Four New Species of Macquartia (Diptera: Oestroidea) from China and Phylogenetic Implications of Tachinidae †
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling Collection and Identification
2.2. DNA Extraction, Mitogenomes Sequencing, Assembly, and Annotation
2.3. Sequence Analysis
2.4. Phylogenetic Analyses
3. Results and Discussion
3.1. Taxonomy
- 1
- Middorsal excavation of abdominal syntergite 1 + 2 extending or nearly extending to its posterior margin. Three or four postsutural dorsocentral setae. Palpi reddish yellow.…………………………………………………………………………………….....2
- -
- Middorsal excavation of abdominal syntergite 1 + 2 not extending to its posterior margin. Three postsutural dorsocentral setae. Palpi dark brown or reddish yellow…………………………………………………………………………………..5
- 2
- Parafacial bare, at most with 3–4 hairs below lowest frontal seta. Aristal hairs at least more than diameter of aristal base. Four or three postsutural dorsocentral setae. Two katepisternal setae. Lower calyptrae divergent from scutellum. Thoracic scutum with gray pruinosity, abdomen black, with sparse pruinosity or without pruinosity, syntergite 1 + 2 with a pair of lateral marginal seta, without median marginal seta; third tergite with two median marginal setae, fourth tergite with a row of marginal setae……………………………………………………………………………………….…3
- -
- Parafacial hairy. Arista almost bare; four postsutural dorsocentral setae; three katepisternal setae. Lower calyptrae not divergent from scutellum. Abdomen black, with gray pruinosity and markings; tergite three with large black marking………………………………………………………….M. tessellum Meigen
- 3
- Pedicel and basal half of flagellomere dark brown. Four postsutural dorsocentral setae. Basicosta reddish yellow. Third tergite with two median discal setae in male and absent in female, fourth tergite with two to a row of discal setae in male and two setae in female, ocelli red …………………………………………………………...M. pubiceps Zetterstedt
- -
- Pedicel and basal half of flagellomere yellow. Three postsutural dorsocentral setae. Third and fouth tergites without median discal setae in both sexes, ocelli yellow.…………………………………………………………………………………4
- 4
- Basicosta dark brown. Legs black. …………………M. flavipedicel Zhang and Li sp. nov.
- -
- Basicosta and legs reddish yellow. Eyes with densely long hairs in male and sparsely short hairs in female. ………………M. flavifemorata Zhang and Li sp. nov.
- 5
- Mid tibia with one anterodorsal seta. Lower calyptrae divergent from scutellum……...……………………………………………………………………………6
- -
- Mid tibia with 2–5 anterodorsal setae. Abdominal third tergite with 2–4 marginal setae………………………………………………………………………………….....7
- 6
- Parafacial bare, at most with 3–4 hairs below lowest frontal seta. Legs black. syntergite 1 + 2 without median marginal and with 1–2 lateral marginal setae ……………………………………………………………M. chinensis Zhang and Li sp. nov.
- -
- Parafacial hairy only on upper half or hairy on whole length ……………………………………………….………………M. macularis Villeneuve
- 7
- Parafacial hairy on whole length …………………………………………..……………...8
- -
- Parafacial bare, at most hairy on upper half..…………………………...……….....9
- 8
- Pedicel and legs black ………………………………………………..……M. dispar Fallén
- -
- Pedicel and tibiae at least reddish yellow. Femora at least reddish yellow on ventral apex. Two katepisternal setae. Frons width at least 1/4 of eye width, frontal vitta about twice as wide as parafrontalia. Prealar seta about as long as hind supra-alar seta in famale. Abdomen covered with dense grayish pruinosity and regular gleaming marking…………………………………………M. viridana R.-D.
- 9
- Lower calyptrae not divergent from scutellum. Palpi reddish yellow, seldom dark at apex…………………………………………………………………………………………10
- -
- Lower calyptrae divergent from scutellum..………………………………………11
- 10
- Hind tibia with three preapical dorsal setae, middle one weaker; three katepisternal setae. Frons 1/7–1/8 of eye width, frontal vitta narrower than parafrontalia. Parafacial hairy on upper half. Abdomen with sparse grayish pruinosity in male and female, or female black, without pruinosity, syntergite 1 + 2 with 2 median marginal and 1–3 lateral marginal setae in both sexes, third and fourth tergites separately with 2 median discal and 1–3 pairs of lateral discal setae in both sexes…………M. tenebricosa Meigen
- -
- Hind tibia with two preapical dorsal setae; syntergite 1 + 2 without median marginal seta. Abdomen of female covered with covered grayish pruinosity, with gleaming markings ………………………………………M. chalconota Meigen
- 11
- Abdominal syntergite 1 + 2 with two strong median marginal setae in male and weak setae in female. fourth tergite with 2–4 median discal setae………….M. nudigena Mesnil
- -
- Abdominal syntergite 1 + 2 without median marginal setae in both sexes.…………….…………..………………M. brunneisquama Zhang and Li sp. nov.
3.2. Mitogenome Organization
3.3. Nucleotide Diversity and Evolutionary Rate Analysis
3.4. Phylogenetic Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- O’Hara, J.; Henderson, S.; Wood, D.M. Preliminary Checklist of the Tachinidae (Diptera) of the World; Version 2.1; PDF Document, 2020; 1039p. Available online: http://www.nadsdiptera.org/Tach/WorldTachs/Checklist/Worldchecklist.html (accessed on 8 January 2022).
- Chao, C. Tachinidae. In Flies of China; Xue, W., Chao, C., Eds.; Liaoning Science and Technology Press: Shenyang, China, 1998; Volume 2, pp. 1661–2206 + pls. 1–30. [Google Scholar]
- O’ Hara, J.; Shima, H.; Zhang, C. Annotated Catalogue of the Tachinidae (Insecta: Diptera) of China. Zootaxa 2009, 2190, 1–236. [Google Scholar] [CrossRef]
- Tschorsnig, H.P. Preliminary Host Catalogue of Palaearctic Tachinidae (Diptera); Version 1; 2017; 480p. Available online: http://www.nadsdiptera.org/Tach/WorldTachs/CatPalHosts/Cat_Pal_tach_hosts_Ver1.pdf (accessed on 28 April 2017).
- Cerretti, P.; O’Hara, J.; Wood, D.M.; Shima, H.; Inclan, D.; Stireman, J., III. Signal through the noise? Phylogeny of the Tachinidae (Diptera) as inferred from morphological evidence. Syst. Entomol. 2014, 39, 335–353. [Google Scholar] [CrossRef]
- Stireman, J., III; Cerretti, P.; O’Hara, J.; Jeremy, D.; Blaschke, J.; Moulton, J. Molecular phylogeny and evolution of world Tachinidae (Diptera). Mol. Phylogenet. Evol. 2019, 139, 106358. [Google Scholar] [CrossRef]
- Herting, B.; Dely-Draskovits, Á. Family Tachinidae. In Catalogue of Palaearctic Diptera; Soós, Á., Papp, L., Eds.; Anthomyiidae—Tachinidae; Hungarian Natural History Museum: Budapest, Hungary, 1993; Volume 13, pp. 118–458, 624p. [Google Scholar]
- Mesnil, L. 64f. Dexiinae. In Die Fliegen der Palaearktischen Region; E. Schweizerbartsche Verlagsbuchhandlung: Stuttgart, Germany, 1980; Volume 9, pp. 1–52. [Google Scholar]
- Tschorsnig, H.P.; Herting, B. Die Raupenfliegen (Diptera: Tachinidae) Mitteleuropas: Bestimmung-stabellen und Angaben zur Verbreitung und kologie der einzelnen Arten. Stuttg. Beitrge Nat. Ser. A Biol. 1994, 560, 1–170. [Google Scholar]
- Tschorsnig, H.P.; Richter, V. Family Tachinidae. In Contributions to a Manual of Palaearctic Diptera (with Special Reference to Flies of Economic Importance); Higher Brachycera; Papp, L., Darvas, B., Eds.; Science Herald: Budapest, Hungary, 1998; Volume 3, pp. 691–827, 880p. [Google Scholar]
- Nie, R.; Vogler, A.; Yang, X.; Lin, M. Higher-level phylogeny of longhorn beetles (Coleoptera: Chrysomeloidea) inferred from mitochondrial genomes. Syst. Entomol. 2021, 46, 56–70. [Google Scholar] [CrossRef]
- McAlpine, J. Morphology and terminology—Adults. In Manual of Nearctic Diptera. Vol. 1. Research Branch, Agriculture Canada, Monograph; McAlpine, J.F., Peterson, B.V., Shewell, G.E., Teskey, H.J., Vockeroth, J.R., Wood, D.M., Eds.; Biosystematics Research Institute: Ottawa, ON, Canada, 1981; Volume 27, pp. 9–63. [Google Scholar]
- Sinclair, B.J. Morphology and terminology of Diptera male terminalia. In Contributions to a Manual of Palaearctic Diptera (with Special Reference to Flies of Economic Importance). Vol. 1. General and Applied Dipterology; Papp, L., Darvas, B., Eds.; Science Herald: Budapest, Hungary, 2000; pp. 53–74. [Google Scholar]
- O’Hara, J. Revision of the Polideini (Tachinidae) of America north of Mexico. Stud. Dipterol. 2002, 10, 1–170. [Google Scholar]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. 1994, 3, 294–299. [Google Scholar]
- Peng, Y.; Leung, H.C.M.; Yiu, S.; Chin, F. IDBA-UD: A de novo assembler for singlecell and metagenomic sequencing data with highly uneven depth. Bioinformatics 2012, 28, 1420–1428. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Crampton-Platt, A.; Timmermans, M.; Gimmel, M.; Kutty, S.; Cockerill, T.; Khen, C.; Vogler, T. Soup to tree: The phylogeny of beetles inferred by mitochondrial metagenomics of a bornean rainforest sample. Mol. Biol. Evol. 2015, 32, 2302–2316. [Google Scholar] [CrossRef]
- Yan, L.P.; Pape, T.; Elgar, M.; Gao, Y.Y.; Zhang, D. Evolutionary history of stomach bot flies in the light of mitogenomics. Syst. Entomol. 2019, 44, 797–809. [Google Scholar] [CrossRef] [Green Version]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Greiner, S.; Lehwark, P.; Bock, R. Organellar genome DRAW (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Jakovli’c, I.; Zou, H.; Zhang, J.; Li, W.; Wang, G. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Perna, N.; Kocher, T. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 1995, 41, 353–358. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sanchez-DelBarrio, J.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.; Sanchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [Green Version]
- Katoh, K.; Standley, D. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Lanfear, R.; Frandsen, P.; Wright, A.; Senfeld, T.; Calcott, B. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular morphological phylogenetic analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [CrossRef] [Green Version]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Hohna, S.; Larget, B.; Liu, L.; Suchard, M.; Huelsenbeck, J. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [Green Version]
- Rambaut, A.; Drummond, A.; Xie, D.; Baele, G.; Suchard, M. Posterior summarization in Bayesian phylogenetics using tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cameron, S. Insect mitochondrial genomics: Implications for evolution and phylogeny. Annu. Rev. Entomol. 2014, 59, 95–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Z.; Su, T.; Chesters, D.; Wang, S.; Ho, S.; Zhu, C.; Chen, X.; Zhang, C.T. The mitochondrial genome of Elodia flavipalpis Aldrich (Diptera: Tachinidae) and the evolutionary timescale of tachinid flies. PLoS ONE 2013, 8, e61814. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.P.; Xu, W.T.; Zhang, D.; Li, J.Q. Comparative analysis of the mitochondrial genomes of flesh flies and their evolutionary implication. Int. J. Biol. Macromol. 2021, 174, 385–391. [Google Scholar] [CrossRef]
- Zhang, D.; Yan, L.P.; Zhang, M.; Chu, H.J.; Cao, J.; Li, K.; Hu, D.F.; Pape, T. Phylogenetic inference of calyptrates, with the first mitogenomes for Gasterophilinae (Diptera: Oestridae) and Paramacronychiinae (Diptera: Sarcophagidae). Int. J. Biol. Sci. 2016, 12, 489–504. [Google Scholar] [CrossRef] [Green Version]
- Kutty, S.; Meusemann, K.; Bayless, K.; Marinho, M.; Pont, A.; Zhou, X.; Misof, B.; Wiegmann, B.; Yeates, D.; Cerretti, P. Phylogenomic analysis of Calyptratae: Resolving the phylogenetic relationships within a major radiation of Diptera. Cladistics 2019, 35, 605–622. [Google Scholar] [CrossRef]
- Yan, L.P.; Buenaventura, E.; Pape, T.; Kutty, S.; Bayless, K.; Zhang, D. A phylotranscriptomic framework for flesh fly evolution (Diptera, Calyptratae, Sarcophagidae). Cladistics 2021, 37, 540–558. [Google Scholar] [CrossRef]
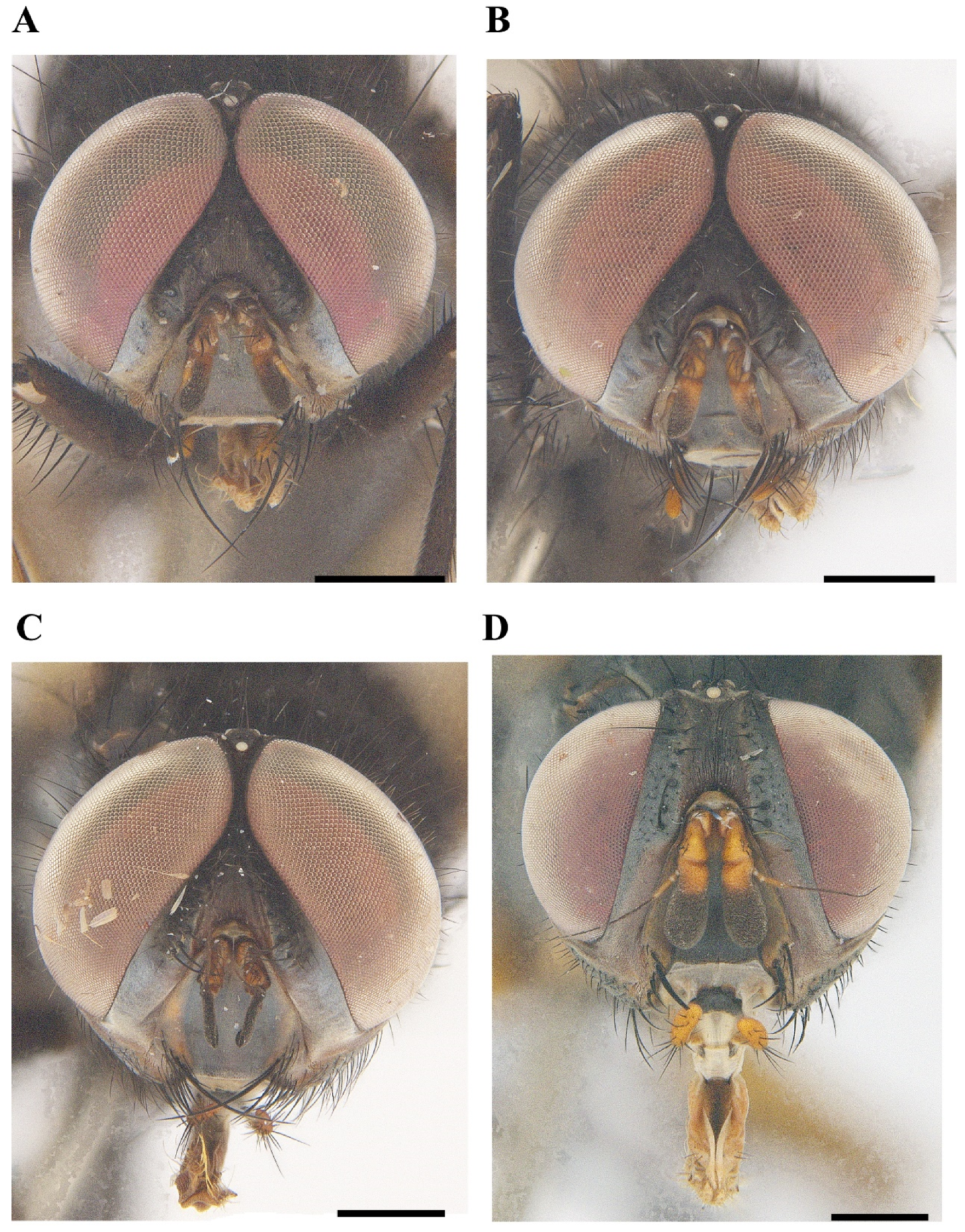










| M. brunneisquama sp. nov. | M. chinensis sp. nov. | M. flavipedicel sp. nov. | M. flavifemorata sp. nov. | |
|---|---|---|---|---|
| Characters | Parafacial hairy at most on upper half | Parafacial bare | Parafacial bare | Parafacial bare |
| Pedicel dark to brown | Pedicel reddish yellow | Pedicel reddish yellow | Pedicel reddish yellow | |
| 2 + 3 dorsocentral setae | 3+3 dorsocentral setae | 3 + 3 dorsocentral setae | 2 + 3 dorsocentral setae | |
| Basicosta dark brown | Basicosta dark brown | Basicosta dark brown | Basicosta reddish yellow | |
| Legs dark brown | Legs dark brown | Legs dark brown | Legs reddish yellow | |
| Mid tibia with 2 anterodorsal setae | Mid tibia with 1 anterodorsal seta | Mid tibia with 1 anterodorsal seta | Mid tibia with 1 anterodorsal seta | |
| Middorsal excavation of abdominal syntergite 1 + 2 not extending to its posterior margin, with 1–2 lateral marginal seta | Middorsal excavation of syntergite 1 + 2 not extending to its posterior margin, without lateral marginal seta | Middorsal excavation of syntergite 1 + 2 extending or nearly to its posterior margin, with a pair of lateral marginal setae | Middorsal excavation of syntergite 1 + 2 nearly to 4/5 of its posterior margin, without lateral marginal setae |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, H.; Zhang, B.; Pei, W.; Sun, H.; Chen, J.; Gao, X.; Peng, H.; Zhang, D.; Zhang, C. Four New Species of Macquartia (Diptera: Oestroidea) from China and Phylogenetic Implications of Tachinidae. Insects 2022, 13, 1096. https://doi.org/10.3390/insects13121096
Li H, Zhang B, Pei W, Sun H, Chen J, Gao X, Peng H, Zhang D, Zhang C. Four New Species of Macquartia (Diptera: Oestroidea) from China and Phylogenetic Implications of Tachinidae. Insects. 2022; 13(12):1096. https://doi.org/10.3390/insects13121096
Chicago/Turabian StyleLi, Henan, Baihui Zhang, Wenya Pei, Haoran Sun, Jinliang Chen, Xinzhang Gao, Honglin Peng, Dong Zhang, and Chuntian Zhang. 2022. "Four New Species of Macquartia (Diptera: Oestroidea) from China and Phylogenetic Implications of Tachinidae" Insects 13, no. 12: 1096. https://doi.org/10.3390/insects13121096
APA StyleLi, H., Zhang, B., Pei, W., Sun, H., Chen, J., Gao, X., Peng, H., Zhang, D., & Zhang, C. (2022). Four New Species of Macquartia (Diptera: Oestroidea) from China and Phylogenetic Implications of Tachinidae. Insects, 13(12), 1096. https://doi.org/10.3390/insects13121096






