006967. [Google Scholar] [CrossRef] [Green Version]
- Mas-Coma, S.; Bargues, M.D. Populations, hybrids and the systematic concepts of species and subspecies in Chagas disease triatomine vectors inferred from nuclear ribosomal and mitochondrial DNA. Acta Trop. 2009, 110, 112–136. [Google Scholar] [CrossRef] [PubMed]
- Bargues, M.D.; Marcilla, A.; Ramsey, J.M.; Dujardin, J.P.; Schofield, C.J.; Mas-Coma, S. Nuclear rDNA-based molecular clock of the evolution of Triatominae (Hemiptera: Reduviidae), vectors of Chagas disease. Mem. Inst. Oswaldo Cruz Rio de Janeiro 2000, 95, 567–573. [Google Scholar] [CrossRef] [PubMed]
- Martinez, F.H.; Villalobos, G.C.; Cevallos, A.M.; De la Torre, P.; Laclette, J.P.; Alejandre-Aguilar, R.; Espinoza, B. Taxonomic study of the Phyllosoma complex and other triatomine (Insecta: Hemiptera: Reduviidae) species of epidemiological importance in the transmission of Chagas disease: Using ITS-2 and mtCytB sequences. Mol. Phylogenet. Evol. 2006, 41, 279–287. [Google Scholar] [CrossRef] [PubMed]
- Pita, S.; Panzera, F.; Vela, J.; Mora, P.; Palomeque, T.; Lorite, P. Complete mitochondrial genome of Triatoma infestans (Hemiptera, Reduviidae, Triatominae), main vector of Chagas disease. Infect. Genet. Evol. 2017, 54, 1580163. [Google Scholar] [CrossRef]
- Rojas de Arias, A.; Messenger, L.A.; Rolon, M.; Vega, M.C.; Acosta, N.; Villalba, C.; Marcet, P.L. Dynamics of Triatoma infestans populations in the Paraguayan Chaco: Population genetic analysis of household reinfestation following vector control. PLoS ONE 2022, 17, e0263465. [Google Scholar] [CrossRef]
- Nascimento, J.D.; da Rosa, J.A.; Salgado-Roa, F.C.; Hernandez, C.; Pardo-Diaz, C.; Alevi, K.C.C.; Ravazi, A.; Oliveira, J.; Oliveira, M.T.V.A.; Salazar, C.; et al. Taxonomical over splitting in the Rhodnius prolixus (Insecta: Hemiptera: Reduviidae) clade: Are R. taquarussuensis (da Rosa et al., 2017) and R. neglectus (Lent, 1954) the same species? PLoS ONE 2019, 14, e0211285. [Google Scholar] [CrossRef] [Green Version]
- Bell-Sakyi, L.; Beliavskaia, A.; Hartley, C.S.; Jones, L.; Luu, L.; Haines, L.R.; Hamilton, J.G.C.; Darby, A.C.; Makepeace, B.L. Isolation in natural host cell lines of Wolbachia strains wPip from the mosquito Culex pipiens and wPap from the sand fly Phlebotomus papatasi. Insects 2021, 12, 871. [Google Scholar] [CrossRef]
- Bell-Sakyi, L.; Ruzek, D.; Gould, E.A. Continuous cell lines from the soft tick Ornithodoros moubata. Exp. Appl. Acarol. 2009, 49, 209–219. [Google Scholar] [CrossRef] [Green Version]
- Sugimoto, T.N.; Watanabe, K.; Akiduki, G.; Imanashi, S.; Mitsuhashi, W. A new continuous cell line from the pest insect, Anomala cuprea (Coleoptera; Scarabaeidae): Emergence of contractile cells. In Vitro Cell. Dev. Biol. Anim. 2022, 58, 610–618. [Google Scholar] [CrossRef]
- Alba Soto, C.D.; Cappa, S.M.G. Trypanosoma cruzi Journey from the Insect Vector to the Host Cell. In Chagas Disease; Altcheh, J.M., Freilij, H.L., Eds.; Springer Nature: Basel, Switzerland, 2019; pp. 25–59. [Google Scholar]
- Minter-Goedbloed, E.; Pudney, M.; Kilgour, V.; Evans, D.A. First record of a reptile trypanosome isolated from Glossina pallipides in Kenya. Z. Parasitenkd. 1983, 69, 17–26. [Google Scholar] [CrossRef]
- Minter-Goedbloed, E.; Leake, C.J.; Minter, D.M.; McNamara, J.; Kimber, C.; Bastien, P.; Evans, D.A.; Le Ray, D. Trypanosoma varani and T. grayi-like trypanosomes: Development in vitro and in insect hosts. Parasitol. Res. 1993, 79, 329–333. [Google Scholar] [CrossRef]
- Kikuchi, S.A.; Sodre, C.L.; Kalume, D.E.; Elias, C.G.R.; Santos, A.J.S.; Soeira, M.N.; Meuser, M.; Chapeaurouge, A.; Perales, J.; Fernandes, O. Proteomic analysis of two Trypanosoma cruzi zymodeme 3 strains. Exp. Parasitol. 2010, 126, 540–551. [Google Scholar] [CrossRef] [PubMed]
- Reduth, D.; Schaub, G.A.; Pudney, M. Cultivation of Blastocrithidia triatomae (Trypanosomatidae) on a cell line of its host Triatoma infestans (Reduviidae). Parasitology 1989, 98, 387–393. [Google Scholar] [CrossRef] [PubMed]
- Neumann, A.S.; Dias, F.A.; Ferreira, J.S.; Fontes, A.N.B.; Rosa, P.S.; Macedo, R.E.; Oliveira, J.H.; Teixeira, R.L.F.; Pessolani, M.C.V.; Moraes, M.O.; et al. Experimental infection of Rhodnius prolixus (Hemiptera, Triatominae) with Mycobacterium leprae indicates potential for leprosy transmission. PLoS ONE 2016, 11, e0156037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vieira, C.B.; Praca, Y.R.; Bentes, K.L.S.; Santiago, P.B.; Silva, S.M.M.; Silva, G.S.; Motta, F.N.; Bastos, I.M.D.; Santana, J.M.; Araujo, C.N. Triatomines: Trypanosomatids, bacteria, and viruses potential vectors? Front. Cell. Infect. Microbiol. 2018, 8, 405. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, J.S.; Oliveira, D.A.S.; Santos, J.P.; Ribeiro, C.C.D.U.; Baêta, B.A.; Teixeira, R.C.; Neumann, A.S.; Rosa, P.S.; Pessolani, M.C.V.; Moraes, M.O.; et al. Ticks as potential vectors of Mycobacterium leprae: Use of tick cell lines to culture the bacilli and generate transgenic strains. PLoS Negl. Trop. Dis. 2018, 12, e0007001. [Google Scholar] [CrossRef] [Green Version]
- Hypša, V.; Dale, C. In vitro culture and phylogenetic analysis of “Candidatus Arsenophonus triatominarum”, an intracellular bacterium from the triatomine bug, Triatoma infestans. Int. J. Syst. Bacteriol. 1997, 47, 1140–1144. [Google Scholar] [CrossRef] [Green Version]
- Calderon, J.M.; Fuya, P.; Santacoloma, L.; Gonzalez, C. Deltamethrin resistance in Chagas disease vectors colonizing oil palm plantations: Implications for vector control strategies in a public health-agriculture interface. Parasit. Vectors 2020, 13, 163. [Google Scholar] [CrossRef] [Green Version]
- Gaspe, M.S.; Cardinal, M.V.; Fernandez, M.P.; Vassena, C.V.; Santo-Orihuela, P.L.; Enriquez, G.F.; Alvedro, A.; Laino, M.A.; Nattero, J.; Alvarado-Otegui, J.A.; et al. Improved vector control of Triatoma infestans limited by emerging pyrethroid resistance across an urban-to-rural gradient in the Argentine Chaco. Parasit. Vectors 2021, 14, 437. [Google Scholar] [CrossRef]
- Flores-Villegas, A.L.; Cabrera-Bravo, M.; De Fuentes-Vicente, J.A.; Jiminez-Cortes, J.G.; Salazar-Schettino, P.M.; Bucio-Torres, M.I.; Cordoba-Aguilar, A. Coinfection by Trypanosoma cruzi and a fungal pathogen increases survival of Chagasic bugs: Advice against a fungal control strategy. Bull. Entomol. Res. 2020, 110, 363–369. [Google Scholar] [CrossRef]
- Querido, J.F.B.; Agirre, J.; Marti, G.A.; Guerin, D.M.A.; Silva, M.S. Inoculation of Triatoma virus (Dicistroviridae: Cripavirus) elicits a non-infective immune response in mice. Parasit. Vectors 2013, 6, 66. [Google Scholar] [CrossRef] [PubMed]
- Brumpt, E. Eclectisme alimentaire des reduvides, vecteurs du Trypanosoma cruzi. Presse Med. 1927, 77, 1161–1162. [Google Scholar]

| Cell Line | Gene Targeted | Sequence Length | Top BLAST Hits (GenBank Accession Number) | Query Cover | % Identity (Identical/Total bp) | Reference |
|---|---|---|---|---|---|---|
| TIE/LULS54 | Euk 18S | 1807 bp | Triatoma sordida, Bolivia (AJ421956) | 100% | 99.89% (1807/1809) | [31] |
| T. infestansb (Y18750) | 99.83% (1807/1810) | [32] | ||||
| ITS-2 a | 533 bp | T. infestans, Brazil (AY860388) | 87% | 98.71% (460/466) | [33] | |
| cytBa | 666 bp | T. infestans, Uruguay (KY640305) | 52% | 84.73% (294/347) | [34] | |
| T. infestans Paraguay (KY654076) | 84.44% (293/347) | [35] | ||||
| RPE/LULS53 | Euk18S | 1775 bp | Rhodnius stali, Bolivia (AJ243335) | 100% | 99.94% (1774/1775) | [32] |
| R. prolixus, Brazil (AJ421962) | 99.83% (1772/1775) | [31] | ||||
| ITS-2 a | 420 bp | R. prolixus, Colombia (AY345868) | 50% | 81.52% (172/211) | Unpublished c | |
| cytBa | 661 bp | R. prolixus, Colombia (MH704763) | 36% | 89.92% (214/238) | [36] | |
| RPE/LULS57 | ITS-2 a | 646 bp | R. prolixus, Colombia (AY345868) | 49% | 67.09% (210/313) | Unpublished c |
| cytBa | 189 bp | R. prolixus, Colombia (MH704763) | 97% | 94.02% (173/184) | [36] |
| Cell Line Sequencing Data | TIE/LULS54 | RPE/LULS53 | RPE/LULS57 | Negative Control |
|---|---|---|---|---|
| Number of reads in first run | 630037 | 453711 | 103105 | 4386 |
| Number of reads in second run | 4124079 | 3874280 | 3464787 | 54662 |
| Mean read length | 751.9 | 782.4 | 940.3 | 802 |
| Mean read quality | 13.4 | 13.4 | 13.4 | 13.3 |
| Number of reads assigned to viruses in first run | 3 (0.0005%) | 16 (0.004%) | 17 (0.02%) | 10 (0.2%) |
| Number of reads assigned to viruses in second run | 4 (0.0001%) | 5 (0.0001%) | 18 (0.0005%) | 39 (0.07%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Penrice-Randal, R.; Hartley, C.; Beliavskaia, A.; Dong, X.; Brandner-Garrod, L.; Whitten, M.; Bell-Sakyi, L. New Cell Lines Derived from Laboratory Colony Triatoma infestans and Rhodnius prolixus, Vectors of Trypanosoma cruzi, Do Not Harbour Triatoma Virus. Insects 2022, 13, 906. https://doi.org/10.3390/insects13100906
Penrice-Randal R, Hartley C, Beliavskaia A, Dong X, Brandner-Garrod L, Whitten M, Bell-Sakyi L. New Cell Lines Derived from Laboratory Colony Triatoma infestans and Rhodnius prolixus, Vectors of Trypanosoma cruzi, Do Not Harbour Triatoma Virus. Insects. 2022; 13(10):906. https://doi.org/10.3390/insects13100906
Chicago/Turabian StylePenrice-Randal, Rebekah, Catherine Hartley, Alexandra Beliavskaia, Xiaofeng Dong, Luke Brandner-Garrod, Miranda Whitten, and Lesley Bell-Sakyi. 2022. "New Cell Lines Derived from Laboratory Colony Triatoma infestans and Rhodnius prolixus, Vectors of Trypanosoma cruzi, Do Not Harbour Triatoma Virus" Insects 13, no. 10: 906. https://doi.org/10.3390/insects13100906





