A Non-Gradual Development Process of Cicada Eyes at the End of the Fifth-Instar Nymphal Stage to Obtain Visual Ability
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
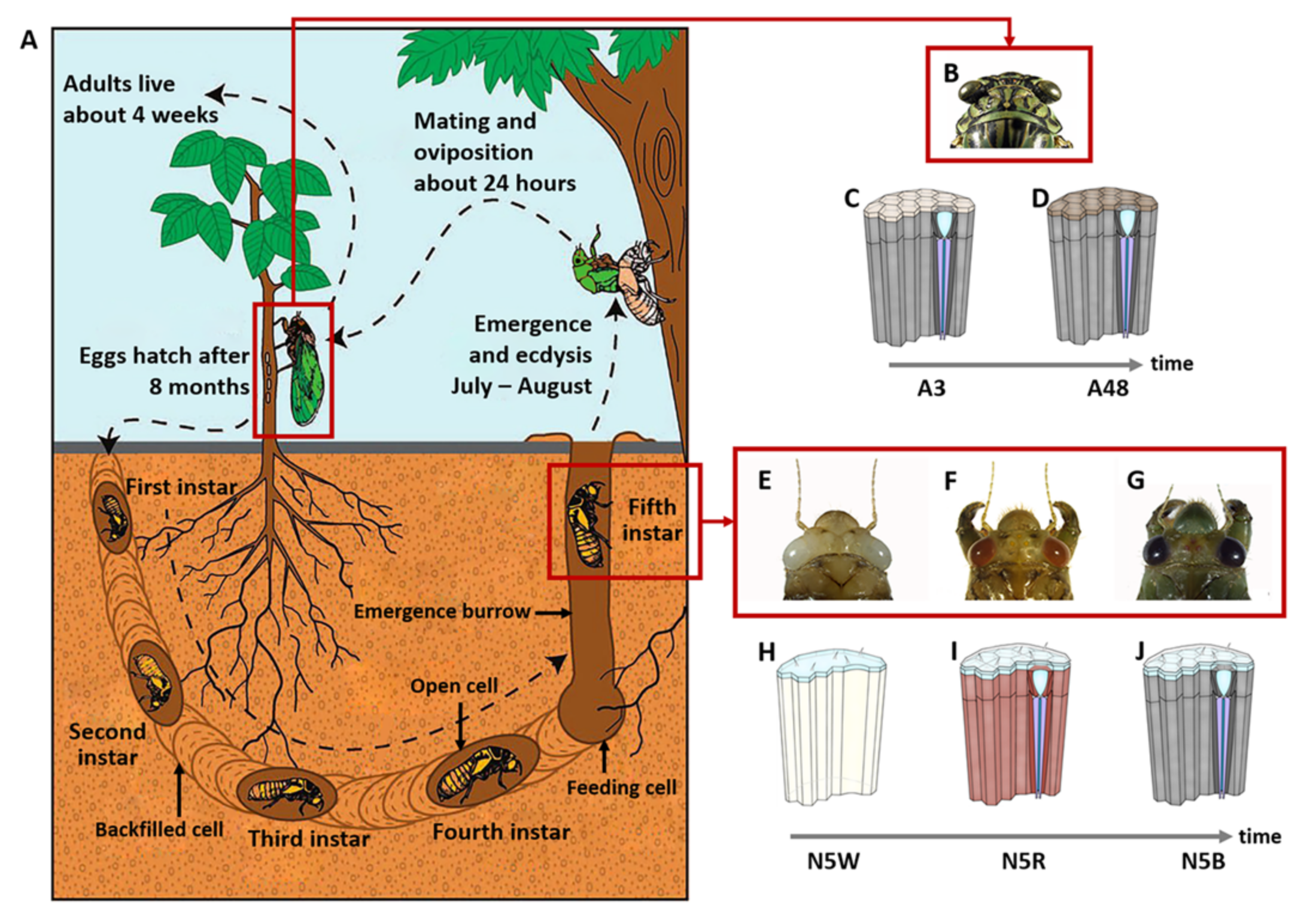
2.1. Sample Preparation
2.2. RNA Extraction, Raw Data Quality Control and cDNA Library Construction
2.3. De Novo Assembly, Unigene Annotation and Functional Classification
2.4. Analysis of Differentially Expressed Genes (DEGs)
2.5. Fuzzy C-Means Clustering
2.6. Enrichment Analysis of GO Enrichment and KEGG Pathway
2.7. Phylogenetic Analysis
2.8. Reverse-Transcription Quantitative PCR (RT-qPCR)
2.9. Accession Number
3. Results
3.1. Stage-Specific RNA-Seq of Compound Eyes of M. mongolica
3.2. Cluster Analyses Based on All Identified Differentially-Expressed Genes
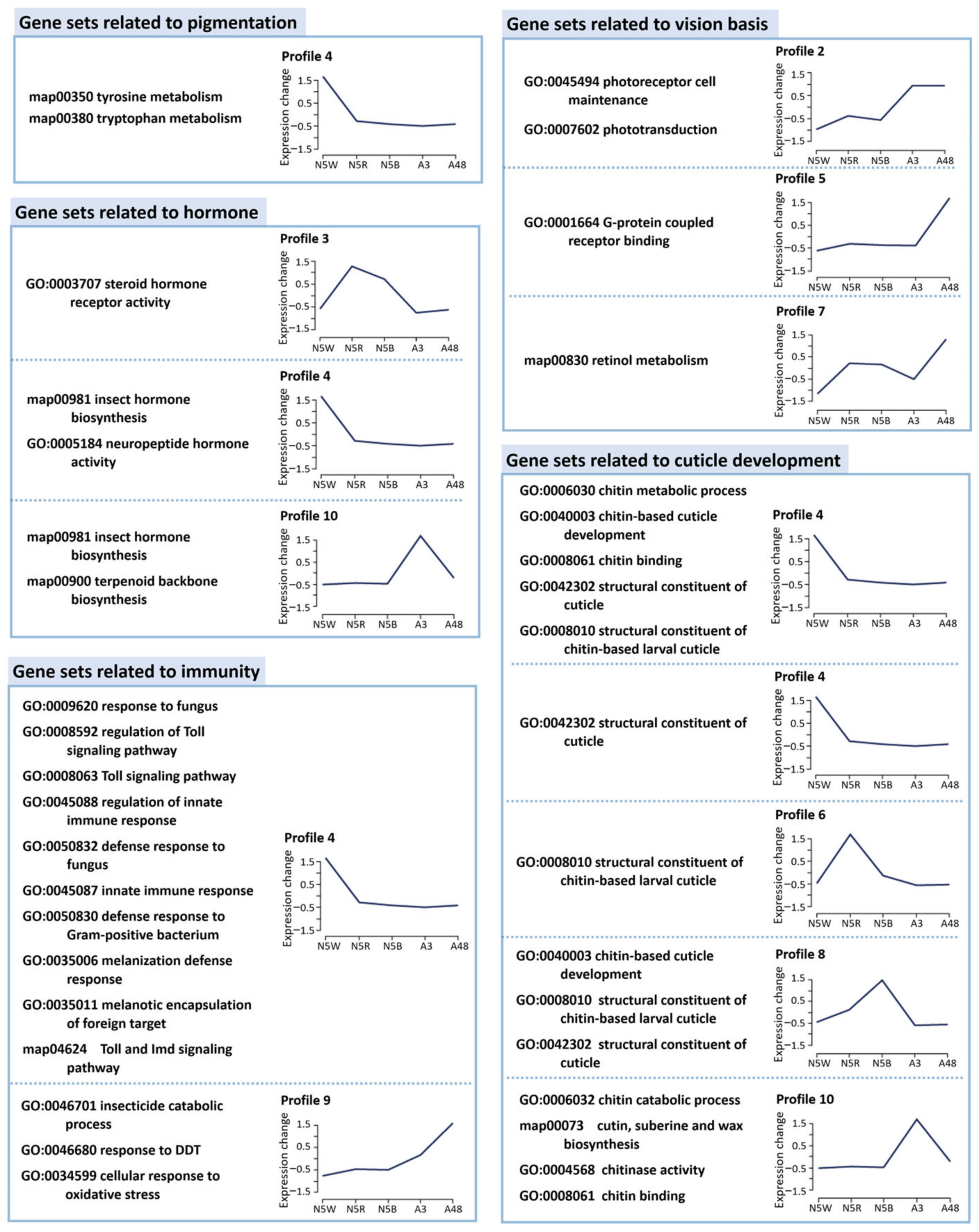
3.3. Functional Enrichment Analysis Based on Stage-Specific and Profile-Specific DEGs
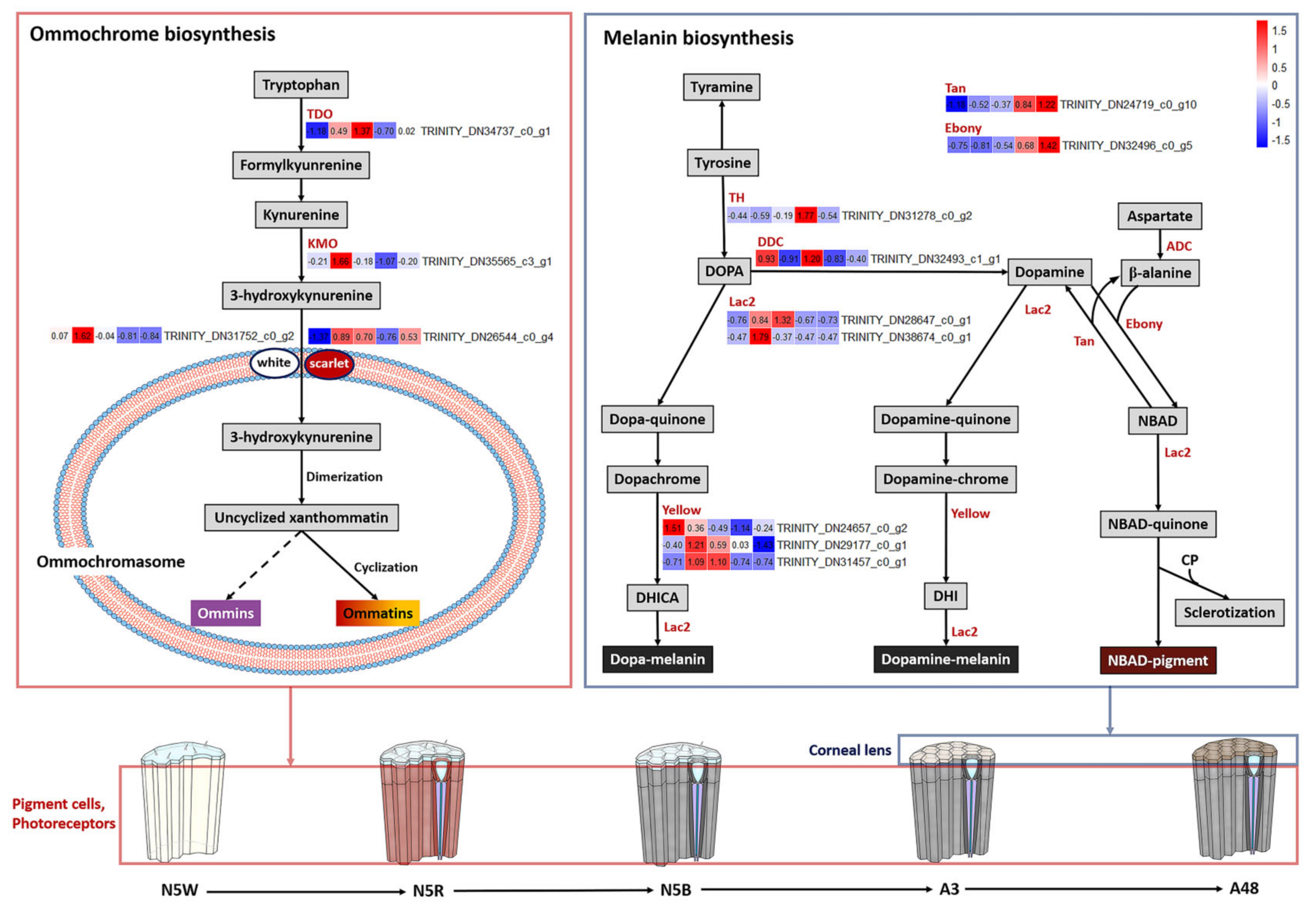
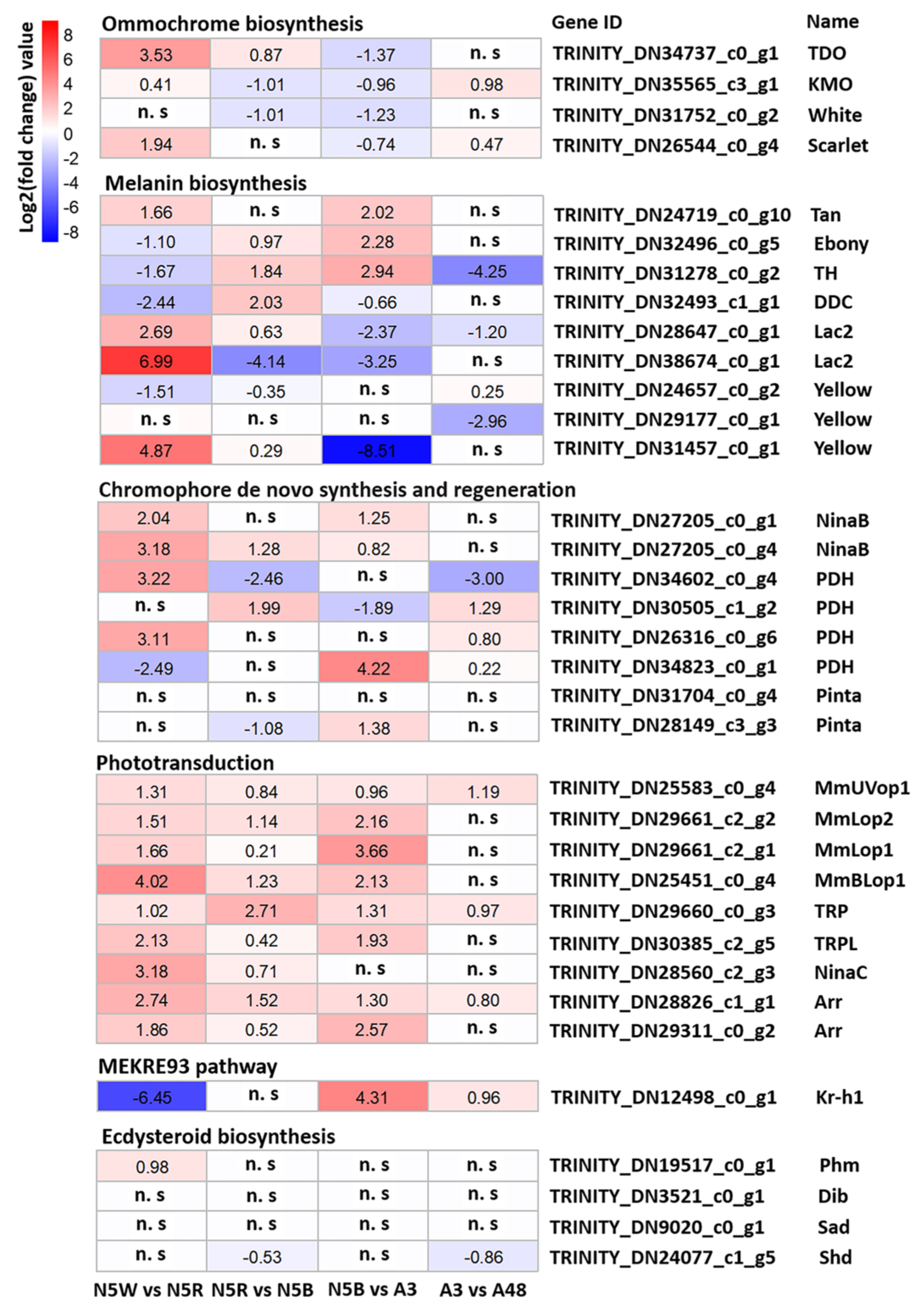
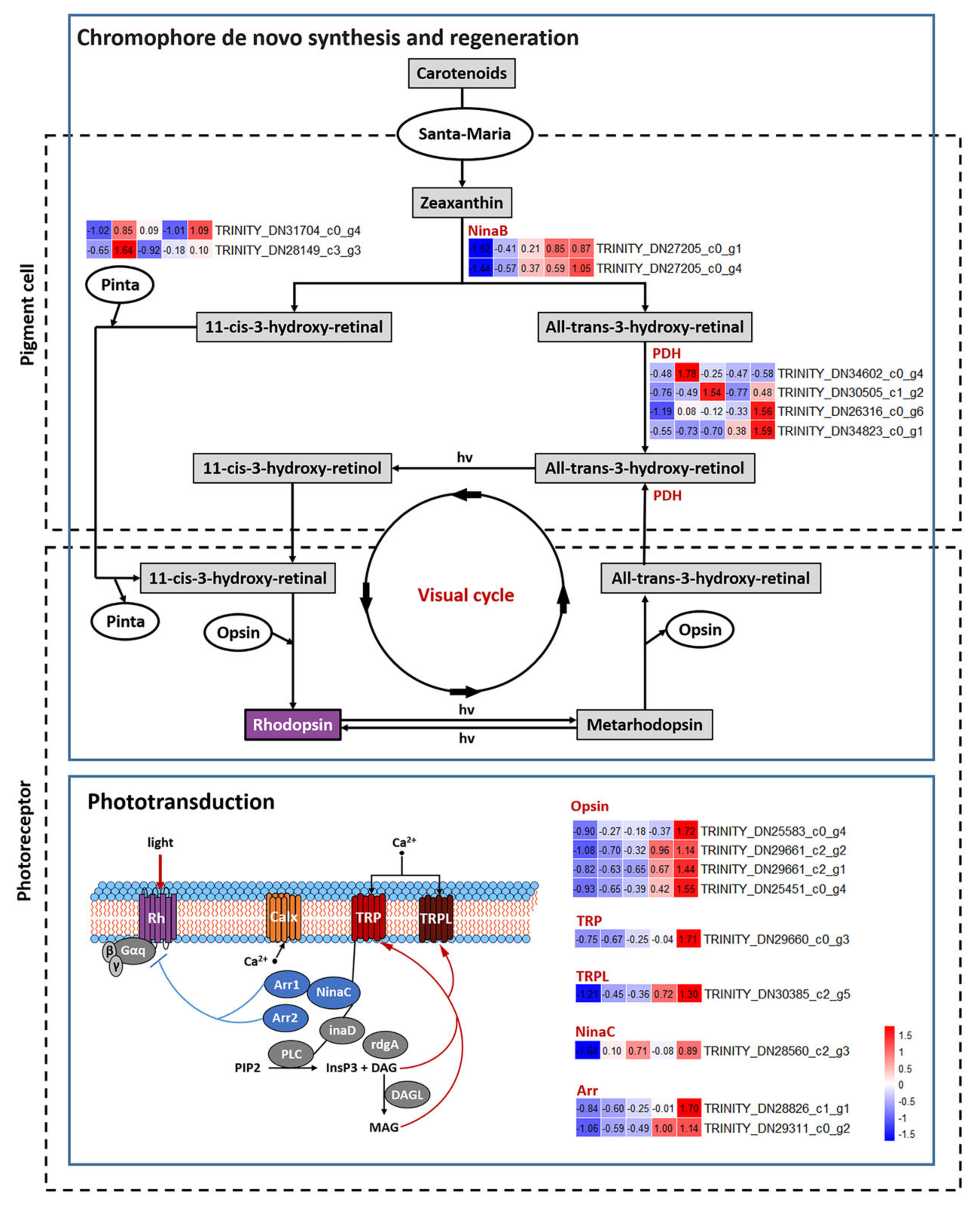
3.4. DEGs Related to Pigmentation
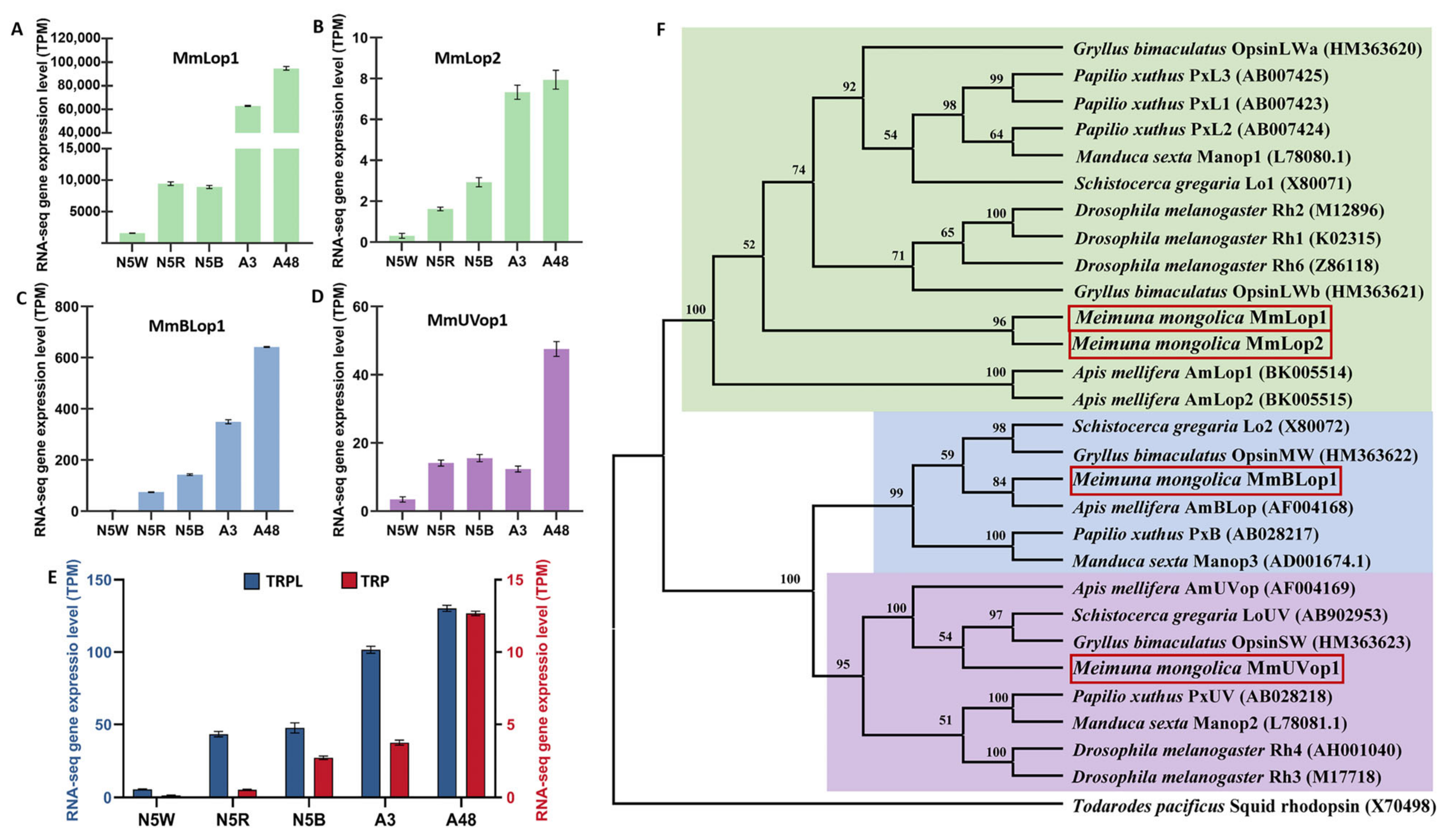
3.5. DEGs Related to Structure of Visual System
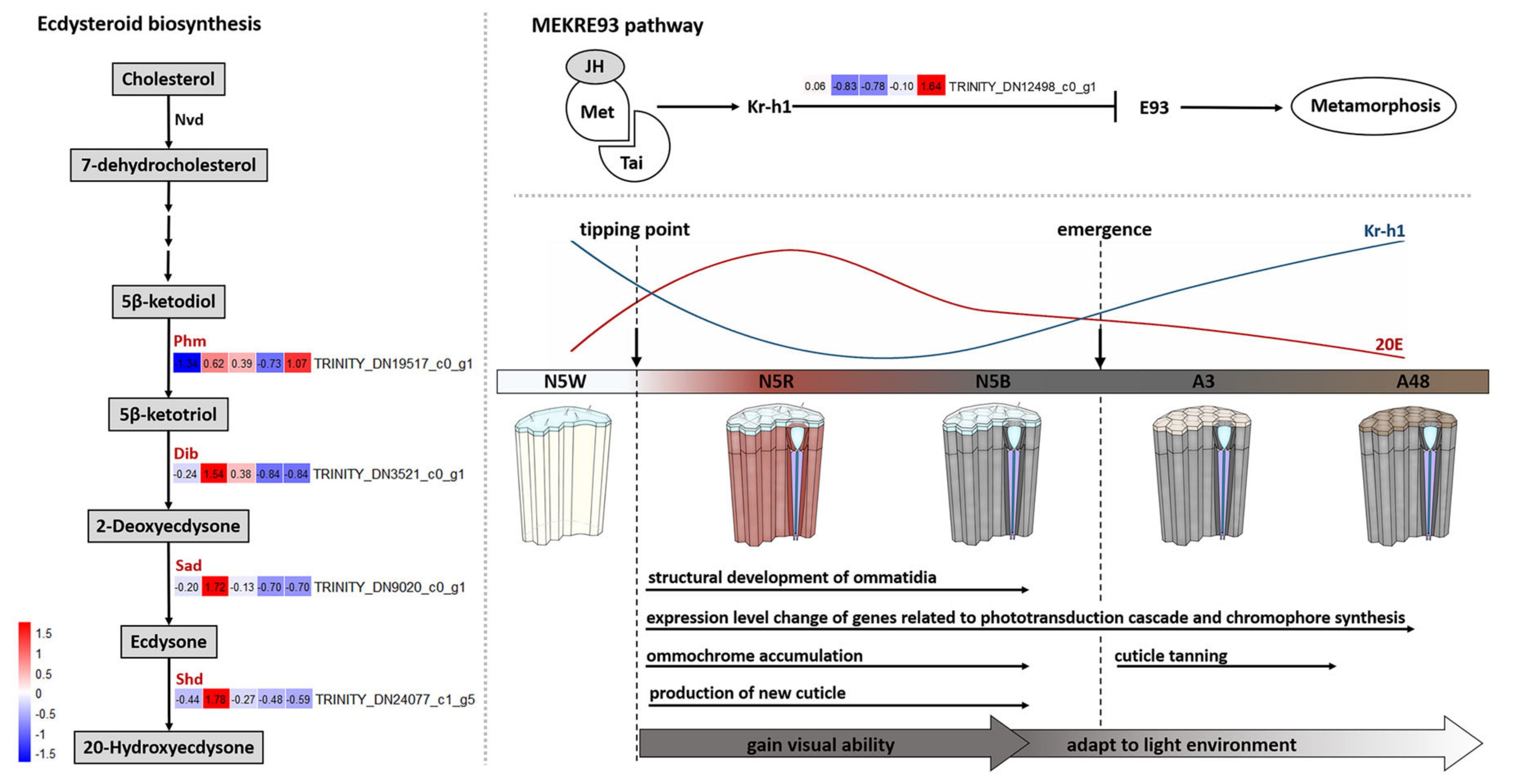
3.6. DEGs Related to Hormone Biosynthesis
3.7. DEGs Related to Immunity
3.8. DEGs Related to Cuticle Development
3.9. Validation of Transcriptome Data Using qRT-PCR
4. Discussion
4.1. Dramatic Development of M. mongolica Eyes before Emergence
4.2. Development of M. mongolica Eyes for Usable Visual Ability after Emergence
4.3. Protective Function of Eye Color Change in Relation to Niche Change
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Honkanen, A.; Immonen, E.V.; Salmela, I.; Heimonen, K.; Weckstrom, M. Insect photoreceptor adaptations to night vision. Philos. Trans. R. Soc. B-Biol. Sci. 2017, 372, 20160077. [Google Scholar] [CrossRef] [PubMed]
- Gullan, P.; Cranston, P. Sensory systems and behavior. In The Insects: An Outline of Entomology, 3rd ed.; Gullan, P.J., Cranston, P.S., Eds.; Wiley-Blackwell, Ltd.: Hoboken, NJ, USA, 2004; pp. 85–112. [Google Scholar]
- Schoenemann, B.; Parnaste, H.; Clarkson, E.N.K. Structure and function of a compound eye, more than half a billion years old. Proc. Natl. Acad. Sci. USA 2017, 114, 13489–13494. [Google Scholar] [CrossRef] [PubMed]
- Buschbeck, E.; Friedrich, M. Evolution of insect eyes: Tales of ancient heritage, deconstruction, reconstruction, remodeling, and recycling. Evol. Educ. Outreach 2008, 1, 448–462. [Google Scholar] [CrossRef]
- Nilsson, H.; Lindström, M. Retinal damage and sensitivity loss of a light-sensitive crustacean compound eye (Cirolana borealis): Electron microscopy and electrophysiology. J. Exp. Biol. 1983, 107, 277–292. [Google Scholar] [CrossRef]
- Shichida, Y.; Matsuyama, T. Evolution of opsins and phototransduction. Philos. Trans. R. Soc. B-Biol. Sci. 2009, 364, 2881–2895. [Google Scholar] [CrossRef]
- Stavenga, D.G. Pigments in compound eyes. In Facets of Vision; Stavenga, D.G., Hardie, R.C., Eds.; Springer, Ltd.: Berlin/Heidelberg, Germany, 1989; pp. 152–172. [Google Scholar]
- Stavenga, D.G. Colour in the eyes of insects. J. Comp. Physiol. A-Neuroethol. Sens. Neural Behav. Physiol. 2002, 188, 337–348. [Google Scholar]
- Dontsov, A.E.; Sakina, N.L.; Yakovleva, M.A.; Bastrakov, A.I.; Bastrakova, I.G.; Zagorinsky, A.A.; Ostrovsky, M.A. Ommochromes from the compound eyes of insects: Physicochemical properties and antioxidant activity. Biochemistry 2020, 85, 668–678. [Google Scholar] [CrossRef]
- Insausti, T.C.; Le Gall, M.; Lazzari, C.R. Oxidative stress, photodamage and the role of screening pigments in insect eyes. J. Exp. Biol. 2013, 216, 3200–3207. [Google Scholar] [CrossRef]
- Ushakova, N.; Dontsov, A.; Sakina, N.; Bastrakov, A.; Ostrovsky, M. Antioxidative properties of melanins and ommochromes from black soldier fly Hermetia illucens. Biomolecules 2019, 9, 408. [Google Scholar] [CrossRef]
- Greiner, B.; Ribi, W.A.; Warrant, E.J. Retinal and optical adaptations for nocturnal vision in the halictid bee Megalopta genalis. Cell Tissue Res. 2004, 316, 377–390. [Google Scholar] [CrossRef]
- French, A.S.; Meisner, S.; Liu, H.; Weckstrom, M.; Torkkeli, P.H. Transcriptome analysis and RNA interference of cockroach phototransduction indicate three opsins and suggest a major role for TRPL channels. Front. Physiol. 2015, 6, 207. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Lu, B.; Xiao, H.; Fu, X.; Murphy, R.; Wu, K. The evolution and expression of the moth visual opsin family. PLoS ONE 2013, 8, e78140. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yan, S.; Zhu, J.; Zhu, W.; Zhang, X.; Li, Z.; Liu, X.; Zhang, Q. The expression of three opsin genes from the compound eye of Helicoverpa armigera (Lepidoptera: Noctuidae) is regulated by a circadian clock, light conditions and nutritional status. PLoS ONE 2014, 9, e111683. [Google Scholar] [CrossRef] [PubMed]
- Futahashi, R.; Kawahara-Miki, R.; Kinoshita, M.; Yoshitake, K.; Yajima, S.; Arikawa, K.; Fukatsu, T. Extraordinary diversity of visual opsin genes in dragonflies. Proc. Natl. Acad. Sci. USA 2015, 112, E1247–E1256. [Google Scholar] [CrossRef] [PubMed]
- Williams, K.S.; Simon, C. The ecology, behavior, and evolution of periodical cicadas. Annu. Rev. Entomol. 1995, 40, 269–295. [Google Scholar] [CrossRef]
- Lan, Y.; Wei, C. Morphology, histology and ultrastructure of the compound eyes of the last instar nymphs and adults of Meimuna mongolica (Hemiptera: Cicadidae). Acta Entomol. Sin. 2020, 63, 1441–1451. [Google Scholar]
- Smith, J.; Hasiotis, S. Traces and burrowing behaviors of the Cicada nymph Cicadetta calliope: Neoichnology and paleoecological significance of extant soil-dwelling insects. Palaios 2008, 23, 503–513. [Google Scholar] [CrossRef]
- Hou, Z.; Li, Q.; Wei, C. Morphology and identification of the final instar nymphs of three cicadas (Hemiptera, Cicadidae) in Guanzhong Plain, China based on comparative morphometrics. ZooKeys 2014, 425, 33–50. [Google Scholar]
- Martin, M. CUTADAPT removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Grabherr, M.; Haas, B.; Yassour, M.; Levin, J.; Thompson, D.; Amit, I.; Regev, A. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2014, 12, 59–60. [Google Scholar] [CrossRef] [PubMed]
- Ioannidou, Z.; Theodoropoulou, M.; Papandreou, N.; Willis, J.; Hamodrakas, S. CutProtFam-Pred: Detection and classification of putative structural cuticular proteins from sequence alone, based on profile Hidden Markov Models. Insect Biochem. Mol. Biol. 2014, 52, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Patro, R.; Duggal, G.; Love, M.; Irizarry, R.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, A.; Williams, B.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.; McCarthy, D.; Smyth, G. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Kumar, L.; Futschik, M. Mfuzz: A software package for soft clustering of microarray data. Bioinformation 2007, 2, 5–7. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. ClusterProfiler: An R package for comparing biological themes among gene clusters. Omics A J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Kirschfeld, K. Activation of visual pigment: Chromophore structure and function. In The Molecular Mechanism of Photoreception; Stieve, H., Ed.; Springer, Ltd.: Berlin/Heidelberg, Germany, 1986; pp. 31–49. [Google Scholar]
- Senthilan, P.R.; Piepenbrock, D.; Ovezmyradov, G.; Nadrowski, B.; Bechstedt, S.; Pauls, S.; Goepfert, M.C. Drosophila auditory organ genes and genetic hearing defects. Cell 2012, 150, 1042–1054. [Google Scholar] [CrossRef]
- Katana, R.; Guan, C.; Zanini, D.; Larsen, M.E.; Giraldo, D.; Geurten, B.R.H.; Goepfert, M.C. Chromophore-independent roles of opsin apoproteins in Drosophila mechanoreceptors. Curr. Biol. 2019, 29, 2961–2969. [Google Scholar] [CrossRef] [PubMed]
- Kumar, J.P.; Ready, D.F. Rhodopsin plays an essential structural role in Drosophila photoreceptor development. Development 1995, 121, 4359–4370. [Google Scholar] [CrossRef] [PubMed]
- Lichtenstein, L.; Gruebel, K.; Spaethe, J. Opsin expression patterns coincide with photoreceptor development during pupal development in the honey bee, Apis mellifera. BMC Dev. Biol. 2018, 18, 1. [Google Scholar] [CrossRef] [PubMed]
- Shamim, G.; Ranjan, S.K.; Pandey, D.M.; Ramani, R. Biochemistry and biosynthesis of insect pigments. Eur. J. Entomol. 2014, 111, 149–164. [Google Scholar] [CrossRef]
- Linzen, B. The tryptophan ommochrome pathway in insects. In Advances in Insect Physiology; Treherne, J.E., Berridge, M.J., Wigglesworth, V.B., Eds.; Academic Press, Ltd.: Cambridge, MA, USA, 1974; pp. 117–246. [Google Scholar]
- Petryk, A.; Warren, J.T.; Marqués, G.; Jarcho, M.P.; Gilbert, L.I.; Kahler, J.; O’Connor, M.B. Shade is the Drosophila P450 enzyme that mediates the hydroxylation of ecdysone to the steroid insect molting hormone 20-hydroxyecdysone. Proc. Natl. Acad. Sci. USA 2003, 100, 13773–13778. [Google Scholar] [CrossRef]
- Rewitz, K.F.; Rybczynski, R.; Warren, J.T.; Gilbert, L.I. Developmental expression of Manduca shade, the P450 mediating the final step in molting hormone synthesis. Mol. Cell. Endocrinol. 2006, 247, 166–174. [Google Scholar] [CrossRef]
- Warren, J.T.; Petryk, A.; Marqués, G.; Parvy, J.P.; Shinoda, T.; Itoyama, K.; Gilbert, L.I. Phantom encodes the 25-hydroxylase of Drosophila melanogaster and Bombyx mori: A P450 enzyme critical in ecdysone biosynthesis. Insect Biochem. Mol. Biol. 2004, 34, 991–1010. [Google Scholar] [CrossRef]
- Belles, X.; Santos, C.G. The MEKRE93 (Methoprene tolerant-Kruppel homolog 1-E93) pathway in the regulation of insect metamorphosis, and the homology of the pupal stage. Insect Biochem. Mol. Biol. 2014, 52, 60–68. [Google Scholar] [CrossRef]
- Konopova, B.; Smykal, V.; Jindra, M. Common and distinct roles of Juvenile Hormone signaling genes in metamorphosis of holometabolous and hemimetabolous insects. PLoS ONE 2011, 6, e28728. [Google Scholar] [CrossRef]
- Lozano, J.; Belles, X. Conserved repressive function of Kruppel homolog 1 on insect metamorphosis in hemimetabolous and holometabolous species. Sci. Rep. 2011, 1, 163. [Google Scholar] [CrossRef]
- Kambris, Z.; Brun, S.; Jang, I.H.; Nam, H.J.; Romeo, Y.; Takahashi, K.; Lee, W.J.; Ueda, R.; Lemaitre, B. Drosophila immunity: A large-scale in vivo RNAi screen identifies five serine proteases required for Toll activation. Curr. Biol. 2006, 16, 808–813. [Google Scholar] [CrossRef] [PubMed]
- Morisato, D.; Anderson, K.V. Signaling pathways that establish the dorsal-ventral pattern of the Drosophila embryo. Annu. Rev. Genet. 1995, 29, 371–399. [Google Scholar] [CrossRef] [PubMed]
- Schwind, R. Zonation of the optical environment and zonation in the rhabdom structure within the eye of the backswimmer, Notonecta glauca. Cell Tissue Res. 1983, 232, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Stange, G. The ocellar component of flight equilibrium control in dragonflies. J. Comp. Physiol. 1981, 141, 335–347. [Google Scholar] [CrossRef]
- Möller, R. Insects could exploit UV-Green contrast for landmark navigation. J. Theor. Biol. 2002, 214, 619–631. [Google Scholar] [CrossRef]
- Reuss, H.; Mojet, M.H.; Chyb, S.; Hardie, R.C. In vivo analysis of the Drosophila light-sensitive channels, TRP and TRPL. Neuron 1997, 19, 1249–1259. [Google Scholar] [CrossRef]
- Leung, H.T.; Geng, C.X.; Pak, W.L. Phenotypes of trpl mutants and interactions between the transient receptor potential (TRP) and TRP-like channels in Drosophila. J. Neurosci. 2000, 20, 6797–6803. [Google Scholar] [CrossRef]
- Oberhauser, V.; Voolstra, O.; Bangert, A.; von Lintig, J.; Vogt, K. NinaB combines carotenoid oxygenase and retinoid isomerase activity in a single polypeptide. Proc. Natl. Acad. Sci. USA 2008, 105, 19000–19005. [Google Scholar] [CrossRef]
- Dewett, D.; Lam-Kamath, K.; Poupault, C.; Khurana, H.; Rister, J. Mechanisms of vitamin A metabolism and deficiency in the mammalian and fly visual system. Dev. Biol 2021, 476, 68–78. [Google Scholar] [CrossRef]
- Wang, X.; Wang, T.; Jiao, Y.; von Lintig, J.; Montell, C. Requirement for an enzymatic visual cycle in Drosophila. Curr. Biol. 2010, 20, 93–102. [Google Scholar] [CrossRef]
- Parker, R.O.; Crouch, R.K. Retinol dehydrogenases (RDHs) in the visual cycle. Exp. Eye Res. 2010, 91, 788–792. [Google Scholar] [CrossRef] [PubMed]
- Boulton, M.; Rozanowska, M.; Rozanowski, B. Retinal photodamage. J. Photochem. Photobiol. B-Biol. 2001, 64, 144–161. [Google Scholar] [CrossRef] [PubMed]
- Tsukahara, N.; Tani, Y.; Kikuchi, H.; Sugita, S. Light transmission of the ocular media in birds and mammals. J. Vet. Med. Sci. 2014, 76, 93–95. [Google Scholar] [CrossRef] [PubMed]
- True, J.R. Insect melanism: The molecules matter. Trends Ecol. Evol. 2003, 18, 640–647. [Google Scholar] [CrossRef]
- Futahashi, R.; Banno, Y.; Fujiwara, H. Caterpillar color patterns are determined by a two-phase melanin gene prepatterning process: New evidence from tan and laccase2. Evol. Dev. 2010, 12, 157–167. [Google Scholar] [CrossRef]
- Riedel, F.; Vorkel, D.; Eaton, S. Megalin-dependent Yellow endocytosis restricts melanization in the Drosophila cuticle. Development 2011, 138, 149–158. [Google Scholar] [CrossRef]
- Matsuoka, Y.; Monteiro, A. Melanin pathway genes regulate color and morphology of butterfly wing scales. Cell Rep. 2018, 24, 56–65. [Google Scholar] [CrossRef]
- Ye, Y.X.; Pan, P.L.; Kang, D.; Lu, J.B.; Zhang, C.X. The multicopper oxidase gene family in the brown planthopper, Nilaparvata lugens. Insect Biochem. Mol. Biol. 2015, 63, 124–132. [Google Scholar] [CrossRef]
- Lemonds, T.R.; Liu, J.; Popadic, A. The contribution of the melanin pathway to overall body pigmentation during ontogenesis of Periplaneta americana. Insect Sci. 2016, 23, 513–519. [Google Scholar] [CrossRef]
- Niu, B.L.; Shen, W.F.; Liu, Y.; Weng, H.B.; He, L.H.; Mu, J.J.; Meng, Z.Q. Cloning and RNAi-mediated functional characterization of MaLac2 of the pine sawyer, Monochamus alternatus. Insect Mol. Biol. 2008, 17, 303–312. [Google Scholar] [CrossRef]
- Pentzold, S.; Grabe, V.; Ogonkov, A.; Schmidt, L.; Boland, W.; Burse, A. Silencing cuticular pigmentation genes enables RNA FISH in intact insect appendages. J. Exp. Biol. 2018, 221, jeb185710. [Google Scholar] [CrossRef] [PubMed]
- Bai, T.; Pei, X.J.; Liu, T.X.; Fan, Y.; Zhang, S.Z. Melanin synthesis genes BgTH and BgDdc affect body color and cuticle permeability in Blattella germanica. Insect Sci. 2022, 29, 1552–1568. [Google Scholar] [CrossRef] [PubMed]
- Sarna, T.; Dulȩba, A.; Korytowski, W.; Swartz, H. Interaction of melanin with oxygen. Arch. Biochem. Biophys. 1980, 200, 140–148. [Google Scholar] [CrossRef]
- Felix, C.C.; Hyde, J.S.; Sarna, T.; Sealy, R.C. Melanin photoreactions in aerated media: Electron spin resonance evidence for production of superoxide and hydrogen peroxide. Biochem. Biophys. Res. Commun. 1978, 84, 335–341. [Google Scholar] [CrossRef] [PubMed]
- Wilson, K.; Cotter, S.; Reeson, A.; Pell, J. Melanin and disease resistance in insects. Ecol. Lett. 2008, 4, 637–649. [Google Scholar] [CrossRef]
- Ortonne, J.P. Photoprotective properties of skin melanin. Br. J. Dermatol. 2002, 146, 7–10. [Google Scholar] [CrossRef] [PubMed]
- Riley, P.A. Melanin. Int. J. Biochem. Cell Biol. 1997, 29, 1235–1239. [Google Scholar] [CrossRef]
- Galván, I.; Solano, F. Melanin chemistry and the ecology of stress. Physiol. Biochem. Zool. 2015, 88, 352–355. [Google Scholar] [CrossRef]
- Vences, M.; Galán, P.; Vieites, D.; Puente, M.; Oetter, K.; Wanke, S. Field body temperatures and heating rates in a montane frog population: The importance of black dorsal pattern for thermoregulation. Ann. Zool. Fenn. 2002, 39, 209–220. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Su, M.; Yuan, F.; Li, T.; Wei, C. A Non-Gradual Development Process of Cicada Eyes at the End of the Fifth-Instar Nymphal Stage to Obtain Visual Ability. Insects 2022, 13, 1170. https://doi.org/10.3390/insects13121170
Su M, Yuan F, Li T, Wei C. A Non-Gradual Development Process of Cicada Eyes at the End of the Fifth-Instar Nymphal Stage to Obtain Visual Ability. Insects. 2022; 13(12):1170. https://doi.org/10.3390/insects13121170
Chicago/Turabian StyleSu, Minjing, Feimin Yuan, Tiantian Li, and Cong Wei. 2022. "A Non-Gradual Development Process of Cicada Eyes at the End of the Fifth-Instar Nymphal Stage to Obtain Visual Ability" Insects 13, no. 12: 1170. https://doi.org/10.3390/insects13121170
APA StyleSu, M., Yuan, F., Li, T., & Wei, C. (2022). A Non-Gradual Development Process of Cicada Eyes at the End of the Fifth-Instar Nymphal Stage to Obtain Visual Ability. Insects, 13(12), 1170. https://doi.org/10.3390/insects13121170





