First Detection of Honeybee Pathogenic Viruses in Butterflies
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Laboratory Investigations
2.3. Data Analysis
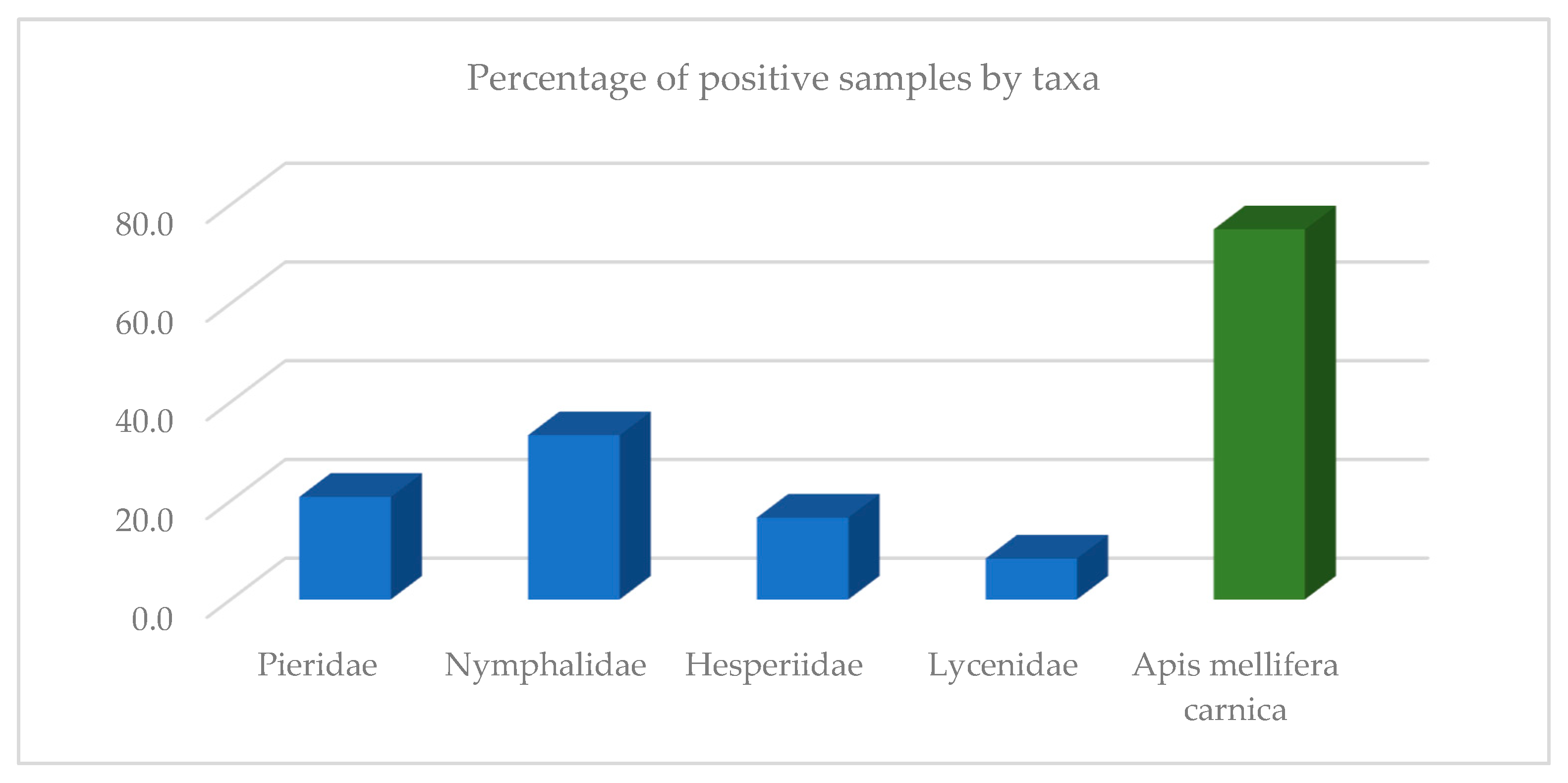
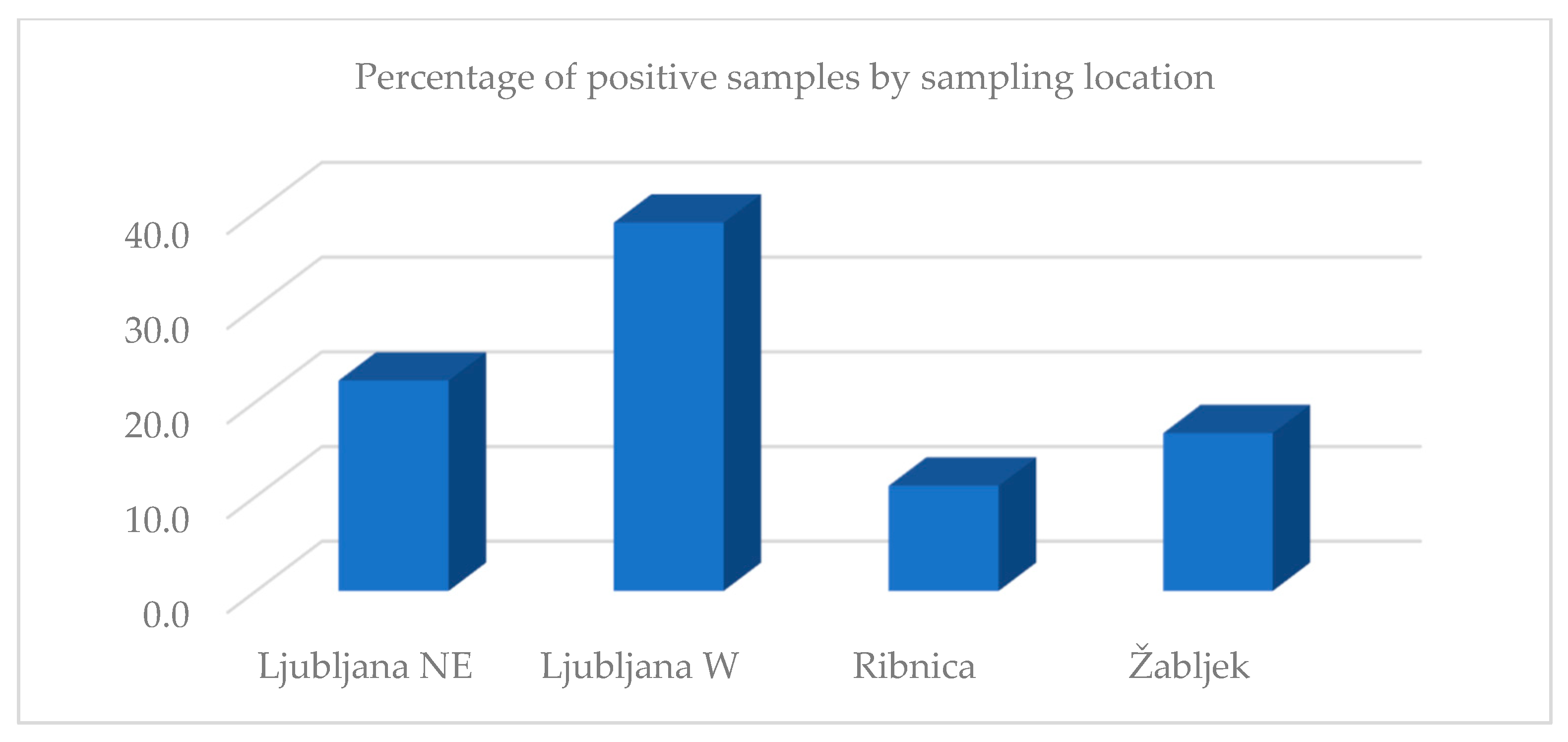
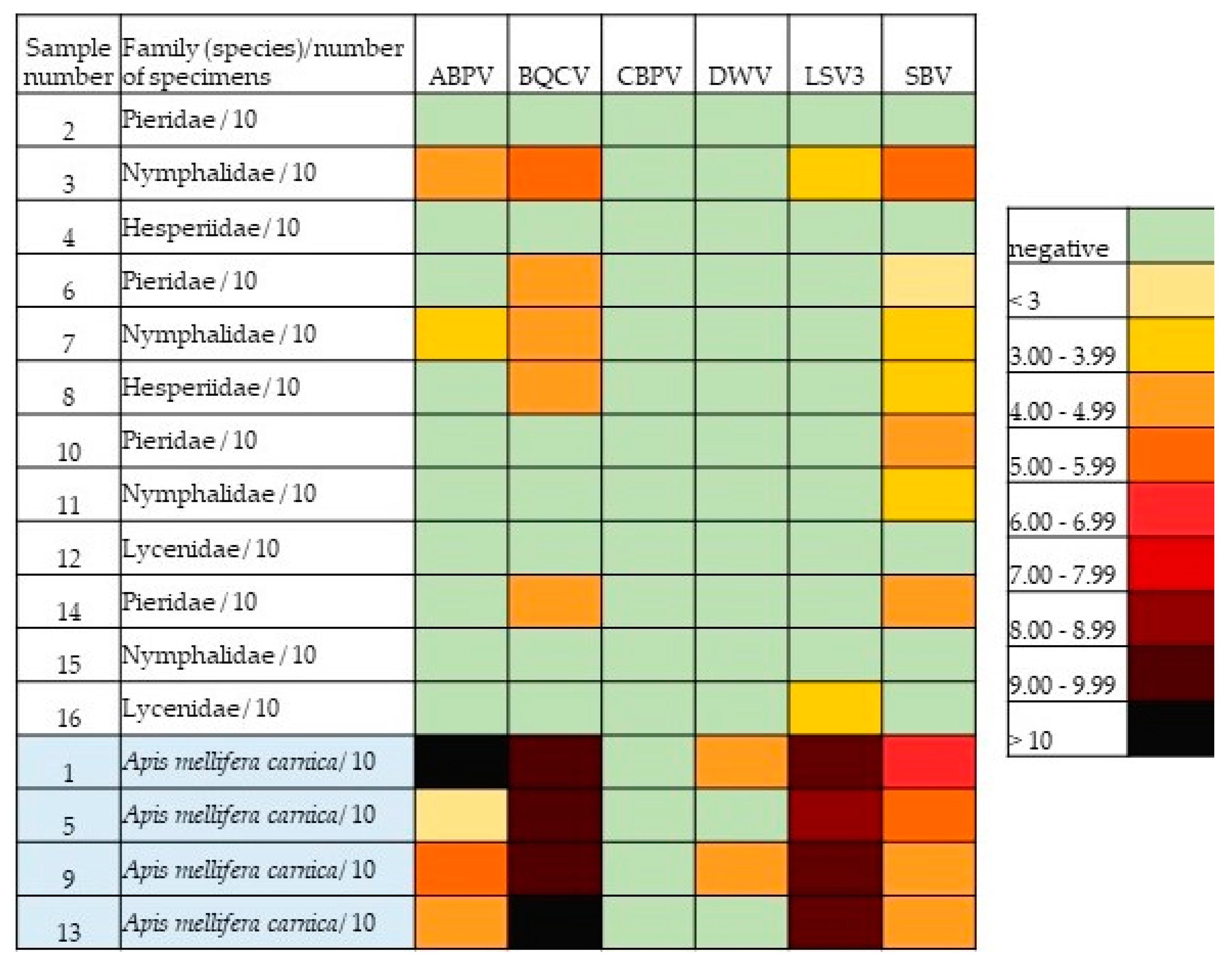
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Potts, S.; Biesmeijer, K.; Bommarco, R.; Breeze, T.; Carvalheiro, L.; Franzen, M.; Gonzalez-Varo, J.P.; Holzschuh, A.; Kleijn, D.; Klein, A.M.; et al. Status and Trends of European Pollinators; Key Findings of the STEP Project; Pensoft Publishers: Sofia, Bulgaria, 2015; 72p. [Google Scholar]
- Schowalter, T.D.; Noriega, J.A.; Tscharntke, T. Insect effects on ecosystem services—Introduction. Basic Appl. Ecol. 2018, 26, 1–7. [Google Scholar] [CrossRef]
- Garibaldi, L.A.; Steffan-Dewenter, I.; Winfree, R.; Aizen, M.A.; Bommarco, R.; Cunningham, S.A.; Kremen, C.; Carvalheiro, L.G.; Harder, L.D.; Afik, O.; et al. Wild Pollinators Enhance Fruit Set of Crops Regardless of Honey Bee Abundance. Science 2013, 339, 1608–1611. [Google Scholar] [CrossRef] [PubMed]
- Ollerton, J. Pollinator diversity: Distribution, ecological function, and conservation. Annu. Rev. Ecol. Evol. Syst. 2017, 48, 353–376. [Google Scholar] [CrossRef] [Green Version]
- Warren, M.S.; Maes, D.; van Swaay, C.A.M.; Goffart, P.; van Dyck, H.; Bourn, N.A.D.; Wynhoff, I.; Hoare, D.; Ellis, S. The decline of butterflies in Europe: Problems, significance, and possible solutions. Proc. Natl. Acad. Sci. USA 2021, 118, e2002551117. [Google Scholar] [CrossRef]
- Thomas, J.A. Monitoring change in the abundance and distribution of insects using butterflies and other indicator groups. Phil. Trans. R. Soc. 2005, 360, 339–357. [Google Scholar] [CrossRef]
- Butterfly Conservation. Available online: https://butterfly-conservation.org/ (accessed on 20 April 2022).
- Wardhaugh, C.W. How many species of arthropods visit flowers? Arthropod-Plant Interact. 2015, 9, 547–565. [Google Scholar] [CrossRef]
- Mertens, J.E.J.; Brisson, L.; Janeček, Š.; Klomberg, Y.; Maicher, V.; Safian, S.; Delabye, S.; Potocky, P.; Kobe, I.N.; Pyrcz, T.; et al. Elevational and seasonal patterns of butterflies and hawkmoths in plant-pollinator networks in tropical rainforests of Mount Cameroon. Sci. Rep. 2021, 11, 9710. [Google Scholar] [CrossRef]
- Banza, P.; Belo, A.D.F.; Evans, D.M. The structure and robustness of nocturnal Lepidopteran pollen-transfer networks in a Biodiversity Hotspot. Insect Conserv. Divers. 2015, 8, 538–546. [Google Scholar] [CrossRef] [Green Version]
- Hahn, M.; Brühl, C.A. The secret pollinators: An overview of moth pollination with a focus on Europe and North America. Arthropod-Plant Interact. 2016, 10, 21–28. [Google Scholar] [CrossRef]
- Macgregor, C.J.; Pocock, M.J.O.; Fox, R.; Evans, D.M. Pollination by nocturnal Lepidoptera, and the effects of light pollution: A review. Ecol. Entomol. 2015, 40, 187–198. [Google Scholar] [CrossRef]
- Rader, R.; Bartomeus, I.; Garibaldi, L.A.; Garratt, M.P.D.; Howlett, B.G.; Winfree, R.; Cunningham, S.A.; Mayfield, M.M.; Arthur, A.D.; Andersson, G.K.S.; et al. Non-bee insects are important contributors to global crop pollination. Proc. Natl. Acad. Sci. USA 2016, 113, 146–151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maes, D.; Verovnik, R.; Wiemers, M.; Brosens, D.; Beshkov, S.; Bonelli, S.; Buszko, J.; Cantu-Salazar, L.; Cassar, L.F.; Collins, S.; et al. Integrating national Red Lists for prioritising conservation actions for European butterflies. J. Insect. Conserv. 2019, 23, 301–330. [Google Scholar] [CrossRef]
- van Swaay, C.; Collins, S.; Dušej, G.; Maes, D.; Munguira, M.; Rakosy, L.; Ryrholm, N.; Šašić, M.; Settele, J.; Thomas, J.; et al. Dos and Don’ts for butterflies of the Habitats Directive of the European Union. Nat. Conserv. 2012, 1, 73–153. [Google Scholar] [CrossRef] [Green Version]
- Wallis de Vries, M.F.; van Swaay, C.A.M.; Plate, C.L. Changes in nectar supply: A possible cause of widespread butterfly decline. Curr. Zool. 2012, 58, 384–391. [Google Scholar] [CrossRef] [Green Version]
- Potts, S.G.; Biesmeijer, J.C.; Kremen, C.; Neumann, P.; Schweiger, O.; Kunin, W.E. Global pollinator declines: Trends, impacts and drivers. Trends Ecol. Evol. 2010, 25, 345–353. [Google Scholar] [CrossRef]
- Gilburn, A.S.; Bunnefeld, N.; McVean Wilson, J.; Botham, M.S.; Brereton, T.M.; Fox, R.; Goulson, D. Are neonicotinoid insecticides driving declines of widespread butterflies? PeerJ 2015, 3, e1402. [Google Scholar] [CrossRef] [Green Version]
- Kurze, S.; Heinken, T.; Fartmann, T. Nitrogen enrichment in host plants increases the mortality of common Lepidoptera species. Oecologia 2018, 188, 1227–1237. [Google Scholar] [CrossRef]
- van Dyck, H.; van Strien, A.J.; Maes, D.; van Swaay, C.A.M. Declines in common, widespread butterflies in a landscape under intense human use. Conserv. Biol. J. 2009, 23, 957–965. [Google Scholar] [CrossRef]
- Parmesan, C.; Ryrholm, N.; Stefanescu, C.; Hill, J.K.; Thomas, C.D.; Descimon, H.; Huntley, B.; Kaila, L.; Kullberg, J.; Tammaru, T.; et al. Poleward shifts in geographical ranges of butterfly species associated with regional warming. Nature 1999, 399, 579–583. [Google Scholar] [CrossRef]
- Devictor, V.; van Swaay, C.; Brereton, T.; Brotons, L.; Chamberlain, D.; Heliölä, J.; Herrando, S.; Julliard, R.; Kuussaari, M.; Lindström, A.; et al. Differences in the climatic debts of birds and butterflies at a continental scale. Nat. Clim. Chang. 2012, 2, 121–124. [Google Scholar] [CrossRef] [Green Version]
- Singer, M.C.; Parmesan, C. Phenological asynchrony between herbivorous insects and their hosts: Signal of climate change or pre-existing adaptive strategy? Phil. Trans. R. Soc. 2010, 365, 3161–3176. [Google Scholar] [CrossRef] [PubMed]
- Hajek, A.E.; McManus, M.L.; Delalibera, I., Jr. A review of introductions of pathogens and nematodes for classical biological control of insects and mites. Biol. Control 2007, 41, 1–13. [Google Scholar] [CrossRef]
- Altizer, S.; de Roode, J. When Butterflies get Bugs: The ABCs of Lepidopteran Disease. Am. Butterflies 2010, 18, 16–26. [Google Scholar]
- Altizer, S.M.; Oberhauser, K.S. Effects of the protozoan parasite ophryocystis elektroscirrha on the fitness of monarch butterflies (Danaus plexippus). J. Invertebr. Pathol. 1999, 74, 76–88. [Google Scholar] [CrossRef] [Green Version]
- Altizer, S.M.; Oberhauser, K.S.; Brower, L.P. Associations between host migration and the prevalence of a protozoan parasite in natural populations of adult monarch butterflies. Ecol. Entomol. 2000, 25, 125–139. [Google Scholar] [CrossRef]
- Satterfield, D.A.; Altizer, S.; Williams, M.K.; Hall, R.J. Environmental Persistence Influences Infection Dynamics for a Butterfly Pathogen. PLoS ONE. 2017, 12, e0169982. [Google Scholar] [CrossRef] [Green Version]
- Myers, J.H.; Cory, J.S. Ecology and evolution of pathogens in natural populations of Lepidoptera. Evol. Appl. 2015, 9, 231–247. [Google Scholar] [CrossRef] [Green Version]
- Dhawan, M.; Joshi, N. Enzymatic comparison and mortality of Beauveria bassiana against cabbage caterpillar Pieris brassicae LINN. Braz. J. Microbiol. 2017, 48, 522–529. [Google Scholar] [CrossRef]
- Ismail, K.S. Study of bacterial contaminants isolated from adult monarch butterfly (Ddanaus plexippus) found on milkweed (Calotropis procera) in the Jazan province of Saudi Arabia. J. Sci. 2014, 4, 36–39. [Google Scholar]
- Monarch Watch. Available online: https://www.monarchwatch.org/biology/pred3.htm (accessed on 20 April 2022).
- Lacey, L.A.; Grzywacz, D.; Shapiro-Ilan, D.I.; Frutos, R.; Brownbridge, M.; Goettel, M.S. Insect pathogens as biological control agents: Back to the future. J. Invertebr. Pathol. 2015, 132, 1–41. [Google Scholar] [CrossRef] [Green Version]
- Hilgenboecker, K.; Hammerstein, P.; Schlattmann, P.; Telschow, A.; Werren, J.H. How many species are infected with Wolbachia? --A statistical analysis of current data. FEMS Microbiol. Lett. 2008, 281, 215–220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmed, M.Z.; Araujo-Jnr, E.V.; Welch, J.J.; Kawahara, A.Y. Wolbachia in butterflies and moths: Geographic structure in infection frequency. Front. Zool. 2015, 12, 16. [Google Scholar] [CrossRef] [PubMed]
- Werren, J.H.; Baldo, L.; Clark, M.E. Wolbachia: Master manipulators of invertebrate biology. Nat. Rev. Microbiol. 2008, 6, 741–751. [Google Scholar] [CrossRef]
- Williams, T.; Virto, C.; Murillo, R.; Caballero, P. Covert Infection of Insects by Baculoviruses. Front. Microbiol. 2017, 8, 1337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Satterfield, D.A.; Maerz, J.C.; Altizer, S. Loss of migratory behaviour increases infection risk for a butterfly host. Proc. Biol. Sci. 2015, 282, 20141734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davis, A.K.; de Roode, J.C. Effects of the parasite, Ophryocystis elektroscirrha, on wing characteristics important for migration in the monarch butterfly. Anim. Migr. 2018, 5, 84–93. [Google Scholar] [CrossRef]
- Sokol, R.; Michalczyk, M.; Micholap, P. Preliminary studies on the occurrence of honeybee pathogens in the national bumblebee population. Ann Parasitol. 2018, 64, 385–390. [Google Scholar] [CrossRef]
- Fürst, M.A.; McMahon, D.P.; Osborne, J.L.; Paxton, R.J.; Brown, M.J.F. Disease associations between honeybees and bumblebees as threat to wild pollinators. Nature 2014, 506, 364–366. [Google Scholar] [CrossRef] [Green Version]
- Evison, S.E.F.; Roberts, K.E.; Laurenson, L.; Pietravalle, S.; Hui, J.; Biesmeijer, J.C.; Smith, J.E.; Budge, G.; Hughes, W.O.H. Pervasiveness of Parasites in Pollinators. PLoS ONE 2012, 7, e30641. [Google Scholar] [CrossRef] [Green Version]
- Colla, S.R.; Otterstatter, M.C.; Gegear, R.J.; Thomson, J.D. Plight of the bumble bee: Pathogen spillover from commercial to wild populations. Biol. Conserv. 2006, 129, 461–467. [Google Scholar] [CrossRef]
- Cameron, S.A.; Lim, H.C.; Lozier, J.D.; Duennes, M.A.; Thorp, R. Test of the invasive pathogen hypothesis of bumble bee decline in North America. Proc. Natl. Acad. Sci. USA 2016, 113, 4386–4391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Graystock, P.; Blane, E.J.; McFrederick, Q.S.; Goulson, D.; Hughes, W.O.H. Do managed bees drive parasite spread and emergence in wild bees? Int. J. Parasitol. Parasites Wildl. 2016, 5, 64–75. [Google Scholar] [CrossRef] [PubMed]
- Graystock, P.; Goulson, D.; Hughes, W.O.H. The relationship between managed bees and the prevalence of parasites in bumblebees. PeerJ 2014, 2, e522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McNeil, D.J.; McCormick, E.; Heimann, A.C.; Kammerer, M.; Douglas, M.R.; Goslee, S.C.; Grozinger, C.M.; Hines, H.M. Bumble bees in landscapes with abundant floral resources have lower pathogen loads. Sci. Rep. 2020, 10, 22306. [Google Scholar] [CrossRef]
- Graystock, P.; Goulson, D.; Hughes, W.O. Parasites in bloom: Flowers aid dispersal and transmission of pollinator parasites within and between bee species. Proc. R. Soc. B 2015, 282, 20151371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McArt, S.H.; Koch, H.; Irwin, R.E.; Adler, L.S. Arranging the bouquet of disease: Floral traits and the transmission of plant and animal pathogens. Ecol. Lett. 2014, 17, 624–636. [Google Scholar] [CrossRef]
- Adler, L.S.; Irwin, R.E.; McArt, S.H.; Vannette, R.L. Floral traits affecting the transmission of beneficial and pathogenic pollinator-associated microbes. Curr. Opin. Insect Sci. 2021, 44, 1–7. [Google Scholar] [CrossRef]
- Figueroa, L.L.; Grab, H.; Ng, W.H.; Myers, C.R.; Graystock, P.; McFrederick, Q.S.; McArt, S.H. Landscape simplification shapes pathogen prevalence in plant-pollinator networks. Ecol. Lett. 2020, 23, 1212–1222. [Google Scholar] [CrossRef]
- Graystock, P.; Ng, W.H.; Parks, K.; Tripodi, A.D.; Muniz, P.A.; Fersch, A.A.; Myers, C.R.; McFrederick, Q.S.; McArt, S.H. Dominant bee species and floral abundance drive parasite temporal dynamics in plant-pollinator communities. Nat. Ecol. Evol. 2020, 4, 1358–1367. [Google Scholar] [CrossRef]
- Alger, S.A.; Burnham, P.A.; Brody, A.K. Flowers as viral hot spots: Honey bees (Apis mellifera) unevenly deposit viruses across plant species. PLoS ONE 2019, 14, e0221800. [Google Scholar] [CrossRef]
- Singh, R.; Levitt, A.L.; Rajotte, E.G.; Holmes, E.C.; Ostiguy, N.; van Engelsdorp, D.; Lipkin, I.W.; de Pamphilis, C.W.; Toth, A.L.; Cox-Foster, D.L. RNA viruses in hymenopteran pollinators: Evidence of inter-taxa virus transmission via pollen and potential impact on non-Apis hymenopteran species. PLoS ONE 2010, 5, e14357. [Google Scholar] [CrossRef] [PubMed]
- McMahon, D.P.; Fürst, M.A.; Caspar, J.; Theodorou, P.; Brown, M.J.; Paxton, R.J. A sting in the spit: Widespread cross-infection of multiple RNA viruses across wild and managed bees. J. Anim. Ecol. 2015, 84, 615–624. [Google Scholar] [CrossRef] [PubMed]
- Toplak, I.; Šimenc, L.; Pislak Ocepek, M.; Bevk, D. Determination of genetically identical strains of four honeybee viruses in bumblebee positive samples. Viruses 2020, 12, 1310. [Google Scholar] [CrossRef]
- Šimenc, L.; Kuhar, U.; Jamnikar-Ciglenečki, U.; Toplak, I. First Complete Genome of Lake Sinai Virus Lineage 3 and Genetic Diversity of Lake Sinai Virus Strains From Honey Bees and Bumble Bees. J. Econ. Entomol. 2020, 113, 1055–1061. [Google Scholar] [CrossRef]
- Pislak Ocepek, M.; Toplak, I.; Zajc, U.; Bevk, D. The Pathogens Spillover and Incidence Correlation in Bumblebees and Honeybees in Slovenia. Pathogens 2021, 10, 884. [Google Scholar] [CrossRef] [PubMed]
- Gomboc, S.; Lasan, M. The Checklist of Slovenian Lepidoptera—Overview and some open questions. In Proceedings of the 1st Slovenian Entomological Symposium, Slovenian Entomological Society of Štefan Michieli and Slovenian Museum of Natural History, Ljubljana, Slovenia, 4–5 November 2006; pp. 20–21. (In Slovenian). [Google Scholar]
- Šimenc, L.; Knific, T.; Toplak, I. The Comparison of Honeybee Viral Loads for Six Honeybee Viruses (ABPV, BQCV, CBPV, DWV, LSV3 and SBV) in Healthy and Clinically Affected Honeybees with TaqMan Quantitative Real-Time RT-PCR Assays. Viruses 2021, 13, 1340. [Google Scholar] [CrossRef] [PubMed]
- Toplak, I.; Jamnikar-Ciglenečki, U.; Aronstein, K.; Gregorc, A. Chronic bee paralysis virus and Nosema ceranae experimental co-infection of winter honey bee workers (Apis mellifera L.). Viruses 2013, 5, 2282–2297. [Google Scholar] [CrossRef] [Green Version]
- Runkel, C.; Flenniken, M.L.; Engel, J.C.; Ruby, J.G.; Ganem, D.; Andino, R.; DeRisi, J.L. Temporal analysis of the honey bee microbiome reveals four novel viruses and seasonal prevalence of known viruses, Nosema and Crithidia. PLoS ONE 2011, 6, e20656. [Google Scholar] [CrossRef] [Green Version]
- Toplak, I.; Rihtarič, D.; Jamnikar Ciglenečki, U.; Hostnik, P.; Jenčič, V.; Barlič-Maganja, D. Detection of six honeybee viruses in clinically affected colonies of Carniolan gray bee (Appis mellifera carnica). Slov. Vet. Res. 2012, 49, 89–96. [Google Scholar]
- Chen, Y.P.; Siede, R. Honey bee viruses. Adv. Virus Res. 2007, 70, 33–80. [Google Scholar] [CrossRef] [Green Version]
- Shen, M.; Cui, L.; Ostiguy, N.; Cox-Foster, D. Intricate transmission routes and interactions between picorna-like viruses (Kashmir bee virus and sacbrood virus) with the honeybee host and the parasitic varroa mite. J. Gen. Virol. 2005, 86, 2281–2289. [Google Scholar] [CrossRef] [PubMed]
- Al-Sherif, A.A.; Mazeed, A.M.; Ewis, M.A.; Nafea, E.A.; Hagag, E.S.E.; Kamel, A.A. Activity of salivary glands in secreting honey-elaborating enzymes in two subspecies of honeybee (Apis mellifera L). Physiol. Entomol. 2017, 42, 397–403. [Google Scholar] [CrossRef]
| Sample Number | Location | Habitat Type | Taxon | Species/Number of Specimens |
|---|---|---|---|---|
| 1 | 1/Ljubljana-NE | dry meadows, shrubs, forest path | Apidae | Apis mellifera carnica/10 |
| 2 | 1/Ljubljana-NE | Pieridae | Leptidea sp./10 | |
| 3 | 1/Ljubljana-NE | Nymphalidae | Maniola jurtina/10 | |
| 4 | 1/Ljubljana-NE | Hesperiidae | Tymelicus sp./10 | |
| 5 | 2/Ljubljana-W | wet meadows, forest edge | Apidae | Apis mellifera carnica/10 |
| 6 | 2/Ljubljana-W | Pieridae | Gonopteryx rhamni/10 | |
| 7 | 2/Ljubljana-W | Nymphalidae | Maniola jurtina/10 | |
| 8 | 2/Ljubljana-W | Hesperiidae | Ochlodes sylvanus/9, Tymelicus sp./1 | |
| 9 | 3/Ribnica | flowering edges along the railway, extensively managed meadows | Apidae | Apis mellifera carnica/10 |
| 10 | 3/Ribnica | Pieridae | Pieris rapae/10 | |
| 11 | 3/Ribnica | Nymphalidae | Melanargia galathea/10 | |
| 12 | 3/Ribnica | Lycenidae | Polyommatus icarus/10 | |
| 13 | 4/Žabljek | cultivated and extensively managed wet meadows | Apidae | Apis mellifera carnica/10 |
| 14 | 4/Žabljek | Pieridae | Pieris sp. (P. napi/1; P. brassicae/1; P. rapae/8) | |
| 15 | 4/Žabljek | Nymphalidae | Coenonympha pamphilus/10 | |
| 16 | 4/Žabljek | Lycenidae | Polyommatus icarus/10 |
| ABPV | BQCV | CBPV | DWV | LSV3 | SBV | ||
|---|---|---|---|---|---|---|---|
| Sample Number | Family (Species)/Number of Specimens | No. of Copies | No. of Copies | No. of Copies | No. of Copies | No. of Copies | No. of Copies |
| 2 | Pieridae/10 | 0 | 0 | 0 | 0 | 0 | 0 |
| 3 | Nymphalidae/10 | 9.5 × 104 | 4.7 × 105 | 0 | 0 | 9.8 × 103 | 1.1 × 105 |
| 4 | Hesperiidae/10 | 0 | 0 | 0 | 0 | 0 | 0 |
| 6 | Pieridae/10 | 0 | 2.1 × 104 | 0 | 0 | 0 | 8.5 × 102 |
| 7 | Nymphalidae/10 | 5.5 × 103 | 5.6 × 104 | 0 | 0 | 0 | 4.7× 103 |
| 8 | Hesperiidae/10 | 0 | 4.4 × 104 | 0 | 0 | 0 | 4.7 × 103 |
| 10 | Pieridae/10 | 0 | 0 | 0 | 0 | 0 | 1.1 × 104 |
| 11 | Nymphalidae/10 | 0 | 0 | 0 | 0 | 0 | 1.2 × 103 |
| 12 | Lycenidae/10 | 0 | 0 | 0 | 0 | 0 | 0 |
| 14 | Pieridae/10 | 0 | 2.5 × 104 | 0 | 0 | 0 | 3.9 × 104 |
| 15 | Nymphalidae/10 | 0 | 0 | 0 | 0 | 0 | 0 |
| 16 | Lycenidae/10 | 0 | 0 | 0 | 0 | 1.7 × 103 | 0 |
| 1 | Apis mellifera carnica/10 | 5.4 × 1012 | 5.0 × 109 | 0 | 1.2 × 104 | 6.6 × 109 | 3.3 × 106 |
| 5 | Apis mellifera carnica/10 | 8.5 × 102 | 1.5 × 109 | 0 | 0 | 4.7 × 108 | 1.7 × 105 |
| 9 | Apis mellifera carnica/10 | 1.7 × 105 | 3.7 × 109 | 0 | 1.0 × 104 | 1.5 × 109 | 6.8 × 104 |
| 13 | Apis mellifera carnica/10 | 2.4 × 104 | 5.1 × 1010 | 0 | 0 | 3.6 × 109 | 3.2 × 104 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pislak Ocepek, M.; Glavan, G.; Verovnik, R.; Šimenc, L.; Toplak, I. First Detection of Honeybee Pathogenic Viruses in Butterflies. Insects 2022, 13, 925. https://doi.org/10.3390/insects13100925
Pislak Ocepek M, Glavan G, Verovnik R, Šimenc L, Toplak I. First Detection of Honeybee Pathogenic Viruses in Butterflies. Insects. 2022; 13(10):925. https://doi.org/10.3390/insects13100925
Chicago/Turabian StylePislak Ocepek, Metka, Gordana Glavan, Rudi Verovnik, Laura Šimenc, and Ivan Toplak. 2022. "First Detection of Honeybee Pathogenic Viruses in Butterflies" Insects 13, no. 10: 925. https://doi.org/10.3390/insects13100925
APA StylePislak Ocepek, M., Glavan, G., Verovnik, R., Šimenc, L., & Toplak, I. (2022). First Detection of Honeybee Pathogenic Viruses in Butterflies. Insects, 13(10), 925. https://doi.org/10.3390/insects13100925






