Reference Genome Sequences of the Oriental Armyworm, Mythimna separata (Lepidoptera: Noctuidae)
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Preparation and Sequencing
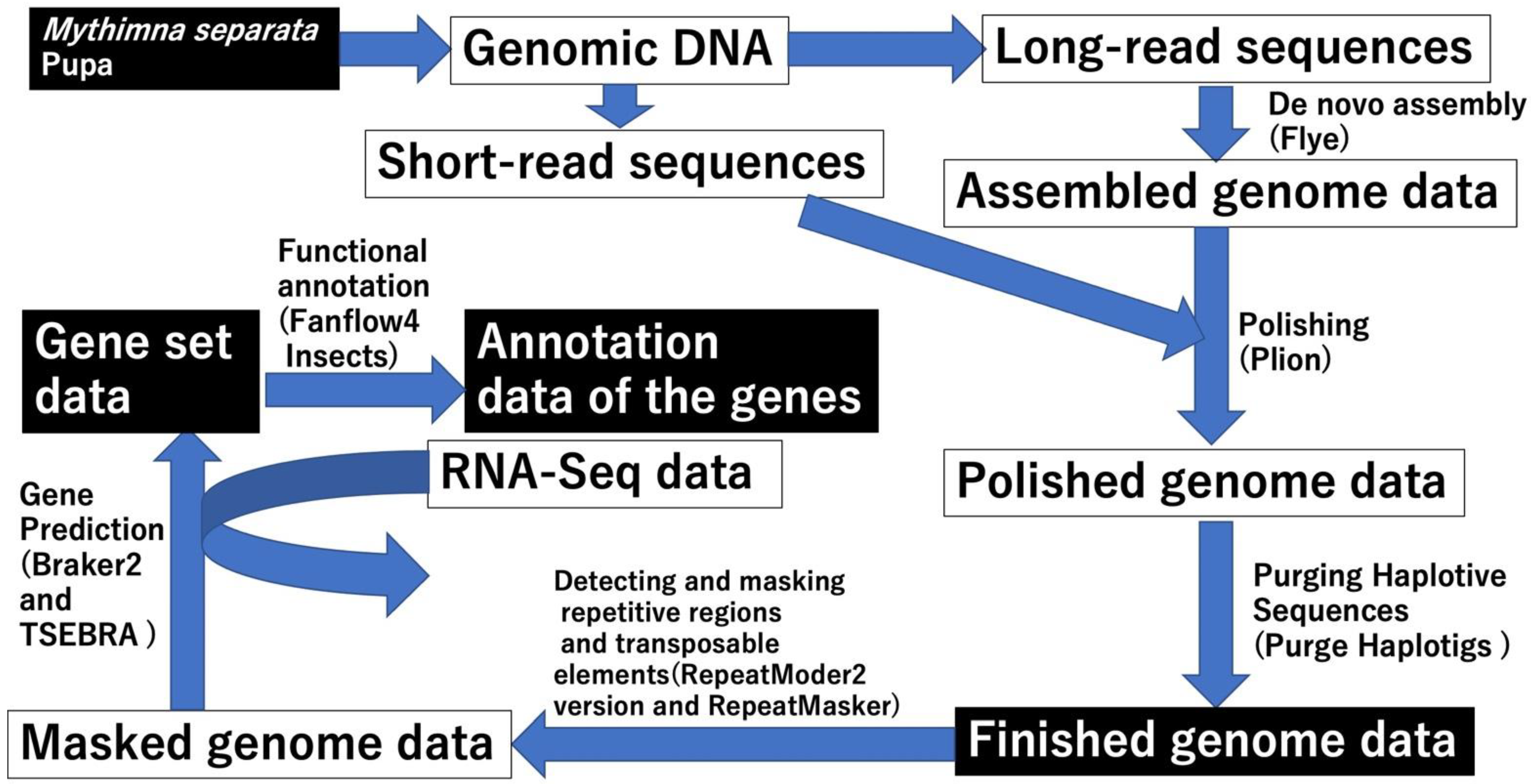
2.2. Genome Assembly and Gene Prediction
2.3. Search for and Sequence Analysis of Immune-Related Genes in M. separata
3. Results
3.1. Construction of the Genome Sequences of M. separata
3.2. Search for Repetitive Region and Gene Prediction
3.3. Immune-Related Genes in M. Separata
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Goergen, G.; Kumar, P.L.; Sankung, S.B.; Togola, A.; Tamò, M. First Report of Outbreaks of the Fall Armyworm Spodoptera frugiperda (J E Smith) (Lepidoptera, Noctuidae), a New Alien Invasive Pest in West and Central Africa. PLoS ONE 2016, 11, e0165632. [Google Scholar] [CrossRef] [PubMed]
- Cheng, T.; Wu, J.; Wu, Y.; Chilukuri, R.V.; Huang, L.; Yamamoto, K.; Feng, L.; Li, W.; Chen, Z.; Guo, H.; et al. Genomic Adaptation to Polyphagy and Insecticides in a Major East Asian Noctuid Pest. Nat. Ecol. Evol. 2017, 1, 1747–1756. [Google Scholar] [CrossRef] [PubMed]
- Sharma, H.C.; Davies, J.C. The Oriental Armyworm, Mythimna separata (Wlk.). Distribution, Biology and Control: A Literature Review. Orient. Armyworm Myth. Sep. Wlk Distrib. Biol. Control Lit. Rev. 1983, 59, 1–24. [Google Scholar]
- Hiruma, K.; Riddiford, L.M. Developmental Expression of MRNAs for Epidermal and Fat Body Proteins and Hormonally Regulated Transcription Factors in the Tobacco Hornworm, Manduca sexta. J. Insect Physiol. 2010, 56, 1390–1395. [Google Scholar] [CrossRef]
- Kanost, M.R.; Jiang, H.; Yu, X.-Q. Innate Immune Responses of a Lepidopteran Insect, Manduca sexta. Immunol. Rev. 2004, 198, 97–105. [Google Scholar] [CrossRef]
- Mita, K.; Kasahara, M.; Sasaki, S.; Nagayasu, Y.; Yamada, T.; Kanamori, H.; Namiki, N.; Kitagawa, M.; Yamashita, H.; Yasukochi, Y. The Genome Sequence of Silkworm, Bombyx mori. DNA Res. 2004, 11, 27–35. [Google Scholar] [CrossRef]
- Xia, Q.; Zhou, Z.; Lu, C.; Cheng, D.; Dai, F.; Li, B.; Zhao, P.; Zha, X.; Cheng, T.; Chai, C. A Draft Sequence for the Genome of the Domesticated Silkworm (Bombyx mori). Science 2004, 306, 1937–1940. [Google Scholar]
- The International Silkworm Genome Consortium The Genome of a Lepidopteran Model Insect, the Silkworm Bombyx mori. Insect Biochem. Mol. Biol. 2008, 38, 1036–1045. [CrossRef]
- Kawamoto, M.; Jouraku, A.; Toyoda, A.; Yokoi, K.; Minakuchi, Y.; Katsuma, S.; Fujiyama, A.; Kiuchi, T.; Yamamoto, K.; Shimada, T. High-Quality Genome Assembly of the Silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 2019, 107, 53–62. [Google Scholar] [CrossRef]
- Gouin, A.; Bretaudeau, A.; Nam, K.; Gimenez, S.; Aury, J.-M.; Duvic, B.; Hilliou, F.; Durand, N.; Montagné, N.; Darboux, I.; et al. Two Genomes of Highly Polyphagous Lepidopteran Pests (Spodoptera frugiperda, Noctuidae) with Different Host-Plant Ranges. Sci. Rep. 2017, 7, 11816. [Google Scholar] [CrossRef]
- Kakumani, P.K.; Malhotra, P.; Mukherjee, S.K.; Bhatnagar, R.K. A Draft Genome Assembly of the Army Worm, Spodoptera frugiperda. Genomics 2014, 104, 134–143. [Google Scholar] [CrossRef] [PubMed]
- Simon, S.; Breeschoten, T.; Jansen, H.J.; Dirks, R.P.; Schranz, M.E.; Ros, V.I.D. Genome and Transcriptome Analysis of the Beet Armyworm Spodoptera exigua Reveals Targets for Pest Control. G3 Genes|Genomes|Genetics 2021, 11, jkab311. [Google Scholar] [CrossRef]
- Ward, C.M.; Perry, K.D.; Baker, G.; Powis, K.; Heckel, D.G.; Baxter, S.W. A Haploid Diamondback Moth (Plutella xylostella L.) Genome Assembly Resolves 31 Chromosomes and Identifies a Diamide Resistance Mutation. Insect Biochem. Mol. Biol. 2021, 138, 103622. [Google Scholar] [CrossRef] [PubMed]
- You, M.; Yue, Z.; He, W.; Yang, X.; Yang, G.; Xie, M.; Zhan, D.; Baxter, S.W.; Vasseur, L.; Gurr, G.M.; et al. A Heterozygous Moth Genome Provides Insights into Herbivory and Detoxification. Nat. Genet. 2013, 45, 220–225. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Yang, X.; Tetreau, G.; Song, X.; Coutu, C.; Hegedus, D.; Blissard, G.; Fei, Z.; Wang, P. A High-Quality Chromosome-Level Genome Assembly of a Generalist Herbivore, Trichoplusia ni. Mol. Ecol. Resour. 2019, 19, 485–496. [Google Scholar] [CrossRef]
- Kanost, M.R.; Arrese, E.L.; Cao, X.; Chen, Y.-R.; Chellapilla, S.; Goldsmith, M.R.; Grosse-Wilde, E.; Heckel, D.G.; Herndon, N.; Jiang, H.; et al. Multifaceted Biological Insights from a Draft Genome Sequence of the Tobacco Hornworm Moth, Manduca sexta. Insect Biochem. Mol. Biol. 2016, 76, 118–147. [Google Scholar] [CrossRef]
- Gershman, A.; Romer, T.G.; Fan, Y.; Razaghi, R.; Smith, W.A.; Timp, W. De Novo Genome Assembly of the Tobacco Hornworm Moth (Manduca sexta). G3 Genes|Genomes|Genetics 2021, 11, jkaa047. [Google Scholar] [CrossRef]
- Liu, G.; Chang, Z.; Chen, L.; He, J.; Dong, Z.; Yang, J.; Lu, S.; Zhao, R.; Wan, W.; Ma, G.; et al. Genome Size Variation in Butterflies (Insecta, Lepidotera, Papilionoidea): A Thorough Phylogenetic Comparison. Syst. Entomol. 2020, 45, 571–582. [Google Scholar] [CrossRef]
- Lemaitre, B.; Hoffmann, J. The Host Defense of Drosophila melanogaster. Annu. Rev. Immunol. 2007, 25, 697–743. [Google Scholar] [CrossRef]
- Oda, Y.; Matsumoto, H.; Kurakake, M.; Ochiai, M.; Ohnishi, A.; Hayakawa, Y. Adaptor Protein Is Essential for Insect Cytokine Signaling in Hemocytes. Proc. Natl. Acad. Sci. USA 2010, 107, 15862–15867. [Google Scholar] [CrossRef]
- Ishihara, T.; Maruyama, Y.; Furukawa, S. Gene Expression and Molecular Characterization of a Novel C-Type Lectin, Encapsulation Promoting Lectin (EPL), in the Rice Armyworm, Mythimna separata. Insect Biochem. Mol. Biol. 2017, 89, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Tian, M.; Dong, Y.; Ren, C.; Du, Y.; Hu, J. The C-Type Lectin IML-10 Promotes Hemocytic Encapsulation by Enhancing Aggregation of Hemocytes in the Asian Corn Borer Ostrinia furnacalis. Insect Biochem. Mol. Biol. 2020, 118, 103314. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Liu, F.-F.; Fu, L.-Q.; Liu, Z.; Zhang, W.-T.; Wang, Q.; Rao, X.-J. Identification of 35 C-Type Lectins in the Oriental Armyworm, Mythimna separata (Walker). Insects 2021, 12, 559. [Google Scholar] [CrossRef] [PubMed]
- Harvey, J.A.; Poelman, E.H.; Tanaka, T. Intrinsic Inter- and Intraspecific Competition in Parasitoid Wasps. Annu. Rev. Entomol. 2013, 58, 333–351. [Google Scholar] [CrossRef]
- Suzuki, M.; Tanaka, T. Development of Meteorus Pulchricornis and Regulation of Its Noctuid Host, Pseudaletia separata. J. Insect Physiol. 2007, 53, 1072–1078. [Google Scholar] [CrossRef]
- Suzuki, M.; Miura, K.; Tanaka, T. The Virus-like Particles of a Braconid Endoparasitoid Wasp, Meteorus Pulchricornis, Inhibit Hemocyte Spreading in Its Noctuid Host, Pseudaletia separata. J. Insect Physiol. 2008, 54, 1015–1022. [Google Scholar] [CrossRef]
- Yamashita, K.; Zhang, K.; Ichiki, R.T.; Nakamura, S.; Furukawa, S. Novel Host Immune Evasion Strategy of the Endoparasitoid Drino inconspicuoides. Bull. Entomol. Res. 2019, 109, 643–648. [Google Scholar] [CrossRef]
- Schwier, N.; Zhang, K.; Nakamura, S.; Furukawa, S. Larvae of the Tachinid Fly, Drino inconspicuoides (Diptera: Tachinidae), Suppress Melanization in Host Lepidopteran Insects. J. Asia-Pac. Entomol. 2021, 24, 1050–1054. [Google Scholar] [CrossRef]
- Yokoi, K.; Sano, T.; Suzuki, M.; Tanaka, T.; Minakuchi, C.; Miura, K. The Major Constituents of the Venom Gland of a Braconid Endoparasitoid, Meteorus pulchricornis (Hymenoptera: Braconidae). Appl. Entomol. Zool. 2017, 52, 271–285. [Google Scholar] [CrossRef]
- Yokoi, K.; Kato, Y.; Suzuki, M.; Miura, K. Molecular Cloning and Functional Analyses of an Adhesion Molecule, Neuroglian, in Mythimna separata (Lepidoptera: Noctuidae). Eur. J. Entomol. 2018, 115, 157–166. [Google Scholar] [CrossRef]
- Kamezaki, M.; Yokoi, K.; Miura, K. RNA Interference Mediated Knockdown of an Inhibitor of Apoptosis Protein Induces Apoptosis in Mythimna separata (Lepidoptera: Noctuidae). Eur. J. Entomol. 2018, 115, 223–231. [Google Scholar] [CrossRef]
- Takabayashi, J.; Noda, T.; Takahashi, S. Effect of Kairomones in the Host Searching Behavior of Apanteles kariyai WATANABE (Hymenoptera : Braconidae), a Parasitoid of the Common Armyworm, Pseudaletia separata WALKER (Lepidoptera : Noctuidae). : I. Presence of Arresting Stimulants Produced by the Host Larvae. Appl. Entomol. Zool. 1985, 20, 484–489. [Google Scholar] [CrossRef]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of Long, Error-Prone Reads Using Repeat Graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef]
- Li, H. Minimap2: Pairwise Alignment for Nucleotide Sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve Years of SAMtools and BCFtools. GigaScience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Manni, M.; Berkeley, M.R.; Seppey, M.; Simão, F.A.; Zdobnov, E.M. BUSCO Update: Novel and Streamlined Workflows along with Broader and Deeper Phylogenetic Coverage for Scoring of Eukaryotic, Prokaryotic, and Viral Genomes. Mol. Biol. Evol. 2021, 38, 4647–4654. [Google Scholar] [CrossRef]
- Flynn, J.M.; Hubley, R.; Goubert, C.; Rosen, J.; Clark, A.G.; Feschotte, C.; Smit, A.F. RepeatModeler2 for Automated Genomic Discovery of Transposable Element Families. Proc. Natl. Acad. Sci. USA 2020, 117, 9451–9457. [Google Scholar] [CrossRef]
- Smit, A.; Hubley, R.; Green, P. RepeatMasker Open-4.0. 2013-2015. Available online: http://www.repeatmasker.org (accessed on 1 September 2022).
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-Based Genome Alignment and Genotyping with HISAT2 and HISAT-Genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Brůna, T.; Hoff, K.J.; Lomsadze, A.; Stanke, M.; Borodovsky, M. BRAKER2: Automatic Eukaryotic Genome Annotation with GeneMark-EP+ and AUGUSTUS Supported by a Protein Database. NAR Genom. Bioinform. 2021, 3, lqaa108. [Google Scholar] [CrossRef]
- Gabriel, L.; Hoff, K.J.; Brůna, T.; Borodovsky, M.; Stanke, M. TSEBRA: Transcript Selector for BRAKER. BMC Bioinform. 2021, 22, 566. [Google Scholar] [CrossRef] [PubMed]
- Bono, H.; Sakamoto, T.; Kasukawa, T.; Tabunoki, H. Systematic Functional Annotation Workflow for Insects. Insects 2022, 13, 586. [Google Scholar] [CrossRef] [PubMed]
- Moore, A.D.; Held, A.; Terrapon, N.; Weiner, J., 3rd; Bornberg-Bauer, E. DoMosaics: Software for Domain Arrangement Visualization and Domain-Centric Analysis of Proteins. Bioinformatics 2014, 30, 282–283. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Teufel, F.; Almagro Armenteros, J.J.; Johansen, A.R.; Gíslason, M.H.; Pihl, S.I.; Tsirigos, K.D.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 6.0 Predicts All Five Types of Signal Peptides Using Protein Language Models. Nat. Biotechnol. 2022, 40, 1023–1025. [Google Scholar] [CrossRef]
- Hallgren, J.; Tsirigos, K.D.; Pedersen, M.D.; Armenteros, J.J.A.; Marcatili, P.; Nielsen, H.; Krogh, A.; Winther, O. DeepTMHMM Predicts Alpha and Beta Transmembrane Proteins Using Deep Neural Networks. bioRxiv 2022. [Google Scholar] [CrossRef]
- Roach, M.J.; Schmidt, S.A.; Borneman, A.R. Purge Haplotigs: Allelic Contig Reassignment for Third-Gen Diploid Genome Assemblies. BMC Bioinform. 2018, 19, 460. [Google Scholar] [CrossRef]
- De Gregorio, E.; Spellman, P.T.; Rubin, G.M.; Lemaitre, B. Genome-Wide Analysis of the Drosophila Immune Response by Using Oligonucleotide Microarrays. Proc. Natl. Acad. Sci. USA 2001, 98, 12590–12595. [Google Scholar] [CrossRef]
- Yi, H.-Y.; Chowdhury, M.; Huang, Y.-D.; Yu, X.-Q. Insect Antimicrobial Peptides and Their Applications. Appl. Microbiol. Biotechnol. 2014, 98, 5807–5822. [Google Scholar] [CrossRef]
- Brown, G.D.; Willment, J.A.; Whitehead, L. C-Type Lectins in Immunity and Homeostasis. Nat. Rev. Immunol. 2018, 18, 374–389. [Google Scholar] [CrossRef]
- Xia, X.; You, M.; Rao, X.-J.; Yu, X.-Q. Insect C-Type Lectins in Innate Immunity. Dev. Comp. Immunol. 2018, 83, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Yokoi, K.; Kimura, K.; Bono, H. Revealing Landscapes of Transposable Elements in Apis Species by Meta-Analysis. Insects 2022, 13, 698. [Google Scholar] [CrossRef] [PubMed]
- Misof, B.; Liu, S.; Meusemann, K.; Peters, R.S.; Donath, A.; Mayer, C.; Frandsen, P.B.; Ware, J.; Flouri, T.; Beutel, R.G.; et al. Phylogenomics Resolves the Timing and Pattern of Insect Evolution. Science 2014, 346, 763–767. [Google Scholar] [CrossRef] [PubMed]
- Zou, Z.; Evans, J.D.; Lu, Z.; Zhao, P.; Williams, M.; Sumathipala, N.; Hetru, C.; Hultmark, D.; Jiang, H. Comparative Genomic Analysis of the Tribolium Immune System. Genome Biol. 2007, 8, R177. [Google Scholar] [CrossRef] [PubMed]
- Yokoi, K.; Uchiyama, H.; Wakamiya, T.; Yoshiyama, M.; Takahashi, J.-I.; Nomura, T.; Furukawa, T.; Yajima, S.; Kimura, K. The Draft Genome Sequence of the Japanese Honey Bee, Apis cerana japonica (Hymenoptera: Apidae). Eur. J. Entomol. 2018, 115, 650–657. [Google Scholar] [CrossRef]
- Evans, J.D.; Aronstein, K.; Chen, Y.P.; Hetru, C.; Imler, J.-L.; Jiang, H.; Kanost, M.; Thompson, G.J.; Zou, Z.; Hultmark, D. Immune Pathways and Defence Mechanisms in Honey Bees Apis mellifera. Insect Mol. Biol. 2006, 15, 645–656. [Google Scholar] [CrossRef]
- Stokes, B.A.; Yadav, S.; Shokal, U.; Smith, L.C.; Eleftherianos, I. Bacterial and Fungal Pattern Recognition Receptors in Homologous Innate Signaling Pathways of Insects and Mammals. Front. Microbiol. 2015, 6, 19. [Google Scholar] [CrossRef]
- Bao, Y.-Y.; Qu, L.-Y.; Zhao, D.; Chen, L.-B.; Jin, H.-Y.; Xu, L.-M.; Cheng, J.-A.; Zhang, C.-X. The Genome- and Transcriptome-Wide Analysis of Innate Immunity in the Brown Planthopper, Nilaparvata lugens. BMC Genom. 2013, 14, 160. [Google Scholar] [CrossRef]
- Nishide, Y.; Kageyama, D.; Yokoi, K.; Jouraku, A.; Tanaka, H.; Futahashi, R.; Fukatsu, T. Functional Crosstalk across IMD and Toll Pathways: Insight into the Evolution of Incomplete Immune Cascades. Proc. R. Soc. B Biol. Sci. 2019, 286, 20182207. [Google Scholar] [CrossRef] [PubMed]
- Koyama, H.; Kato, D.; Minakuchi, C.; Tanaka, T.; Yokoi, K.; Miura, K. Peptidoglycan Recognition Protein Genes and Their Roles in the Innate Immune Pathways of the Red Flour Beetle, Tribolium castaneum. J. Invertebr. Pathol. 2015, 132, 86–100. [Google Scholar] [CrossRef]
- Yokoi, K.; Koyama, H.; Minakuchi, C.; Tanaka, T.; Miura, K. Antimicrobial Peptide Gene Induction, Involvement of Toll and IMD Pathways and Defense against Bacteria in the Red Flour Beetle, Tribolium castaneum. Results Immunol. 2012, 2, 72–82. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.-Q.; Kanost, M.R. Manduca Sexta Lipopolysaccharide-Specific Immulectin-2 Protects Larvae from Bacterial Infection. Dev. Comp. Immunol. 2003, 27, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, A.; Yoshizawa, Y.; Tanaka, S.; Kitami, M.; Sato, R. Extra- and Intracellular Signaling Pathways Regulating Nodule Formation by Hemocytes of the Silkworm, Bombyx mori (Lepidoptera: Bombicidae). J. Insect Biotechnol. Sericology 2011, 80, 49–56. [Google Scholar] [CrossRef]
- Sawa, T.; Tanaka, T.; Kato, Y.; Nakamatsu, Y. Cky811 Protein Expressed by Polydnavirus and Venom Gland of Cotesia kariyai Regulates the Host Mythimna separata Larvae Immune Response Function of C-Type Lectin Responsible for Foreign Substance Recognition Which Suppresses Its Melanization and Encapsulation. Arch. Insect Biochem. Physiol. 2021, 107, e21786. [Google Scholar] [CrossRef] [PubMed]

| Genome Sequence Name | Polished Genome Data | Finished Genome Data (Reference Genome Data) |
|---|---|---|
| Total length (bp) | 842,875,911 | 681,943,315 |
| Contig number | 991 | 569 |
| Contig N50 (bp) | 2,220,816 | 2,745,150 |
| Largest contig length (bp) | 12,409,004 | 12,409,004 |
| Average of contig length (bp) | 850,530.69 | 1,198,494.40 |
| Genome Sequence Name (BUSCO Data Set) | Polished Genome Data (eukaryota_odb10) | Finished Genome Data (eukaryota_odb10) | Finished Genome Data (lepidoptera_odb10) |
|---|---|---|---|
| Complete BUSCOs | 253 (99.2%) | 252 (98.8%) | 5213 (98.6%) |
| Complete and single-copy BUSCOs | 178 (69.8%) | 251 (98.4%) | 5185 (98.1%) |
| Complete and duplicated BUSCOs | 75 (29.4%) | 1 (0.4%) | 28 (0.5%) |
| Fragmented BUSCOs | 1 (0.4%) | 2 (0.8%) | 20 (0.4%) |
| Missing BUSCOs | 1 (0.4%) | 1 (0.4%) | 53 (1.0%) |
| Total BUSCO groups searched | 255 | 255 | 5286 |
| Total Length: | 681,943,315 bp | ||
| Bases Masked: | 317,706,294 bp (46.59%) | ||
| Number of Elements * | Length Occupied | Percentage of Sequence | |
| Retroelements | 483,943 | 102,500,601 bp | 15.03% |
| SINEs: | 69 | 3659 bp | 0.00% |
| Penelope | 0 | 0 bp | 0.00% |
| LINEs: | 466,443 | 94,162,645 bp | 13.81% |
| CRE/SLACS | 11,265 | 2,482,075 bp | 0.36% |
| L2/CR1/Rex | 47,274 | 15,582,975 bp | 2.29% |
| R1/LOA/Jockey | 125,283 | 28,136,850 bp | 4.13% |
| R2/R4/NeSL | 8079 | 2,985,258 bp | 0.44% |
| RTE/Bov-B | 184,015 | 31,548,956 bp | 4.63% |
| L1/CIN4 | 0 | 0 bp | 0.00% |
| LTR elements: | 17,431 | 8,334,297 bp | 1.22% |
| BEL/Pao | 1963 | 3,457,738 bp | 0.51% |
| Ty1/Copia | 2715 | 1,358,093 bp | 0.20% |
| Gypsy/DIRS1 | 6472 | 2,769,678 bp | 0.41% |
| Retroviral | 160 | 76,839 bp | 0.01% |
| DNA transposons | 80,267 | 20,701,313 bp | 3.04% |
| hobo-Activator | 6121 | 1,085,735 bp | 0.16% |
| Tc1-IS630-Pogo | 24,800 | 10,838,060 bp | 1.59% |
| En-Spm | 0 | 0 bp | 0.00% |
| MuDR-IS905 | 0 | 0 bp | 0.00% |
| PiggyBac | 166 | 236,668 bp | 0.03% |
| Tourist/Harbinger | 2055 | 550,091 bp | 0.08% |
| Other (Mirage, P-element, Transib) | 110 | 177,033 bp | 0.03% |
| Rolling-circles | 281,101 | 46,800,852 bp | 6.86% |
| Unclassified: | 880,600 | 140,919,871 bp | 20.66% |
| Total interspersed repeats: | 264,121,785 bp | 38.73% | |
| Small RNA: | 204 | 23,041 bp | 0.00% |
| Satellites: | 177 | 69,651 bp | 0.01% |
| Simple repeats: | 102,349 | 5,964,291 bp | 0.87% |
| Low complexity: | 15,392 | 726,674 bp | 0.11% |
| Numbers of the Hit Proteins (among 24,453 Proteins) | Percentage of the Hit Proteins (%) | |
|---|---|---|
| H. sapience | 14,037 | 57.4039995 |
| M. musculus | 13,699 | 56.021756 |
| C. elegans | 11,235 | 45.9452828 |
| D. melanogaster | 12,879 | 52.6683842 |
| B. mori | 19,704 | 80.5790701 |
| M. sexta | 17,996 | 73.594242 |
| A. mellifera | 14,427 | 58.9988958 |
| T. castaneum | 16,496 | 67.4600254 |
| Unigene | 16,086 | 65.7833395 |
| Pfam | 13,244 | 54.1610436 |
| Total Number of CTLs | Single CTLD * | Dual CTLD (Immune-Lectin Group or Dual-CTLD Type Lectins) * | More than Three CTLD | CTL-X Group * | Signal Peptide (SignalP) | Signal Peptide (DeepTMHMM) | Transmembrane Domain |
|---|---|---|---|---|---|---|---|
| 105 | 12 | 81 | 7 | 5 | 77 | 84 | 4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yokoi, K.; Furukawa, S.; Zhou, R.; Jouraku, A.; Bono, H. Reference Genome Sequences of the Oriental Armyworm, Mythimna separata (Lepidoptera: Noctuidae). Insects 2022, 13, 1172. https://doi.org/10.3390/insects13121172
Yokoi K, Furukawa S, Zhou R, Jouraku A, Bono H. Reference Genome Sequences of the Oriental Armyworm, Mythimna separata (Lepidoptera: Noctuidae). Insects. 2022; 13(12):1172. https://doi.org/10.3390/insects13121172
Chicago/Turabian StyleYokoi, Kakeru, Seiichi Furukawa, Rui Zhou, Akiya Jouraku, and Hidemasa Bono. 2022. "Reference Genome Sequences of the Oriental Armyworm, Mythimna separata (Lepidoptera: Noctuidae)" Insects 13, no. 12: 1172. https://doi.org/10.3390/insects13121172
APA StyleYokoi, K., Furukawa, S., Zhou, R., Jouraku, A., & Bono, H. (2022). Reference Genome Sequences of the Oriental Armyworm, Mythimna separata (Lepidoptera: Noctuidae). Insects, 13(12), 1172. https://doi.org/10.3390/insects13121172







