Simple Summary
We sequenced the complete mitochondrial genomes (mitogenomes) of Gryllotalpa henana Cai & Niu, 1998 and the Chinese G. orientalis Burmeister, 1838 for the first time, and reconstructed the mitogenomic phylogeny of the infraorder Gryllidea. The results show that the two new mitogenomes are double-stranded circular molecules with a typical gene complement, gene arrangement and base composition, the same as those of other gryllotalpids and ancestral insects. Tandem repeats of the control region were discovered in Gryllotalpidae for the first time. Considering both the high nucleotide divergence and the elevated ratio of Ka/Ks, the genes nad2 and nad6 may be evaluated as potential markers for future phylogeny and species delimitation in Gryllotalpidae. The results of phylogenetic analyses provide supports for the mitogenomic and transcriptomic trees, but partially contradict those of the multilocus phylogenies.
Abstract
Owing to limited molecular data, the phylogenetic position of the family Gryllotalpidae is still controversial in the infraorder Gryllidea. Mitochondrial genome (mitogenome) plays a crucial role in reconstructing phylogenetic relationships and revealing the molecular evolution of insects. However, only four mitogenomes have been reported in Gryllotalpidae to date. Herein, we obtained the first mitogenomes of Gryllotalpa henana Cai & Niu, 1998 and the Chinese G. orientalis Burmeister, 1838, made a detailed comparison of all mitogenomes available in Gryllotalpidae and reconstructed the phylogeny of Gryllidea based on mitogenomes using Bayesian inference (BI) and maximum likelihood (ML) methods. The results show that the complete mitogenome sequences of G. henana (15,504 bp) and G. orientalis (15,497 bp) are conserved, both exhibiting the double-stranded circular structure, typical gene content and the ancestral insect gene arrangement. The complete mitogenome of G. henana exhibits the lowest average AT content ever detected in Gryllotalpidae, and even Gryllidea. The gene nad2 of both species has atypical initiation codon GTG. All tRNAs exhibit typical clover-leaf structure, except for trnS1 lacking the dihydrouridine (DHU) arm. A potential stem–loop structure, containing a (T)n(TC)2(T)n sequence, is detected in the control region of all gryllotalpids investigated and is likely related to the replication initiation of the minority strand. The phylogenetic analyses recover the six families of Gryllidea as Gryllotalpidae + (Myrmecophilidae + (Mogoplistidae + (Trigonidiidae + (Phalangopsidae + Gryllidae)))), similar to the trees based on transcriptomic and mitogenomic data. However, the trees are slightly different from the multilocus phylogenies, which show the sister-group relationship of Gryllotalpidae and Myrmecophilidae. The contradictions between mitogenomic and multilocus trees are briefly discussed.
1. Introduction
The mitochondrial genomes (or mitogenomes) of insects are double-stranded circular molecules with lengths ranging from approximately 15 kb to 20 kb, and generally comprise 37 genes with 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), two ribosome RNA genes (rRNAs) and a non-coding control region (CR) [1]. Mitogenomes are one of the most information-rich characteristics, and are useful in phylogeny, evolutionary history, species delimitation and population genetics [2,3,4,5]. Such studies have been well documented in many insect groups, and greatly contributed to understanding their phylogeny and evolution [6,7,8,9]. In Gryllotalpidae, however, only four species have available mitogenomes in GenBank to date [10,11,12,13].
Gryllotalpidae is a small family of mole crickets and currently consists of more than 100 species in eight extant genera worldwide [14,15]. Gryllotalpids comprise an exclusive group that possesses a pair of digging forelegs, a tumescent pronotum, short antennae and hind legs lacking jumping ability [16]. Owing to the limited molecular data, the phylogenetic position of Gryllotalpidae in Gryllidea is still controversial. Gryllotalpidae is sister to Myrmecophilidae based on multilocus analysis [17], but has conflicting phylogenetic positions in mitogenome-based trees [8,18].
Gryllotalpa Latreille, 1802, characterized by forelegs with four tibial dactyls, is the largest and most widespread genus in Gryllotalpidae and comprises more than ½ the species of the family recorded from all zoogeographical regions, with only 11 species distributed in China [19,20]. The species of Gryllotalpa are similar in external morphology, but exhibit complicated variations intraspecifically in morphology of wing venation and male genitalia, leading to difficulties in species delimitation [21,22]. The application of additional characteristics is necessary to resolve the taxonomic problem.
In this study, we present the first complete mitogenomes of G. henana and the Chinese G. orientalis, make a detailed comparison of gryllotalpid mitogenomes, and reconstruct the phylogeny of the infraorder Gryllidea, in an attempt to contribute the mitogenomic data of Gryllotalpidae for future phylogenetic studies of Orthoptera.
2. Materials and Methods
2.1. Sample Collection and Processing
An adult female of G. henana and an adult male of G. orientalis were collected at the Danjiang River Beach (33°5′ N, 111°13′ E, elevation 220–240 m) in Xichuan County, Henan Province, China, from late May to middle June 2021. The middle leg on one side of each specimen was stored in dry ice and sent to Biomarker Technologies, Inc. (Beijing, China) for extraction and sequencing. The complete mitogenome sequences were generated using the Illumina HiSeq™ 4000 system. The rest of the specimens were preserved in 75% ethanol and placed in the Laboratory of Agricultural Entomology and Pest Control, College of Agriculture, Ningxia University.
2.2. Sequence Analyses
The mitochondrial invertebrate genetic code was selected as the general code for all the programs used in the present study. The raw paired reads were retrieved and quality trimmed by CLC Genomics Workbench v7.0.4 (CLC Bio, Aarhus, Denmark) with default parameters, using the mitogenomic sequence of G. unispina (KC894752) and G. orientalis (AY660929) as references, respectively. The mitochondrial genomes of G. henana and the Chinese G. orientalis were annotated with Geneious 8.1.3 [23] with the same references. All 13 PCGs were determined by comparing with the ORF Finder and the homologous sequences of reference mitogenomes. Twenty-two tRNAs and two rRNAs were identified using the MITOS Web Server (http://mitos2.bioinf.uni-leipzig.de/index.py, accessed on 20 June 2022) [24]. Transfer RNAs were manually plotted, according to the secondary structure predicted by MITOS, using Adobe Illustrator CS5 (Adobe Inc., San Jose, CA, USA). Tandem Repeats Finder server (https://tandem.bu.edu/trf/trf.html, accessed on 17 May 2022) [25] and Mfold Web Server (http://www.mfold.org/, accessed on 17 May 2022) [26] were used to identify tandem repeats and to infer the stem-loop structure, respectively. Mitogenome maps were drawn using OGDRAW [27].
The base composition, codon usage and relative synonymous codon usage (RSCU) were all calculated using PhyloSuite [28]. DnaSP 6.0 [29] was used to conduct the nucleotide diversity (Pi), and non-synonymous (Ka) and synonymous (Ks) substitutions of each PCG among the species of Gryllotalpidae. Sliding window analyses with a window of 100 bp and a step size of 25 bp were performed to estimate the sequence diversity for each independent PCG, using DnaSP 6.0. Genetic distances based on 13 PCGs were estimated using MEGA 7.0 with Kimura-2-parameter (K2P) [30]. AT-content (the proportion of A + T out of the total) was used to assess the overall composition of the double-stranded molecule [31]. Strand asymmetry was calculated according to the formula: AT-skew = (A − T)/(A + T) and GC-skew = (G − C)/(G + C) [32]. The AT-content, AT-skew and GC-skew were graphically plotted by Origin 2018 (OriginLab Corp., Northampton, MA, USA). The Pi values were graphically plotted by CorelDRAW 2020 (Corel Corp., Ottawa, ON, Canada). The genetic distance and Ka/Ks ratios were graphically plotted by Microsoft Excel spreadsheet.
2.3. Phylogenetic Analyses
Twenty-eight species from six families of Gryllidea were chosen as the ingroup, and four species in Tettigoniidea and one species in Schizodactyloidea were selected as outgroups. The detailed information of species used in phylogenetic analyses were listed in Table 1. Statistics for the basic characteristics of the mitogenome and the extraction of PCGs and rRNAs were produced by PhyloSuite. The alignment of all 13 PCGs was conducted in batches with MAFFT integrated into PhyloSuite with codon alignment mode setting [33,34]. Two rRNAs were aligned using the Q-INS-i algorithm incorporated into MAFFT-with-extensions software (http://mafft.cbrc.jp/alignment/server/, accessed on 29 March 2022) [33]. Ambiguous sites of alignments of all genes were manually removed, and the modified alignments were concatenated using PhyloSuite [34].
Phylogenetic analyses were conducted using four different datasets: (1) P123: 13 PCGs (10,899 bp), (2) P123R: 13 PCGs + 2 rRNAs (13,550 bp), (3) P12: 13 PCGs excluding the third codon position (7266 bp), (4) P12R: 13 PCGs excluding the third codon position + 2 rRNAs (9917 bp). Phylogenetic trees were reconstructed using Bayesian inference (BI) and maximum likelihood (ML) analyses, with partition strategies for analyzing mitogenome data according to Leavitt [35]. The best-fit partition schemes and models for BI analyses were inferred using PartitionFinder 2 [36] integrated into PhyloSuite [34], and are shown in Supplementary Table S1. BI trees were conducted using MrBayes 3.2.6 [37] with 10 million MCMC generations, sampling every 1000 generations. The convergence was considered to be reached when the average standard deviation of the split frequencies was lower than 0.01. The first 25% were discarded as “burn-in”, and the remaining samples were used to generate the majority consensus trees and estimate the posterior probabilities (PPs). The best-fit substitution models for ML analyses were selected by ModelFinder [38], and shown in Supplementary Table S2. ML trees were reconstructed using IQ-TREE integrated into PhyloSuite under Ultrafast bootstrap. Bootstrap supports (BSs) were evaluated with 1000 replicates.

Table 1.
Details of the species investigated and the relative information.
Table 1.
Details of the species investigated and the relative information.
| Superfamily | Family | Species | Locality | Size (bp) | Accession No. | Resource |
|---|---|---|---|---|---|---|
| Gryllotalpoidea | Gryllotalpidae | Gryllotalpa henana Cai & Niu, 1998 | China | 15,504 | ON243749 | This study |
| G. orientalis Burmeister, 1838 | China | 15,497 | ON210982 | This study | ||
| G. orientalis Burmeister, 1838 | Korea | 15,521 | AY660929 | [12] | ||
| G. pluvialis (Mjöberg, 1913) | Australia | 15,525 | EU938371 | [11] | ||
| Gryllotalpa sp. | China | 15,506 | MK903562 | [10] | ||
| G. unispina Saussure, 1874 | China | 15,513 | KC894752 | [13] | ||
| Myrmecophilidae | Myrmecophilus kubotai Maruyama, 2004 | Japan | 15,345 | MZ440658 | [18] | |
| M. manni Schimmer, 1911 | USA | 15,323 | EU938370 | [11] | ||
| Myrmecophilus sp. | Japan | 15,341 | MZ440659 | [18] | ||
| Grylloidea | Phalangopsidae | Meloimorpha japonica (Haan, 1844) | China | 15,880 | MH580273 | [39] |
| Cacoplistes rogenhoferi Saussure, 1877 | China | 16,018 | MH580272 | [39] | ||
| Mogoplistidae | Ornebius bimaculatus (Shiraki, 1930) | China | 16,136 | MH580274 | [39] | |
| O. fuscicerci (Shiraki, 1930) | China | 16,368 | MH580275 | [39] | ||
| O. kanetataki (Matsumura, 1904) | China | 16,589 | MH580276 | [39] | ||
| Trigonidiidae | Dianemobius fascipes (Walker, 1869) | China | 15,363 | MK303550 | [40] | |
| D. furumagiensis (Ohmachi & Furukawa, 1929) | China | 15,350 | MK303551 | [40] | ||
| Homoeoxipha nigripes Xia & Liu, 1993 | China | 15,679 | MK303553 | [40] | ||
| Natula pravdini (Gorochov, 1985) | China | 15,817 | MG701239 | [40] | ||
| Svistella anhuiensis He, Li & Liu, 2009 | China | 16,494 | MG701238 | [40] | ||
| Polionemobius taprobanensis (Walker, 1869) | China | 16,641 | MK303552 | [40] | ||
| Gryllidae | Gryllodes sigillatus (Walker, 1869) | China | 16,369 | MW365703 | [41] | |
| Gryllus bimaculatus De Geer, 1773 | Korea | 16,075 | MT993975 | [42] | ||
| Loxoblemmus doenitzi Stein, 1881 | - | 15,620 | KX673202 | [43] | ||
| Oecanthus sinensis Walker, 1869 | China | 16,142 | KY783908 | [44] | ||
| Truljalia hibinonis (Matsumura, 1917) | China | 15,120 | KY783909 | [44] | ||
| Turanogryllus eous Bey-Bienko, 1956 | China | 16,045 | MK656322 | [45] | ||
| Velarifictorus hemelytrus (Saussure, 1877) | China | 16,123 | KU562918 | [46] | ||
| Acheta domesticus (Linnaeus, 1758) | Japan | 16,071 | MZ440654 | [18] | ||
| Outgroup | Tettigoniidae | Alloxiphidiopsis emarginata (Tinkham, 1944) | China | 16,207 | MN562488 | [47] |
| Tettigonia chinensis Willemse, 1933 | - | 16,244 | KX057727 | [48] | ||
| Gryllacrididae | Camptonotus carolinensis (Gerstaecker, 1860) | - | 15,211 | KM657333 | [49] | |
| Schizodactylidae | Comicus campestris Irish, 1986 | - | 15,691 | KM657337 | [49] | |
| Prophalangopsidae | Tarragoilus diuturnus Gorochov, 2001 | China | 16,144 | JQ999995 | [50] |
3. Results and Discussion
3.1. Genome Structure and Base Composition
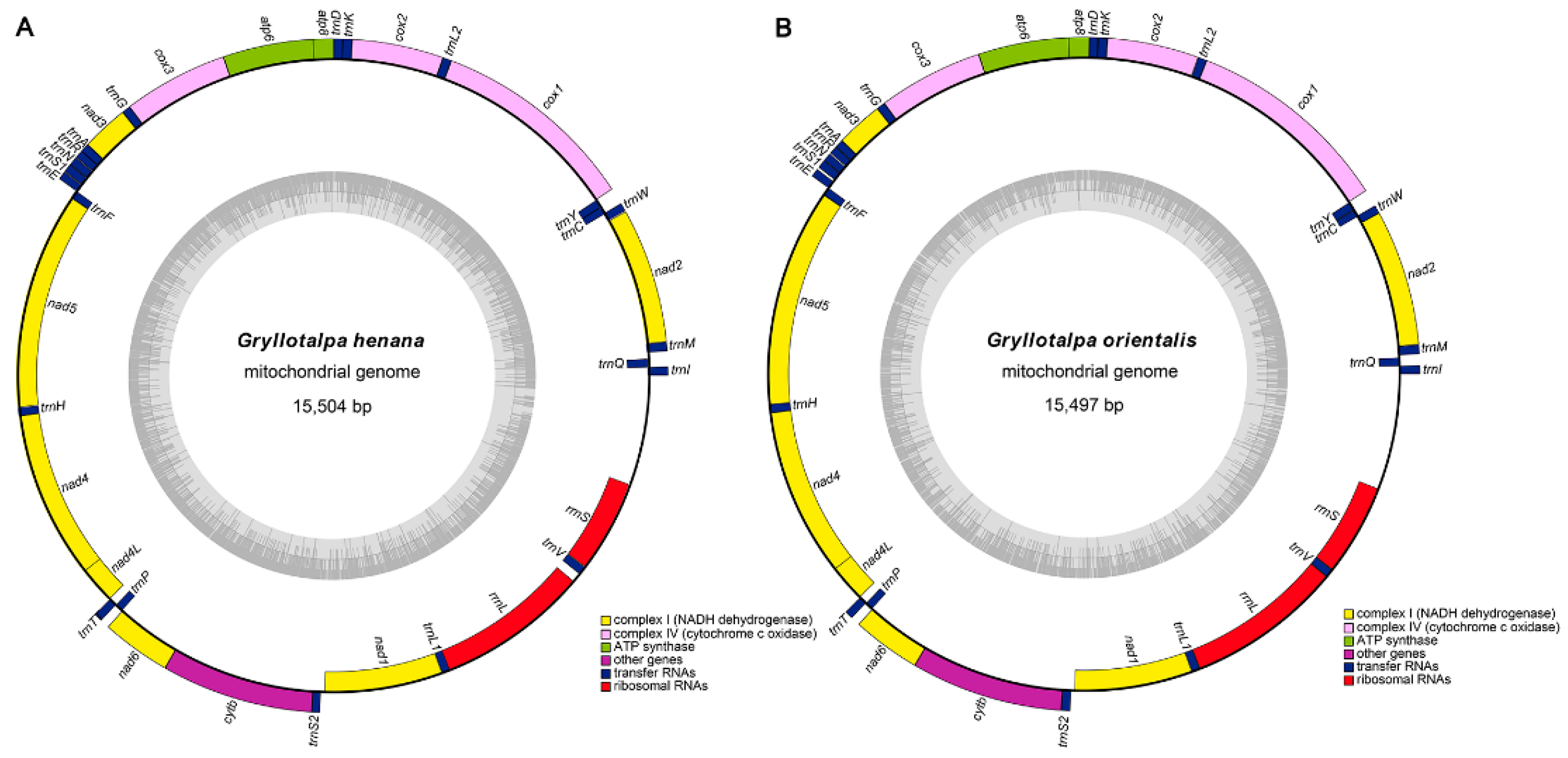
The complete mitogenomes of G. henana (Figure 1A) and the Chinese G. orientalis (Figure 1B) are 15,504 bp and 15,497 bp representing the smallest sizes known in Gryllotalpidae (Table 1). The lengths of two new mitogenomes are quite conserved, and within the size range of orthopteran mitogenomes (14–17 kb) [35,48,51]. Size differences of the mitogenomes in Gryllotalpidae are mainly due to variations in the length of the control region (CR) and the intergenic spaces between some of the tRNAs [10,11,12,13]. The mitogenomes of both species, similar to those of other gryllotalpids, are circular double-stranded molecules and contain the complete set of 37 genes (13 PCGs, 22 tRNAs and two rRNAs) and a non-coding CR (AT-rich region) (Figure 1). The majority strand (J-strand) encodes 23 genes, including nad2, nad3, nad6, cytb, cox1, cox2, cox3, atp8, atp6, trnI, trnM, trnW, trnL2, trnK, trnD, trnG, trnA, trnR, trnN, trnS1, trnE, trnT and trnS2. The remaining 14 genes (nad1, nad4, nad4L, nad5, trnQ, trnC, trnY, trnF, trnH, trnP, trnL1, trnV, rrnL and rrnS) are encoded on the minority strand (N-strand). The mitogenomes obtained herein are identical to those of other gryllotalpids in gene order and gene orientation, which are the hypothesized ancestral arrangements found in several insect orders [1].
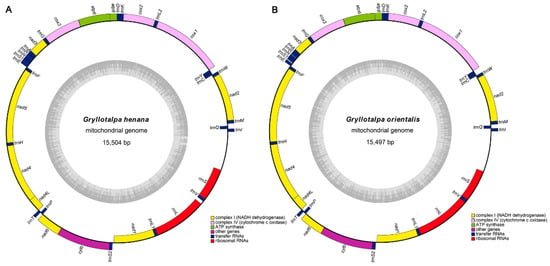
Figure 1.
Mitochondrial genome arrangements. (A) Mitochondrial genome of Gryllotalpa henana. (B) Mitochondrial genome of Gryllotalpa orientalis. The J-strand is visualized on the outer circle and the N-strand on the inner circle. The dark and light areas of the grey inner circle represent the GC- and AT-content, respectively.
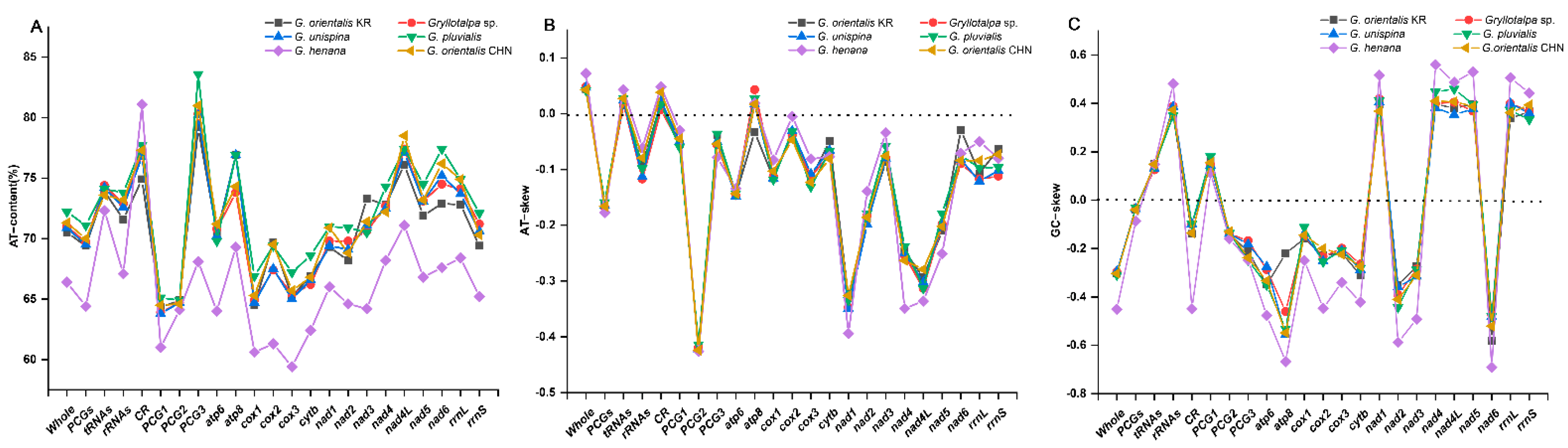
Two separated features, base proportion (AT-content) and strand asymmetry (AT- and GC-skew), are used to assess the base compositional bias of mitogenomes [31,32]. The AT-content of the whole mitogenomes ranges from 66.4% in G. henana to 72.2% in G. pluvialis (Figure 2A, Table 2), indicating that the overall composition is biased towards A and T in Gryllotalpidae. The CR in all six species exhibits a higher value of the AT-content (74.9–81.1%), followed successively by tRNAs (72.3–74.4%), rRNAs (67.1–73.8%) and PCGs (64.4–71.1%). In PCGs, the third codon positions have the highest AT-content (68.1–83.6%), compared with the first (61.0–65.1%) and second codon positions (64.1–64.9%) in all gryllotalpids. The AT-contents of four genomic regions are generally lower in G. henana than in other five gryllotalpids (Figure 2A, Table 2). The entire mitogenomes of all gryllotalpids exhibit the typical skew pattern of insects with positive AT-skew (0.042–0.072) and negative GC-skew (−0.451–−0.295), indicating that the majority strand of mitogenomes is biased in favor of A and C (Figure 2B,C, Table 2). The skew patterns of the four genomic regions are conserved in Gryllotalpa, and exhibit negative AT- and GC-skew in PCGs, positive AT- and GC-skew in tRNAs, negative AT-skew and positive GC-skew in rRNAs, and positive AT-skew and negative GC-skew in CR. The values of AT-skew are small and not significantly different from zero in the four genomic regions except for the PCGs (−0.177–−0.159). The GC-skew values are also low, with the exception of the increased ones for rRNAs in all the species of Gryllotalpa (0.347–0.482) and the decreased one for CR in G. henana (−0.450).
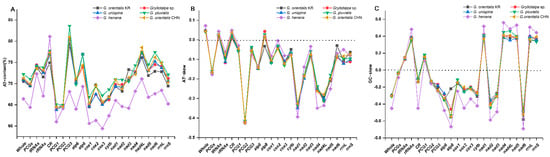
Figure 2.
Comparison of AT content and nucleotide skewness of six species in Gryllotalpidae. (A) AT-content. (B) AT-skew. (C) GC-skew. CHN, China; KR, Korea.

Table 2.
Nucleotide composition of the mitogenomes of six species in Gryllotalpidae.
3.2. Protein-Coding Genes and Codon Usage
The concatenated sequence of the PCGs is 11,097 bp in G. henana and 11,109 bp in the Chinese G. orientalis, accounting for 71.6% and 71.7% of their whole mitogenomes, respectively (Table 2 and Table 3). The 13 PCGs of the two new mitogenomes, similar to those of the other four gryllotalpids, contain two ATPase subunits (atp6 and atp8), three cytochrome c oxidase subunits (cox1–3), one cytochrome b gene (cytb), and seven NADH dehydrogenase subunits (nad1–6 and nad4L) (Figure 1, Table 3). The lengths of the 13 PCGs range from 156 bp of atp8 to 1723 bp of nad5 in both mitogenomes newly sequenced. The shortest atp8 and longest nad5, also found in other four gryllotalpids, are common features in metazoan mitogenomes [52,53].

Table 3.
Mitogenomic organization of six species in Gryllotalpidae.
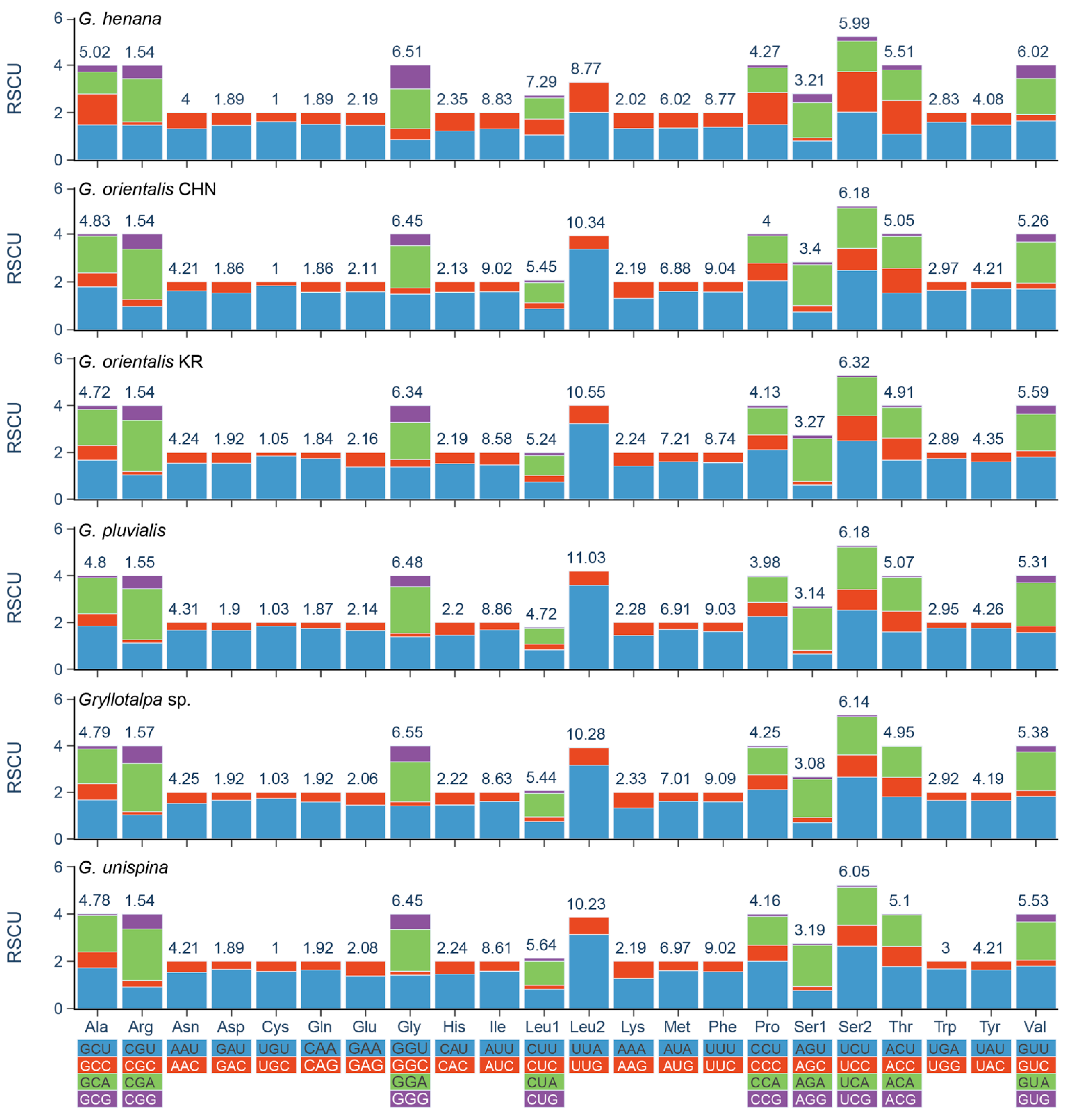
All PCGs of the two new mitogenomes have the typical initiation codon (ATN), except for nad2 starting with GTG (Table 3). The atypical initiation codon is also present in the mitogenomes of two other mole crickets (the Korean G. orientalis and G. unispina) and two katydids (Kuwayamaea brachyptera Gorochov & Kang, 2002 and Ruidocollaris obscura Liu & Jin, 1999) [12,13,54]. The termination codons are relatively conserved in Gryllotalpidae. Most of them are complete triplet bases TAA/TAG, and others are incomplete T/TA immediately followed by or partially overlapped with a tRNA gene. Incomplete stop codons are fairly common in the orthopteran mitogenomes and can be converted into a potential stop codon via polyadenylation to TAA [18,39,40,55]. The results of RSCU analyses show that the PCGs exhibit strong biases toward the nucleotides A and U in the codon usage. The four most frequent codons (UUU/Phe, UUA/Leu2, AUU/Ile, and AUA/Met) are the same in Gryllotalpidae, and all composed wholly of A or U (Figure 3, Supplementary Table S3). The codons ending with A/U occur more frequently than that with G/C, suggesting that the AU composition at third position of codons positively influences the nucleotide AT (or AU) bias of the PCGs in Gryllotalpidae.
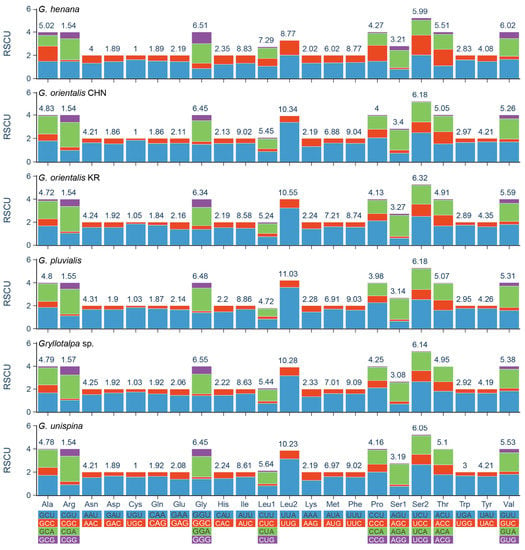
Figure 3.
Relative synonymous codon usage (RSCU) of the mitochondrial genomes of six species in Gryllotalpidae. CHN, China; KR, Korea. The numbers above the colored columns indicate the frequencies of amino acids.
3.3. Transfer and Ribosomal RNA Genes
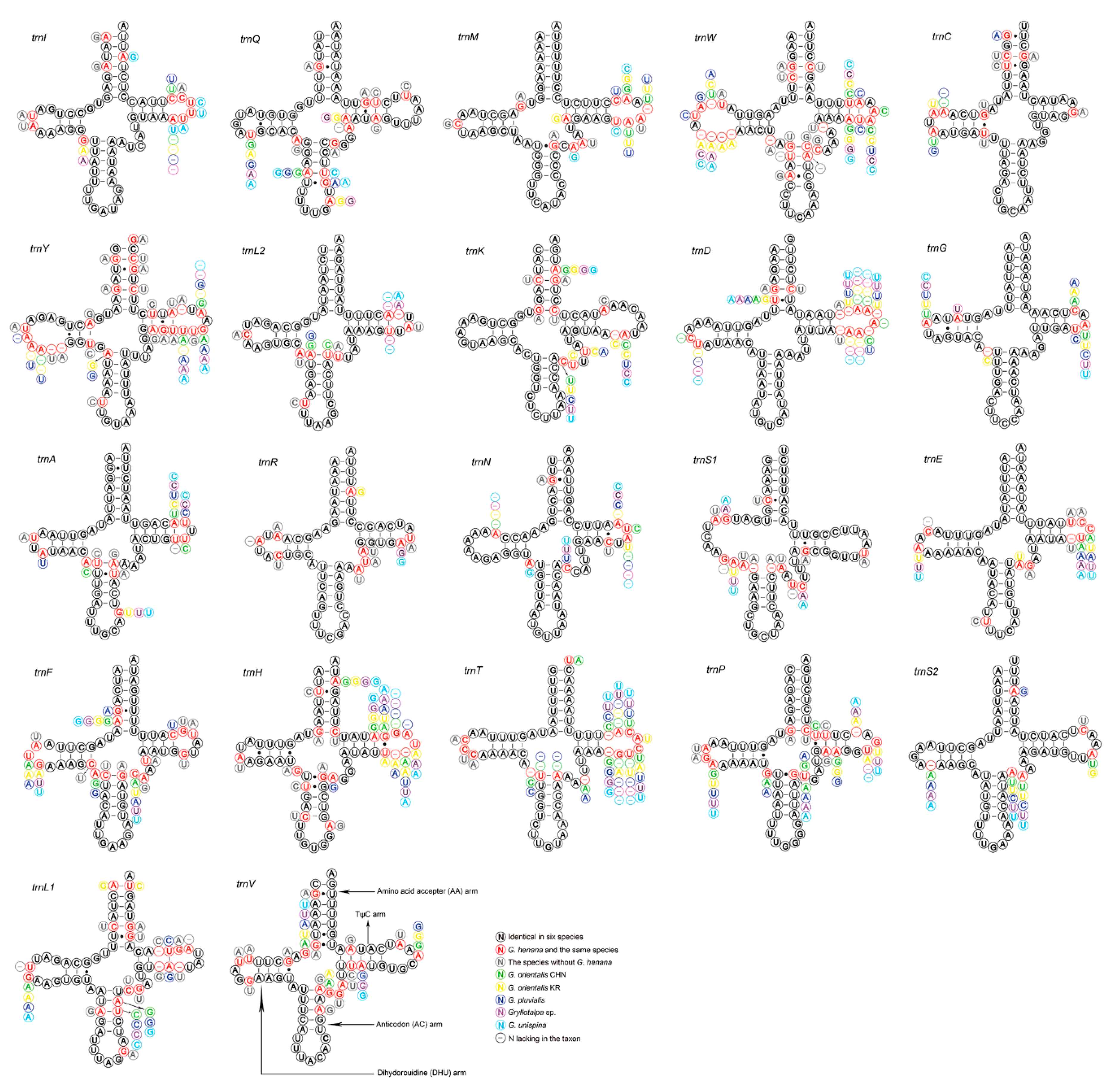
The 22 tRNAs of the two new mitogenomes are scattered around the circular DNA molecule, and are arranged identically in order and direction (Figure 1). The tRNAs of gryllotalpids retain the ancestral gene order [10,11,12,13], whereas multiple patterns of tRNA rearrangements have been detected in many other ensiferans [39,48] and most caeliferans [35]. All tRNAs exhibit typical clover–leaf structure, except for trnS1 (Figure 4). The dihydrouridine (DHU) arm of trnS1 forms a simple loop as in many other metazoans including gryllotalpids [10,11,12,13,52,56]. The length of tRNAs varies from 62 bp (trnC) to 71 bp (trnK) in G. henana and from 61 bp (trnC) to 71 bp (trnK) in the Chinese G. orientalis (Table 2), both within the variation range in Gryllotalpidae. The trnG gene of gryllotalpids generally exhibits the lowest nucleotide substitutions, while trnL1, trnW and trnY genes tend to be more variable among 22 tRNA genes (Figure 4). All tRNAs in the mitogenomes of Gryllotalpidae possess invariable length of 7 bp for both the acceptor stem and the anticodon loop. The length of anticodon stem is relatively conservative, varying from 4 bp in trnK and trnM to 5 bp in the rest of tRNAs. Most of the size variations among tRNAs stemmed from the length variation in DHU and TψC arms, within which the size of loops (all 3–10 bp) is more variable than that of stems (all 3–5 bp).
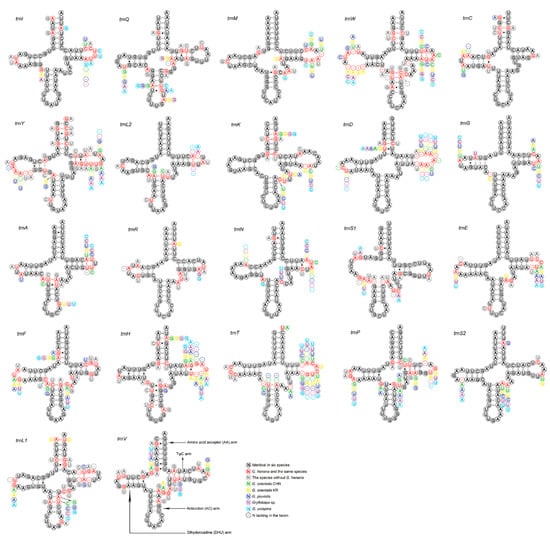
Figure 4.
Secondary structure for the tRNAs of six species in Gryllotalpidae. CHN, China; KR, Korea.
The tRNAs of G. henana possess a total of 36 unmatched base pairs, including 31 GU mismatches in most tRNAs, three AC mismatches in the anticodon stem of trnS1 and trnW and the TψC stem of trnN, and two UU mismatches in the acceptor stem of trnD and the anticodon stem of trnA (Figure 4, Supplementary Table S4). A total of 30 mismatches were detected in the Chinese G. orientalis. Twenty-seven of them are GU pairs, two are UU mismatches in the DHU stem of trnC and the acceptor stem of trnL1, and one is AA pair in the anticodon arm of trnS1. The mismatch number in the Chinese G. orientalis is lower than that in the Korean one (36 mismatches) (Supplementary Table S4), suggesting that the mitogenomes are differentiated intraspecifically.
The two rRNA genes (rrnL and rrnS) are located in the conserved positions as in mitogenomes of other gryllotalpids [10,11,12,13]. rrnL is present between trnL1 and trnV, while rrnS between trnV and the CR (Figure 1). The two genes rrnL and rrnS are 1214 and 733 bp long in G. henana, and 1236 and 730 bp long in the Chinese G. orientalis, respectively (Table 3). The lengths range from 1214 to 1247 bp for rrnL, and from 719 to 783 bp for rrnS in Gryllotalpidae. The AT content of rRNAs is 67.1% in G. henana and 73.2% in the Chinese G. orientalis. The value of AT content is lower in G. henana than those in the other gryllotalpids and many other orthopterans [12,48,57,58,59,60,61,62,63].
3.4. Intergenic Spacers and Gene Overlaps
In G. henana, intergenic spacers are distributed in 12 regions and range in size from 1 to 71 bp with a total of 162 bp (Table 3). Eleven intergenic spacers exist in the mitogenome of the Chinese G. orientalis, ranging from 1 to 25 bp and adding up to 73 bp. The largest has 71 bp located between rrnL and trnV in G. henana, whereas there are 25 bp located between trnS2 and nad1 in the Chinese G. orientalis. Two identical intergenic spacers were detected in the mitogenomes of all gryllotalpids. One is between nad4L and trnT (2 bp), and the other is between nad1 and trnL1 (1 bp). In most cases, the intergenic spacers consist of only 1 or 2 bp.
The gene overlaps of G. henana are distributed in 13 locations with a total of 52 bp, whereas those of the Chinese G. orientalis are in 10 locations with a total of 34 bp (Table 3). The longest gene overlap is 17 bp between trnL1 and rrnL in G. henana, and 8 bp between trnW and trnC in the Chinese G. orientalis. All six gryllotalpids have five identical overlapping regions, including trnK-trnD (1 bp), trnE-trnF (2 bp), trnI-trnQ (3 bp), nad4-nad4L (7 bp) and trnW-trnC (8 bp). In general, the variability of gene overlaps is lower than that of intergenic spacers.
3.5. Control Region
The CR, also called AT-rich region, is located in the conserved position between rrnS and trnI (Figure 1, Table 2). The AT-content of this region is 81.1% in G. henana and 77.3% in the Chinese G. orientalis. In all six gryllotalpids, the Korean G. orientalis shows the lowest AT content of 74.9%, whereas G. henana exhibits the highest 81.1%. The CRs of Gryllotalpidae show low variations in lengths, which range from 863 bp in G. henana to 920 bp in the Korean G. orientalis. The low variations of CR in length are likely attributed to the lacking of conspicuous repeats, which are often found in other insects [6,54,64,65,66]. Two kinds of short repeats were detected in Gryllotalpidae for the first time (Supplementary Table S5). One is the microsatellite (TA)n element found in G. henana and the Chinese G. orientalis. The other recognized in G. pluvalis is the duplicated tandem repeat, containing 18 bp (ATATAATTAAATATTTAA) with 2.3 copies. A potential stem–loop structure, containing (T)n(TC)2(T)n sequences, was detected in the CR near the trnI gene of G. henana and the Chinese G. orientalis, same as the findings in other gryllotalpids (Figure S1). Similar structures were also found in many crickets of Gryllidea [46,67], and likely related to replication initiation of the N-strand [68].
3.6. Genetic Diversity and Selective Constraints
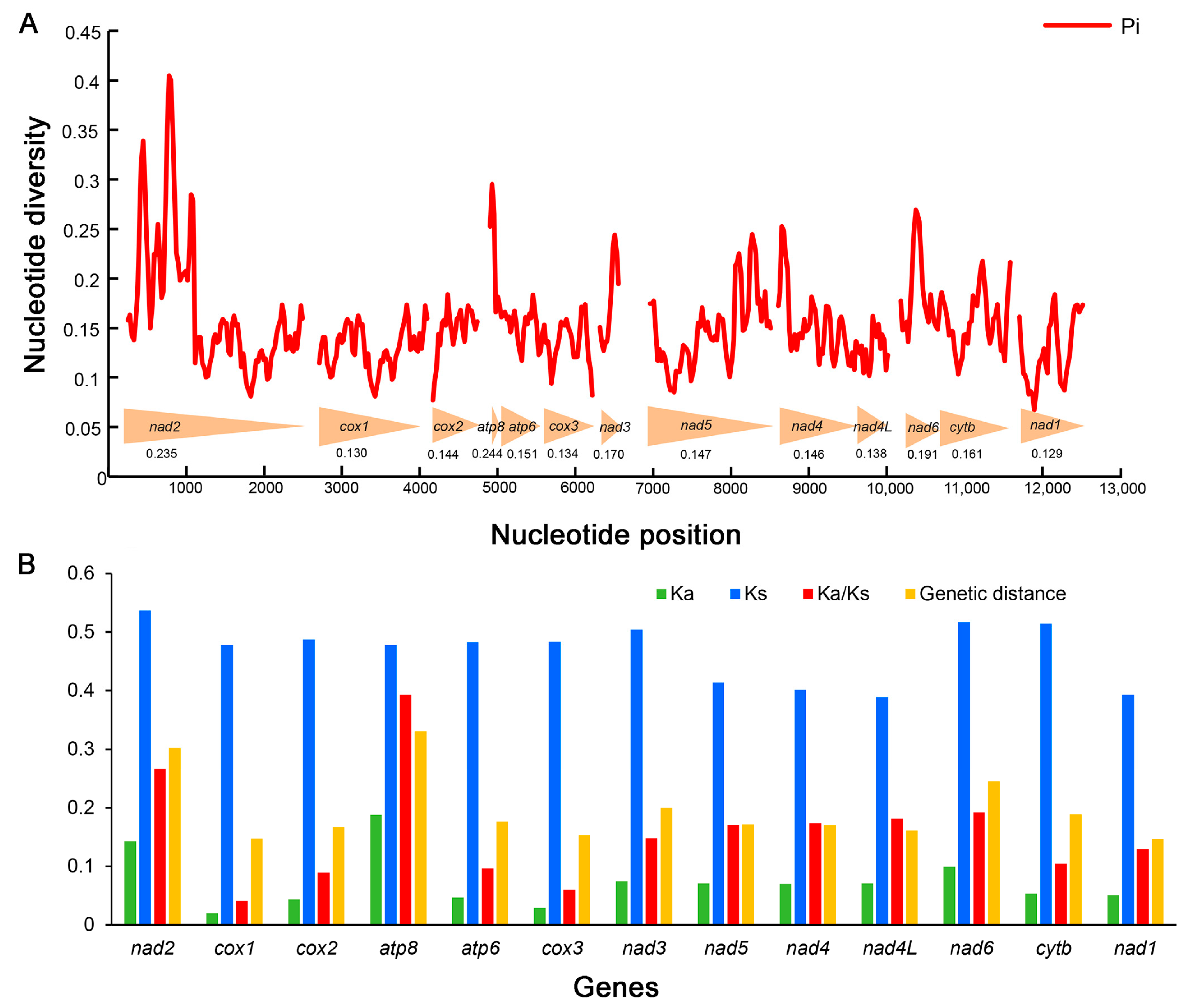
Sliding window analyses exhibit the estimations of nucleotide diversity (Pi) for each PCG of the six mitogenomes (Figure 5A, Supplementary Table S6). The gene atp8 has the highest nucleotide diversity (Pi = 0.244), followed by nad2 (Pi = 0.235) and nad6 (Pi = 0.191). The genes cox3 (Pi = 0.134), cox1 (Pi = 0.130) and nad1 (Pi = 0.129) are the lower variable. A similar pattern was also detected in terms of mean genetic distances (Figure 5B). atp8, nad2 and nad6 show high distances with 0.331, 0.302 and 0.245, whereas cox3, cox1 and nad1 exhibit low distances with 0.154, 0.147 and 0.146, respectively.
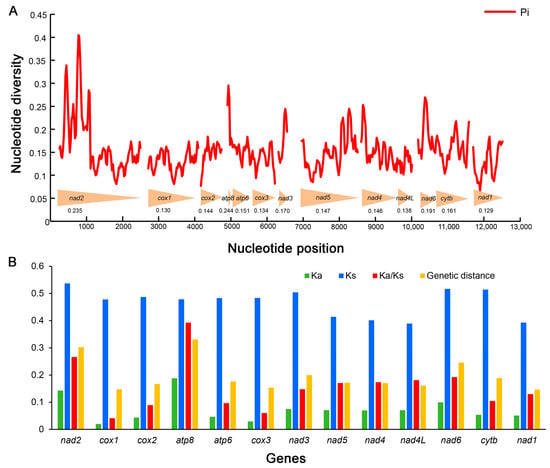
Figure 5.
Genetic diversity and selection pressure among 13 protein-coding genes (PCGs) in Gryllotalpidae. (A) Sliding window analyses with a window of 100 bp and a step size of 25 bp for 13 PCGs. The red curve shows the values of nucleotide diversity (Pi). The arrowheads at the bottom illustrates the position of each PCG. The Pi value of each PCG is shown below the arrowheads. (B) Genetic distance and Ka/Ks ratio of each PCG in Gryllotalpidae. Ka, non-synonymous substitution rates; Ks, synonymous substitution rates.
Ka/Ks ratio (ω) is an important indicator for detecting molecular adaptation correlated to the biological evolution [69,70]. The Ka/Ks ratios of 13 PCGs are all lower than 1 in all mitogenomes of Gryllotalpidae (Figure 5B, Supplementary Table S6), indicating that these PCGs are evolving under purifying selection and suitable for phylogenetic reconstructions in Gryllotalpidae. The Ka/Ks of atp8 (ω = 0.393), nad2 (ω = 0.266) and nad6 (ω = 0.192) are much higher than those of other PCGs, suggesting that the former three genes experience more relaxed evolutionary constraints and retain more non-synonymous mutations. The gene cox1 exhibits the lowest Ka/Ks ratio (ω = 0.041) implying the greatest evolutionary limitation on cox1 among 13 PCG genes. The strong evolutionary constraints (ω << 1) of mitochondrial PCGs suggest that the deleterious mutations are eliminated by purifying selection to maintain highly conserved genes that encode core subunits of the respiratory chain complexes [10,71].
The species of Gryllotalpidae are similar in external morphology but exhibit complicated variations in genitalia, leading to taxonomic difficulties based solely on morphological characters [21,22]. Designing species-specific markers is crucial for resolving such problems. The cox1 gene has long been used as a universal DNA marker for species identification in insects [72,73,74,75], but is the most conservative protein coding gene in mitogenomes of gryllotalpids. Considering both the high nucleotide divergence and the elevated ratio of Ka/Ks, the genes nad2 and nad6 may be evaluated as potential markers for species delimitation in Gryllotalpidae.
3.7. Phylogenetic Analyses
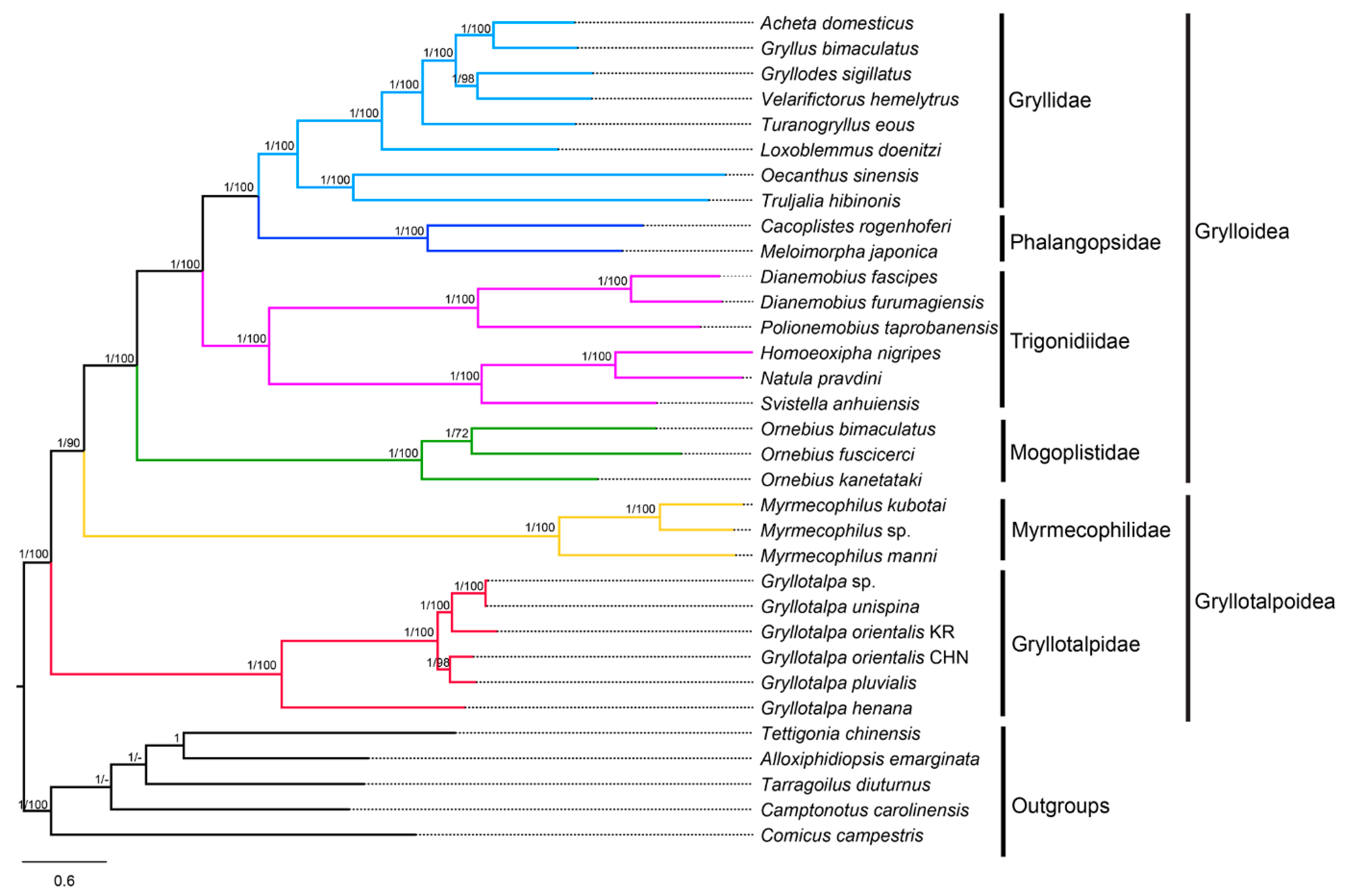
The phylogenetic trees based on the four datasets (P12, P12R, P123 and P123R) are highly consistent, except for the positions of the Korean G. orientalis and Velarifictorus hemelytrus (Saussure, 1877) (Figure 6, Supplementary Figures S2–S4). For the same dataset, the nodal support values in BI trees are generally higher than those in ML trees. For the same inference method (BI or ML), different data combinations slightly affected the topology and support values. The P123 trees are markedly more resolved, and have overall higher supports at nodes than the others. The ingroup topologies between BI and ML trees are identical based on the P123 and P123R datasets, but are inconsistent based on the P12 and P12R datasets, indicating that the inclusion of the third codon positions make topologies more stable in both ML and BI trees. The nodal supports of phylogenetic trees based on P123 dataset are higher than that of P123R dataset. A similar situation was observed between P12 and P12R trees, reflecting that the exclusion of the rRNA genes can improve branch supports of phylogenetic trees. The monophyly of the infraorder Gryllidea was well supported by all datasets with high nodal supports (PPs = 1; BSs = 100), and consistent with the results proposed by Chintauan-Marquier et al. [17], Zhou et al. [48], Chang et al. [10], Song et al. [8] and Sanno et al. [18].
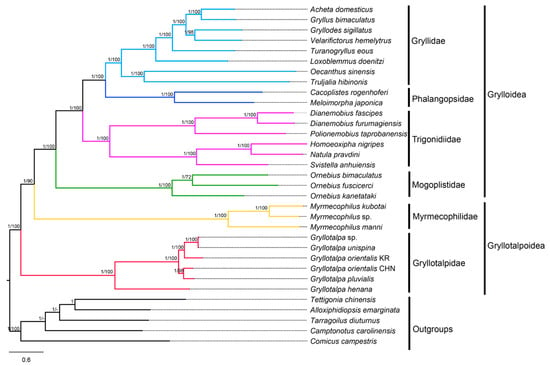
Figure 6.
Phylogenetic tree produced by Bayesian inference (BI) based on the P123 dataset. Numerals at nodes are Bayesian posterior probabilities (PPs) and bootstrap support values (BSs), respectively. “-” indicates that the positions of Tarragoilus diuturnus and Camptonotus carolinensis in the maximum likelihood (ML) tree are slightly different from those in the BI tree.
The monophyletic Grylloidea was confirmed and the relationships within this superfamily were present as Mogoplistidae + (Trigonidiidae + (Phalangopsidae + Gryllidae)). This finding corroborates the generally accepted classification schemes [15] as well as mostly recent studies [8,17,18,39,40,76]. The monophyly of the superfamily Gryllotalpoidea, however, was rejected in the present study. Gryllotalpidae formed the sister taxon to the clade of Myrmecophilidae + Grylloidea rather than solely to Myrmecophilidae. This result is similar to the mitogenome-based trees [8,18], but conflicts with the multilocus-based phylogeny proposed by Chintauan-Marquier et al. [17], which is adopted prevalently as a reference classification. Mitogenomes may experience selective pressures in some insects with peculiar ecological and morphological traits [77,78]. The small and wingless crickets in Myrmecophilidae inhabit subterranean ant nests of low oxygen levels [79,80], whereas mole crickets have larger sizes and short wings, and usually hide in horizontal burrows near the soil surface [81]. The positively selective sites associated with hypoxic adaptability were identified in the cox1 genes of Myrmecophilidae, but were failed to be detected in those of Gryllotalpidae [18], suggesting that the mitogenomes of Myrmecophilidae and Gryllotalpidae have different evolutionary properties. Therefore, we speculated that the contradictions between mitogenomic and multilocus trees are partially attributed to the evolutionary differences of mitogenomes of the two families. In addition, the inconsistent trees may also be influenced by the lack of nuclear genes, which are important for reconstructing deep-level phylogenetic relationships [82,83,84]. The present investigation improved the resolution of the phylogram by Sanno et al. [18], although more species and markers are necessary for future studies.
In Gryllotalpidae, G. henana first split from the remaining gryllotalpids (BSs = 100, PPs = 1) (Figure 6; Supplementary Figures S2–S4). Interestingly, in the second clade, the two specimens of G. orientalis were failed to be clustered in one branch. The Korean G. orientalis was clustered with the clade of Gryllotalpa sp. + G. unispina based on P123 and P123R datasets (Figure 6, Supplementary Figure S2), but was placed with the clade of the Chinese G. orientalis + G. pluvialis based on P12 and P12R datasets (Supplementary Figures S3 and S4). Moreover, the K2P genetic distance of the two specimens of G. orientalis (0.145) is relatively high compared with the interspecific distances of Gryllotalpa (0.022–0.321) (Supplementary Table S7). We speculate that the so-called G. orientalis in China is likely a new species, and further morphological and biological evidences are needed to confirm this inference.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/insects13100919/s1, Table S1: The best partitioning schemes and models for the Bayesian inference (BI) method in different datasets selected by PartitionFinder. Table S2: The best partitioning schemes and models for the Maximum likelihood (ML) method in different datasets selected by ModelFinder. Table S3: The count and relative synonymous codon usage (RSCU) of six species in Gryllotalpidae. Table S4: The numbers of mismatched base pairs in the secondary structure of the tRNAs in the six species of Gryllotalpidae. Table S5: Tandem repeat regions in the control region of six species in Gryllotalpidae. Table S6: Nucleotide diversity (Pi), non-synonymous substitutions rates (Ka), synonymous substitutions rates (Ks), Ka/Ks ratio and genetic distance of six species in Gryllotalpidae. Table S7: The K2P genetic distances in Gryllotalpidae. Figure S1: Location and structure of the potential stem-loops in Gryllotalpidae. (A) The location of the predicted stem-loops in the mitogenome. (B) The structures of potential stem-loops. Figure S2: Phylogenetic tree produced by Bayesian inference (BI) based on the dataset of P123R. Bayesian posterior probabilities (PPs) and bootstrap support values (BSs) are present at nodes. Figure S3: Phylogenetic tree produced by Bayesian inference (BI) based on the P12 dataset. Numerals at nodes are Bayesian posterior probabilities (PPs) and bootstrap support values (BSs). The divergent dotted lines showed the topological tree based on Maximum likelihood (ML) method with underlined bootstrap support values at nodes. Figure S4: Phylogenetic tree produced by Bayesian inference (BI) based on the P12R dataset. Numerals at nodes are Bayesian posterior probabilities (PPs) and bootstrap support values (BSs). The divergent dotted lines showed the topological tree based on Maximum likelihood (ML) method with underlined bootstrap support values.
Author Contributions
Conceptualization, funding acquisition, methodology, project administration, resources, supervision, writing—review and editing, Y.M. (Ying Miao); data curation, formal analysis, investigation, validation, visualization, writing—original draft, Y.M. (Yan Ma). All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Natural Science Foundation of Ningxia Province (grant no. 2022AAC03085).
Data Availability Statement
The data supporting the findings of this study are openly available in National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov, accessed on 12 April 2022), accession numbers are ON243749 and ON210982.
Acknowledgments
We are grateful to Xin-Pu Wang, Lu Jiang, Chao Gao, Yu-Chen Zhao and Shuang Xue for assistance with specimen collection, and to Ning Li, Le-Le He and Yi-Xin Huang for assistance in software operation. We also thank the anonymous reviewers for helpful suggestions that improved this manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Cameron, S.L. Insect mitochondrial genomics: Implications for evolution and phylogeny. Annu. Rev. Entomol. 2014, 59, 95–117. [Google Scholar] [CrossRef]
- Drosopoulou, E.; Syllas, A.; Goutakoli, P.; Zisiadis, G.A.; Konstantinou, T.; Pangea, D.; Sentis, G.; van Sauers-Muller, A.; Wee, S.L.; Augustinos, A.A.; et al. Τhe complete mitochondrial genome of Bactrocera carambolae (Diptera: Tephritidae): Genome description and phylogenetic implications. Insects 2019, 10, 429. [Google Scholar] [CrossRef]
- Du, H.C.; Liu, M.; Zhang, S.F.; Liu, F.; Zhang, Z.; Kong, X.B. Lineage divergence of Dendrolimus punctatus in southern China based on mitochondrial genome. Front. Genet. 2020, 11, 65. [Google Scholar] [CrossRef]
- Liu, Y.Q.; Li, H.; Song, F.; Zhao, Y.S.; Wilson, J.J.; Cai, W.Z. Higher-level phylogeny and evolutionary history of Pentatomomorpha (Hemiptera: Heteroptera) inferred from mitochondrial genome sequences. Syst. Entomol. 2019, 44, 810–819. [Google Scholar] [CrossRef]
- Zheng, B.Y.; Han, Y.Y.; Yuan, R.Z.; Liu, J.X.; van Achterberg, C.; Tang, P.; Chen, X.X. Comparative mitochondrial genomics of 104 Darwin wasps (Hymenoptera: Ichneumonidae) and its implication for phylogeny. Insects 2022, 13, 124. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Hu, G.L.; Hua, B.Z. Complete mitochondrial genomes of Bittacus strigosus and Panorpa debilis and genomic comparisons of Mecoptera. Int. J. Biol. Macromol. 2019, 140, 672–681. [Google Scholar] [CrossRef]
- Nie, R.; Vogler, A.P.; Yang, X.K.; Lin, M. Higher-level phylogeny of longhorn beetles (Coleoptera: Chrysomeloidea) inferred from mitochondrial genomes. Syst. Entomol. 2021, 46, 56–70. [Google Scholar] [CrossRef]
- Song, H.; Bethoux, O.; Shin, S.; Donath, A.; Letsch, H.; Liu, S.; McKenna, D.D.; Meng, G.; Misof, B.; Podsiadlowski, L.; et al. Phylogenomic analysis sheds light on the evolutionary pathways towards acoustic communication in Orthoptera. Nat. Commun. 2020, 11, 4939. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.J.; Zhu, R.Q.; Xue, H.J.; Li, Y.F.; Bu, W.J. Mitogenomics of chinch bugs from China and implications for its coevolutionary relationship with grasses. Insects 2022, 13, 643. [Google Scholar] [CrossRef]
- Chang, H.H.; Qiu, Z.Y.; Yuan, H.; Wang, X.Y.; Li, X.J.; Sun, H.M.; Guo, X.Q.; Lu, Y.C.; Feng, X.L.; Majid, M.; et al. Evolutionary rates of and selective constraints on the mitochondrial genomes of Orthoptera insects with different wing types. Mol. Phylogenet. Evol. 2020, 145, 106734. [Google Scholar] [CrossRef]
- Fenn, J.D.; Song, H.; Cameron, S.L.; Whiting, M.F. A preliminary mitochondrial genome phylogeny of Orthoptera (Insecta) and approaches to maximizing phylogenetic signal found within mitochondrial genome data. Mol. Phylogenet. Evol. 2008, 49, 59–68. [Google Scholar] [CrossRef]
- Kim, I.; Cha, S.Y.; Yoon, M.H.; Hwang, J.S.; Lee, S.M.; Sohn, H.D.; Jin, B.R. The complete nucleotide sequence and gene organization of the mitochondrial genome of the oriental mole cricket, Gryllotalpa orientalis (Orthoptera: Gryllotalpidae). Gene 2005, 353, 155–168. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.L.; Shao, D.D.; Cai, M.; Yin, H.; Zhang, D.C. The complete mitochondrial genome of Gryllotalpa unispina Saussure, 1874 (Orthoptera: Gryllotalpoidea: Gryllotalpidae). Mitochondrial DNA A 2014, 27, 159–160. [Google Scholar] [CrossRef]
- Cadena-Castañeda, O.J. The phylogeny of mole crickets (Orthoptera: Gryllotalpoidea: Gryllotalpidae). Zootaxa 2015, 3985, 451–490. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cigliano, M.M.; Braun, H.; Eades, D.C.; Otte, D. Orthoptera Species File. Version 5.0/5.0. Available online: http://Orthoptera.SpeciesFile.org (accessed on 24 August 2022).
- Cadena-Castañeda, O.J. Two new species of mole crickets (Orthoptera: Gryllotalpidae: Scapteriscinae) from the Colombian Amazon and Orinoquia rainforests. Zootaxa 2011, 3126, 62–68. [Google Scholar] [CrossRef]
- Chintauan-Marquier, I.C.; Legendre, F.; Hugel, S.; Robillard, T.; Grandcolas, P.; Nel, A.; Zuccon, D.; Desutter-Grandcolas, L. Laying the foundations of evolutionary and systematic studies in crickets (Insecta, Orthoptera): A multilocus phylogenetic analysis. Cladistics 2016, 32, 54–81. [Google Scholar] [CrossRef] [PubMed]
- Sanno, R.; Kataoka, K.; Hayakawa, S.; Ide, K.; Nguyen, C.N.; Nguyen, T.P.; Le, B.T.N.; Kim, O.T.P.; Mineta, K.; Takeyama, H.; et al. Comparative analysis of mitochondrial genomes in Gryllidea (Insecta: Orthoptera): Implications for adaptive evolution in ant-loving crickets. Genome Biol. Evol. 2021, 13, evab222. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.K. Annotated checklist and key to species of Gryllotalpa (Orthoptera: Gryllotalpidae) from the Oriental region. Zootaxa 2016, 4132, 77–86. [Google Scholar] [CrossRef] [PubMed]
- He, Z.Q. A checklist of Chinese crickets (Orthoptera: Gryllidea). Zootaxa 2018, 4369, 515–535. [Google Scholar] [CrossRef]
- Kang, L. The species Gryllotalpa africana Palisot from China should be a synonym of Gryllotalpa orientalis Burmeister. Entomol. Knowl. 1993, 30, 124–127. [Google Scholar]
- Townsend, B.C. A revision of the Afrotropical mole-crickets (Orthoptera: Gryllotalpidae). Bull. Br. Mus. Nat. Hist. 1983, 46, 175–203. [Google Scholar]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Bernt, M.; Donath, A.; Juhling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Putz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved De Novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef] [PubMed]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef] [PubMed]
- Lohse, M.; Drechsel, O.; Kahlau, S.; Bock, R. OrganellarGenomeDraw–a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 2013, 41, W575–W581. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Xu, H.Y.; Gu, J.X.; Mao, B.Y.; Chen, Z.L.; Huang, Y.; Huang, J.H. Phylogenetic position of the genera Caryandoides, Paratoacris, Fer and Longchuanacris (Orthoptera:Acrididae) revealed by complete mitogenome sequences. Invertebr. Syst. 2021, 35, 725–741. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sanchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sanchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Zhu, J.C.; Tang, P.; Zheng, B.Y.; Wu, Q.; Wei, S.J.; Chen, X.X. The first two mitochondrial genomes of the family Aphelinidae with novel gene orders and phylogenetic implications. Int. J. Biol. Macromol. 2018, 118, 386–396. [Google Scholar] [CrossRef] [PubMed]
- Perna, N.T.; Kocher, T.D. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 1995, 41, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gao, F.L.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Leavitt, J.R.; Hiatt, K.D.; Whiting, M.F.; Song, H. Searching for the optimal data partitioning strategy in mitochondrial phylogenomics: A phylogeny of Acridoidea (Insecta: Orthoptera: Caelifera) as a case study. Mol. Phylogenet. Evol. 2013, 67, 494–508. [Google Scholar] [CrossRef]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2016, 34, 772–773. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Ma, C.; Li, J.K. Comparative analysis of mitochondrial genomes of the superfamily Grylloidea (Insecta, Orthoptera) reveals phylogenetic distribution of gene rearrangements. Int. J. Biol. Macromol. 2018, 120, 1048–1054. [Google Scholar] [CrossRef]
- Ma, C.; Wang, Y.Y.; Zhang, L.C.; Li, J.K. Mitochondrial genome characterization of the family Trigonidiidae (Orthoptera) reveals novel structural features and nad1 transcript ends. Sci. Rep. 2019, 9, 19092. [Google Scholar] [CrossRef]
- Yang, J.K.; Dong, H.L.; He, M.Q.; Gao, J.G. Mitochondrial Genome Characterization of Gryllodes sigillatus (Orthoptera: Gryllidae) and its phylogenetic implications. Mitochondrial DNA B 2021, 6, 1056–1058. [Google Scholar] [CrossRef]
- Park, B.; Choi, E.H.; Kim, G.; Shin, C.R.; Hwang, J.; Baek, S.Y.; Hwang, U.W. The complete mitochondrial genome of the two-spotted cricket Gryllus bimaculatus (Orthoptera: Gryllidae) from South Korea. Mitochondrial DNA B 2021, 6, 1144–1146. [Google Scholar] [CrossRef]
- Song, N.; Li, H.; Song, F.; Cai, W.Z. Molecular phylogeny of Polyneoptera (Insecta) inferred from expanded mitogenomic data. Sci. Rep. 2016, 6, 36175. [Google Scholar] [CrossRef] [PubMed]
- Li, J.J.; Chen, Q.; Wen, M.; Wang, J.T.; Wang, Y.L.; Ren, B.Z. Phylogeny and acoustic signal evolution of a pure tone song katydid Pseudophyllus titan (Orthoptera: Tettigoniidae) based on the complete mitogenome. Mitochondrial DNA A 2019, 30, 385–396. [Google Scholar] [CrossRef]
- Ma, C.; Zhang, L.C.; Li, J.K. The complete mitochondrial genome of a field cricket Turanogryllus eous (Insecta: Orthoptera). Mitochondrial DNA B 2019, 4, 3852–3853. [Google Scholar] [CrossRef]
- Yang, J.; Ren, Q.L.; Zhang, Q.; Huang, Y. Complete mitochondrial genomes of three crickets (Orthoptera: Gryllidae) and comparative analyses within Ensifera mitogenomes. Zootaxa 2016, 4092, 529–547. [Google Scholar] [CrossRef] [PubMed]
- Mao, S.L.; Lu, Y.; Xun, L.L.; Zhou, Y.F. Characterization of the mitochondrial genome of Alloxiphidiopsis emarginata (Orthoptera, Tettigoniidae, Meconematinae). Mitochondrial DNA B 2019, 4, 4192–4193. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.J.; Zhao, L.; Liu, N.; Guo, H.F.; Guan, B.; Di, J.X.; Shi, F. Towards a higher-level Ensifera phylogeny inferred from mitogenome sequences. Mol. Phylogenet. Evol. 2017, 108, 22–33. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Amédégnato, C.; Cigliano, M.M.; Desutter-Grandcolas, L.; Heads, S.W.; Huang, Y.; Otte, D.; Whiting, M.F. 300 million years of diversification: Elucidating the patterns of orthopteran evolution based on comprehensive taxon and gene sampling. Cladistics 2015, 31, 621–651. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.J.; Shi, F.M.; Zhao, L. The first mitochondrial genome for the superfamily Hagloidea and implications for its systematic status in Ensifera. PLoS ONE 2014, 9, e86027. [Google Scholar] [CrossRef]
- Zhou, Z.; Huang, Y.; Shi, F. The mitochondrial genome of Ruspolia dubia (Orthoptera: Conocephalidae) contains a short A+T-rich region of 70 bp in length. Genome 2007, 50, 855–866. [Google Scholar] [CrossRef]
- Bernt, M.; Braband, A.; Schierwater, B.; Stadler, P.F. Genetic aspects of mitochondrial genome evolution. Mol. Phylogenet. Evol. 2013, 69, 328–338. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.M.; Zhang, L.H.; Lin, Y.J.; Zheng, Y.M.; Jin, W.T.; Storey, K.B.; Yu, D.N.; Zhang, J.Y. The genetic diversity in Thereuonema tuberculata (Wood, 1862) (Scutigeromorpha: Scutigeridae) and the phylogenetic relationship of Scutigeromorpha using the mitochondrial genome. Insects 2022, 13, 620. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Ye, F.; Huang, Y. Mitochondrial genomes of four katydids (Orthoptera: Phaneropteridae): New gene rearrangements and their phylogenetic implications. Gene 2016, 575, 702–711. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Wang, Y.Q.; Shu, X.H.; Meng, L.; Li, B.P. Complete mitochondrial genomes of three Oxya grasshoppers (Orthoptera) and their implications for phylogenetic reconstruction. Genomics 2020, 112, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Wolstenholme, D.R. Animal mitochondrial DNA: Structure and evolution. Int. Rev. Cytol. 1992, 141, 173–216. [Google Scholar] [CrossRef]
- Li, R.; Shu, X.H.; Li, X.D.; Meng, L.; Li, B.P. Comparative mitogenome analysis of three species and monophyletic inference of Catantopinae (Orthoptera: Acridoidea). Genomics 2019, 111, 1728–1735. [Google Scholar] [CrossRef]
- Li, R.; Ying, X.; Deng, W.; Rong, W.; Li, X. Mitochondrial genomes of eight Scelimeninae species (Orthoptera) and their phylogenetic implications within Tetrigoidea. PeerJ 2021, 9, e10523. [Google Scholar] [CrossRef]
- Qiu, Z.; Chang, H.H.; Yuan, H.; Huang, Y.; Lu, H.M.; Li, X.; Gou, X.C. Comparative mitochondrial genomes of four species of Sinopodisma and phylogenetic implications (Orthoptera, Melanoplinae). Zookeys 2020, 969, 23–42. [Google Scholar] [CrossRef]
- Yin, H.; Zhi, Y.C.; Jiang, H.D.; Wang, P.X.; Yin, X.C.; Zhang, D.C. The complete mitochondrial genome of Gomphocerus tibetanus Uvarov, 1935 (Orthoptera: Acrididae: Gomphocerinae). Gene 2012, 494, 214–218. [Google Scholar] [CrossRef]
- Yuan, H.; Huang, Y.; Mao, Y.; Zhang, N.; Nie, Y.M.; Zhang, X.; Zhou, Y.F.; Mao, S.L. The evolutionary patterns of genome size in Ensifera (Insecta: Orthoptera). Front. Genet. 2021, 12, 693541. [Google Scholar] [CrossRef]
- Zhang, D.C.; Zhi, Y.C.; Yin, H.; Li, X.J.; Yin, X.C. The complete mitochondrial genome of Thrinchus schrenkii (Orthoptera: Caelifera, Acridoidea, Pamphagidae). Mol. Biol. Rep. 2011, 38, 611–619. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.J.; Shi, F.M.; Huang, Y. The complete mitogenome of the Chinese bush cricket, Gampsocleis gratiosa (Orthoptera: Tettigonioidea). J. Genet. Genom. 2008, 35, 341–348. [Google Scholar] [CrossRef]
- Voronova, N.V.; Levykina, S.; Warner, D.; Shulinski, R.; Bandarenka, Y.; Zhorov, D. Characteristic and variability of five complete aphid mitochondrial genomes: Aphis fabae mordvilkoi, Aphis craccivora, Myzus persicae, Therioaphis tenera and Appendiseta robiniae (Hemiptera; Sternorrhyncha; Aphididae). Int. J. Biol. Macromol. 2020, 149, 187–206. [Google Scholar] [CrossRef]
- Wang, W.Q.; Huang, Y.X.; Bartlett, C.R.; Zhou, F.M.; Meng, R.; Qin, D.Z. Characterization of the complete mitochondrial genomes of two species of the genus Aphaena Guérin-Méneville (Hemiptera: Fulgoridae) and its phylogenetic implications. Int. J. Biol. Macromol. 2019, 141, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Gao, Z.M.; Yin, J.; Zhang, T.T.; Zhang, X.Y.; Yuan, D.W.; Li, T.; Zhong, Y.; Ma, B.E.; Ren, Z.M. Complete mitochondrial genome of two Thitarodes species (Lepidoptera, Hepialidae), the host moths of Ophiocordyceps sinensis and phylogenetic implications. Int. J. Biol. Macromol. 2019, 140, 794–807. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Huang, Y. Complete mitochondrial genome sequence of Acrida cinerea (Acrididae: Orthoptera) and comparative analysis of mitochondrial genomes in Orthoptera. Comp. Funct. Genom. 2010, 2010, 319486. [Google Scholar] [CrossRef] [PubMed]
- Saito, S.; Tamura, K.; Aotsuka, T. Replication origin of mitochondrial DNA in insects. Genetics 2005, 171, 1695–1705. [Google Scholar] [CrossRef] [PubMed]
- Hurst, L.D. The Ka/Ks ratio: Diagnosing the form of sequence evolution. Trends Genet. 2002, 18, 486–487. [Google Scholar] [CrossRef]
- Yang, Z.H.; Bielawski, J.P. Statistical methods for detecting molecular adaptation. Trends Ecol. Evol. 2000, 15, 496–503. [Google Scholar] [CrossRef]
- Popadin, K.Y.; Nikolaev, S.I.; Junier, T.; Baranova, M.; Antonarakis, S.E. Purifying selection in mammalian mitochondrial protein-coding genes is highly effective and congruent with evolution of nuclear genes. Mol. Biol. Evol. 2012, 30, 347–355. [Google Scholar] [CrossRef]
- Hebert, P.D.; Cywinska, A.; Ball, S.L.; deWaard, J.R. Biological identifications through DNA barcodes. Proc. R. Soc. Lond. B 2003, 270, 313–321. [Google Scholar] [CrossRef]
- Hu, G.L.; Gao, K.; Wang, J.S.; Hebert, P.D.; Hua, B.Z. Molecular phylogeny and species delimitation of the genus Dicerapanorpa (Mecoptera: Panorpidae). Zool. J. Linn. Soc. 2019, 187, 1173–1195. [Google Scholar] [CrossRef]
- Ricarte, A.; Nedeljkovic, Z.; Aguado-Aranda, P.; Marcos-Garcia, M.A. Assessing the diversity and systematics of Brachyopini hoverflies (Diptera: Syrphidae) in the Iberian Peninsula, including the descriptions of two new species†. Insects 2022, 13, 648. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.D.; Wang, H.J.; Zhou, Z.J.; Shi, F.M. Phylogeny and integrative taxonomy of the genera Gymnaetoides and Pseudotachycines (Orthoptera: Rhaphidophoridae). Insects 2022, 13, 628. [Google Scholar] [CrossRef]
- Yu, Z.Y.; Xie, H.C.; Liu, Y.J.; Li, K.; He, Z.Q. The complete mitochondrial genome of cricket Sclerogryllus punctatus (Orthoptera: Gryllidae) and phylogenetic analysis. J. Asia-Pac. Entomol. 2022, 25, 101933. [Google Scholar] [CrossRef]
- Li, X.D.; Jiang, G.F.; Yan, L.Y.; Li, R.; Mu, Y.; Deng, W.A. Positive selection drove the adaptation of mitochondrial genes to the demands of flight and high-altitude environments in grasshoppers. Front. Genet. 2018, 9, 605. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.L.; Zhang, L.; Zhao, T.X.; Wang, J.; Zhu, Q.H.; Chen, J.Y.; Yuan, M.L. Gene sequence variations and expression patterns of mitochondrial genes are associated with the adaptive evolution of two Gynaephora species (Lepidoptera: Lymantriinae) living in different high-elevation environments. Gene 2017, 610, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Bollazzi, M.; Forti, L.C.; Roces, F. Ventilation of the giant nests of Atta leaf-cutting ants: Does underground circulating air enter the fungus chambers? Insect. Soc. 2012, 59, 487–498. [Google Scholar] [CrossRef]
- Hsu, P.W.; Hugel, S.; Wetterer, J.K.; Tseng, S.P.; Ooi, C.S.M.; Lee, C.Y.; Yang, C.C.S. Ant crickets (Orthoptera: Myrmecophilidae) associated with the invasive yellow crazy ant Anoplolepis gracilipes (Hymenoptera: Formicidae): Evidence for cryptic species and potential co-introduction with hosts. Myrmecol. News 2020, 30, 103–129. [Google Scholar] [CrossRef]
- Li, T.C.; Shao, M.A.; Jia, Y.H.; Jia, X.X.; Huang, L.M. Small-scale observation on the effects of the burrowing activities of mole crickets on soil erosion and hydrologic processes. Agr. Ecosyst. Environ. 2018, 261, 136–143. [Google Scholar] [CrossRef]
- Misof, B.; Liu, S.; Meusemann, K.; Peters, R.S.; Donath, A.; Mayer, C.; Frandsen, P.B.; Ware, J.; Flouri, T.; Beutel, R.G.; et al. Phylogenomics resolves the timing and pattern of insect evolution. Science 2014, 346, 763–767. [Google Scholar] [CrossRef] [PubMed]
- Springer, M.S.; DeBry, R.W.; Douady, C.; Amrine, H.M.; Madsen, O.; de Jong, W.W.; Stanhope, M.J. Mitochondrial versus nuclear gene sequences in deep-level mammalian phylogeny reconstruction. Mol. Biol. Evol. 2001, 18, 132–143. [Google Scholar] [CrossRef] [PubMed]
- Hawlitschek, O.; Ortiz, E.M.; Noori, S.; Webster, K.C.; Husemann, M.; Pereira, R.J. Transcriptomic data reveals nuclear-mitochondrial discordance in Gomphocerinae grasshoppers (Insecta: Orthoptera: Acrididae). Mol. Phylogenet. Evol. 2022, 170, 107439. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).