Simple Summary
Gene rearrangement is an additional type of data to support relationships of taxa, with rearrangement synapomorphies identified across multiple orders and at many different taxonomic levels. The concept to use mitochondrial gene rearrangements as phylogenetic markers has been proposed since the mid-1980s, the synapomorphic gene rearrangements have been identified from many lineages. However, mitochondrial gene rearrangement has never been observed in the non-biting midges (Diptera: Chironomidae). Here, seven new mitogenomes of the genus Stenochironomus were sequenced and analyzed. Coupled with published data, phylogenetic analyses were performed within Chironominae. The present study showed that mitogenomes of Stenochironomus are showing a higher A+T bias than other chironomid species. A synapomorphic gene rearrangement that the gene order rearranges from trnI-trnQ-trnM to trnI-trnM-trnQ was identified within Stenochironomus, which is the first instance of mitochondrial gene rearrangement discovered in the Chironomidae. The monophyly of the genus Stenochironomus was strongly supported by mitogenomes. Our study provides new insights into the mitochondrial gene order of Chironomidae, and provides a valuable resource for understanding synapomorphic gene rearrangements.
Abstract
(1) Background: Gene rearrangement of mitochondrial genome, especially those with phylogenetic signals, has long fascinated evolutionary biologists. The synapomorphic gene rearrangements have been identified across multiple orders and at many different taxonomic levels, supporting the monophyletic or systematic relationships of related lineages. However, mitochondrial gene rearrangement has never been observed in the non-biting midges (Diptera: Chironomidae); (2) methods: in this study, the complete mitogenomes of seven Stenochironomus species were sequenced and analyzed for the first time; (3) results: each mitogenome of Stenochironomus contains 37 typical genes and a control region. The whole mitogenomes of Stenochironomus species exhibit a higher A+T bias than other published chironomid species. The gene order rearranges from trnI-trnQ-trnM to trnI-trnM-trnQ in all the seven mitogenomes of Stenochironomus, which might be act as a synapomorphy of the genus, supporting the monophyletic of Stenochironomus species. In addition, another derived gene cluster: trnA-trnG-ND3-trnR exists in Stenochironomus tobaduodecimus. The derived gene orders described above are the first case of mitochondrial gene rearrangement in Chironomidae. Coupled with published data, phylogenetic relationships were reconstructed within Chironominae, and strongly supported the monophyly of Stenochironomus; (4) conclusions: our study provides new insights into the mitochondrial gene order of Chironomidae, and provides a valuable resource for understanding the synapomorphic gene rearrangements.
1. Introduction
Gene rearrangement of mitochondrial genome (mitogenome) has long fascinated evolutionary biologists, and insects are one of the most studied organisms [1,2,3]. The typical mitogenome of insect is a 14–20 kb circular molecule, containing 37 genes (13 protein-coding genes, two ribosomal RNA genes, and 22 transfer RNA genes) and a control region on a single chromosome [4] characterized by the small genome size, maternal inheritance, low sequence recombination, and fast evolution rates [1,5,6]. Benefited by the high-throughput sequencing technology, nucleotide sequences of mitogenomes are now widely used in phylogenetic and evolutionary studies [7,8,9]. Besides, gene rearrangement is an additional type of data to support relationships of taxa, with rearrangement synapomorphies identified across multiple orders and at many different taxonomic levels [1,10,11]. Since the concept of using mitochondrial gene rearrangements as phylogenetic markers has been proposed in the mid-1980s [3], the synapomorphic gene rearrangements have been identified in many taxa, supporting the monophyletic or systematic relationships of related lineages [9,12,13]. In insect mitogenomes, patterns of gene arrangement are usually conserved within lineages [6], but gene rearrangements have also been observed involving tRNA and PCG within many orders, such as Blattodea [14], Ephemeroptera [15,16], Hemiptera [17,18], Hymenoptera [12,19], Lepidoptera [20], Mantodea [21,22], Orthoptera [23,24], Phthiraptera [25], Psocoptera [9], and Thysanoptera [26]. For the mitogenomes of Diptera, gene rearrangements have been detected within several families, e.g., Calliphoridae [27], Cecidomyiidae [28], and Mycetophilidae [29]. Published mitogenome data of Chironomid are relatively rare compared with other insects of Diptera. Previous studies have focused on the genome organization and the application of nucleotide sequence information in phylogenetic analysis [30,31,32,33,34,35,36,37]. However, mitochondrial gene rearrangement has never been reported in the Dipteran family Chironomidae. In the current study, we found mitochondrial gene rearrangements in the genus Stenochironomus Kieffer for the first time in Chironomidae. Stenochironomus is species-diverse, and occurs in all zoogeographic regions except Antarctica, with more than 100 named species [38]. The larvae of Stenochironomus (Figure 1) are found mining leaves or immersed wood in standing and flowing waters [38]. Due to its strictly mining habit which contributes to decomposition of wood, leaves, and aquatic macrophytes, Stenochironomus is regarded as important bioindicator in freshwater ecosystems [39,40,41].

Figure 1.
Larva of Stenochironomus okialbus in immersed wood from Zhejiang, China.
Prior to this study, the mitogenomic characteristics of Stenochironomus have never been studied. We sequenced and annotated the complete mitogenomes of seven species of Stenochironomus. The mitogenomic organization, evolutionary rates, and gene rearrangement pattern within Stenochironomus were revealed. Coupled with published data, phylogenetic relationships of Chironominae were reconstructed based on mitogenomes to explore the monophyly of Stenochironomus.
2. Materials and Methods
2.1. Taxon Sampling and DNA Extraction
Seven species of Stenochironomus were used for mitogenome sequencing (Table 1). Specimens were preserved in ethanol (85% for adults, 95% for immature) and stored in a freezer at −20 °C in the College of Life Sciences at Nankai University (Tianjin, China). The total genomic DNA was the extracted thorax of adult and larva using a Qiagen DNA Blood and Tissue Kit (Qiagen, Hilden, Germany) following the manufacturer’s protocol. The DNA and vouchers of the species are deposited at the College of Life Sciences, Nankai University, Tianjin, China.

Table 1.
Taxonomic information, sampling metadata, GenBank accession numbers and references of mitochondrial genomes used in the study.
2.2. Sequencing and Mitogenome Assembly
Whole mitogenomes were sequenced individually using the Illumina NovaSeq 6000 platform with a 350-bp insert size and a paired-end 150-bp sequencing strategy at Novogene Co., Ltd. (Beijing, China). Adapter sequences and low-quality reads were trimmed with Trimmomatic 0.36 [43], and finally about two Gb of clean data were obtained for each sample. A de novo assembly was performed using IDBA-UD [44] with minimum and maximum k values of 40 and 120 bp, respectively. The partial COI sequence for each species was downloaded from GenBank, and served as the “bait” reference to acquire the targeted mitogenome sequence from the pooled sequencing file. To check the accuracy of the assembly, clean reads were mapped onto the obtained mitogenome sequences using Geneious 2020.2.1 [45].
2.3. Genome Annotation and Sequence Analyses
The MITOS2 webserver (available at http://mitos2.bioinf.uni-leipzig.de/index.py, access on 24 May 2021) was used to identify transfer RNA (tRNA) genes based on the invertebrate mitochondrial genetic code. Protein coding genes (PCGs) and ribosomal RNA (rRNA) genes were annotated by aligning with homologous regions of other published chironomid mitogenomes using Geneious 2020.2.1. Newly sequenced mitogenomes were submitted to GenBank (accession numbers: OL742440, OL742441, OL753645–753649). The CG View server V 1.0 [46] was used to draw the mitogenome maps. Nucleotide compositions of the whole mitogenome and individual genes were calculated by MEGA X [47]. The bias of the nucleotide composition was measured by AT-skew [(A-T)/(A+T)] and GC-skew [(G-C)/(G+C)]. The codon usage of PCGs were computed by MEGA. The non-synonymous substitution rate (Ka) and synonymous substitution rate (Ks) of PCGs were calculated with DnaSP 6.12.03 [48].
2.4. Phylogenetic Analyses
Phylogenetic analyses were conducted using the seven newly sequenced Stenochironomus mitogenomes and five Chironominae species available in GenBank as ingroup taxa (Table 1). One Orthocladiinae species (Rheocricotopus villiculus) was chosen as an outgroup. Individual genes of PCGs were aligned using muscle implemented in MEGA based on amino acid sequences, and then concatenated using SequenceMatrix v1.7.8 [49]. A total of three datasets were prepared for phylogenetic analyses: PCG123 (all three codon positions of the 13 PCGs), PCG12 (the 1st and 2nd codon positions of the 13 PCGs), and AA (amino acid sequences of the 13 PCGs). PartitionFinder 2.0 [50] was used to select the best partitioning scheme and best-fit substitution model. Bayesian inference (BI) and maximum likelihood (ML) methods were used for phylogenetic analyses. BI analysis was conducted using MrBayes 3.2.7a [51] with substitution model in Supplementary Table S1. Two simultaneous Markov chain Monte Carlo (MCMC) runs of 10,000,000 generations were conducted, trees were sampled every 1000 generations and the first 25% of trees discarded as burn-in. The convergence of runs was checked using Tracer 1.7 [52]. The ML analysis was performed using IQ-TREE 1.6.10 [53] with the best-fit substitution model and 1000 bootstrap replicates.
3. Results and Discussion
3.1. General Features of Stenochironomus Mitogenomes
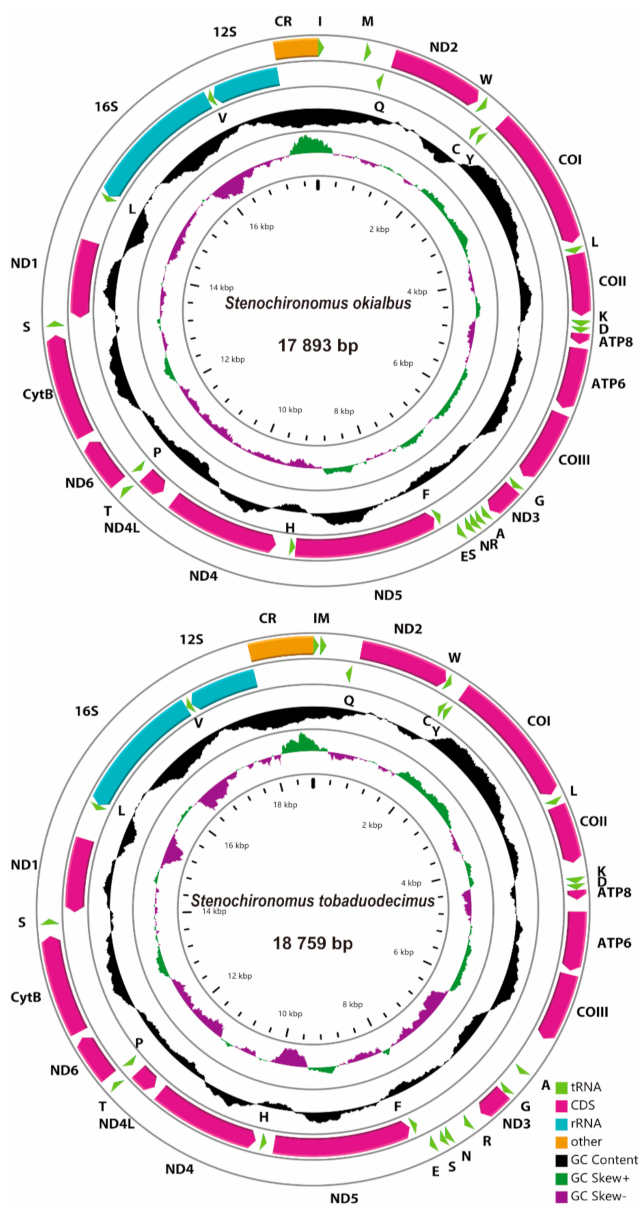
Seven mitogenomes of Stenochironomus were newly sequenced with the length range from 17,694 bp in Stenochironomus sp. 2CZ to 18,759 bp in Stenochironomus tobaduodecimus (Figure 2). Among them, the control regions of Stenochironomus gibbus and Stenochironomus sp. 2CZ failed to complete sequencing due to the complicated structure and high AT content. The entire length of mitogenomes of the Stenochironomus species is larger than other published chironomid species, mainly due to the large number of intergenic spacers [30,31,32,33]. Each mitogenome of Stenochironomus contains 37 typical genes (13 PCGs, two rRNAs, and 22 tRNAs) and one control region. Among these genes, four PCGs, eight tRNAs, and two rRNAs are coded on the minority strand (N strand), while the other genes are coded on the majority strand (J strand) (Figure 2).
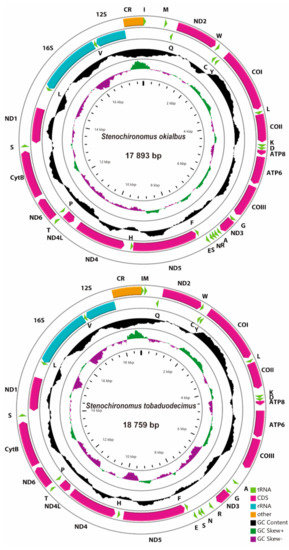
Figure 2.
Mitogenome maps of the represented species of Stenochironomus. The names of PCGs and rRNAs are indicated by standard abbreviations, while names of tRNAs are represented by a single letter abbreviation. The first circle shows the gene arrangement and arrows indicate the orientation of gene transcription. Red, blue, green, and yellow arrows refer to PCGs, rRNAs, tRNAs, and the control region, respectively. The second circle indicates the GC content, which is plotted as the deviation from the average GC content of the entire sequence. The third circle shows the GC-skew, which is plotted as the deviation from the average GC-skew of the entire sequence. The innermost circle shows the sequence length.
The whole mitogenomes of Stenochironomus are significantly biased toward A and T with the A+T content range from 81.7% in Stenochironomus tobaduodecimus and Stenochironomus sp. 1CZ to 83.6% in Stenochironomus sp. 3CZ (Table 2), showing a stronger A+T bias than other chironomid species [30,31,32,33]. Among the mitogenomes of Stenochironomus, the 3rd codon of PCGs and the control region exhibit the highest A+T content while the 1st and 2nd codons of PCGs have the lowest A+T content (Table 2). The whole mitogenomes of all the seven Stenochironomus species exhibit positive AT-skew (0.01 to 0.02) and negative GC-skew (−0.34 to −0.21), except for Stenochironomus tobaduodecimus and Stenochironomus zhengi (Table 2).

Table 2.
Nucleotide composition of mitochondrial genomes of the seven Stenochironomus species.
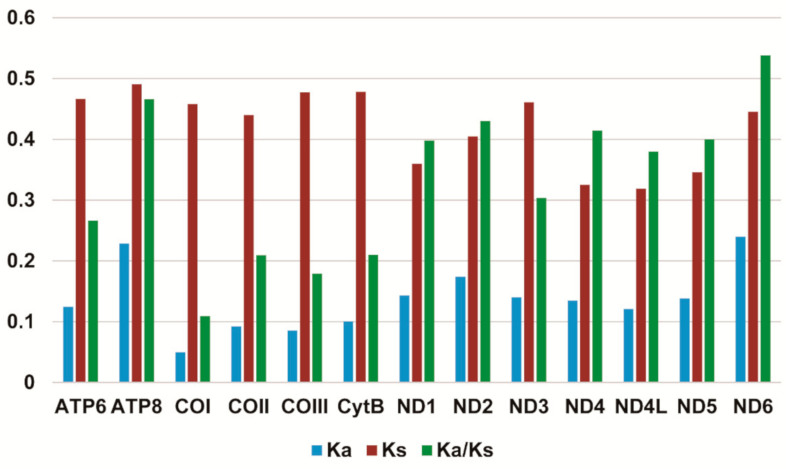
Most PCGs in Stenochironomus mitogenomes initiate with a standard start codon ATN (N represents one of four nucleotides, A, T, C, G). While the start codon of COI is TTG in Stenochironomus zhengi and Stenochironomus sp. 1CZ. The start codon of ND5 in Stenochironomus zhengi, Stenochironomus sp. 1CZ, and Stenochironomus sp. 3CZ is GTG (Supplementary Table S2). All PCGs in Stenochironomus mitogenomes end with TAA or TAG as the termination codon (Supplementary Table S2). The total codon numbers, except the termination codons among Stenochironomus mitogenomes range from 3707 in Stenochironomus sp. 1CZ to 3716 in Stenochironomus sp. 3CZ (Supplementary Table S3). The highest and lowest frequent codon families are Phe and Cys, respectively (Supplementary Figure S1), which is congruent with those of previously published chironomid species [32,33,34,35]. The Ka/Ks value (ω) is used to test for signatures of natural selection. The ω value of all PCGs in Stenochironomus mitogenomes is less than 1 (Figure 3), suggesting that they are under purifying selection. ND6 exhibits the highest ω value among the 13 PCGs of Stenochironomus mitogenomes (Figure 3), while ATP8 evolves at the fastest rate in previously published chironomid species [32,33].
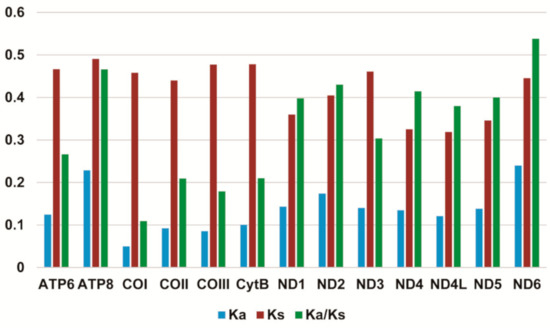
Figure 3.
Evolution rate of each PCG of the seven Stenochironomus mitogenomes. Ka refers to non-synonymous substitution rate, Ks refers to synonymous substitution rate, Ka/Ks refers to evolution rate of each PCG.
Each mitogenome of Stenochironomus contains 22 typical tRNA genes, with A+T content ranging from 83.4% to 85.1% (Table 2). The nucleotide skew of tRNA genes among Stenochironomus mitogenomes is consistent, the concatenated tRNA genes exhibit a positive AT-skew, and negative GC-skew (Table 2). The A+T content of 12S rRNA and 16S rRNA genes range from 86.1% to 89.4%, and 87.2% to 89.4%, respectively. The 12S rRNA exhibits a negative AT-skew and negative GC-skew, while the 16S rRNA exhibits a positive AT-skew and negative GC-skew in most Stenochironomus mitogenomes (Table 2).
3.2. Gene Rearrangement
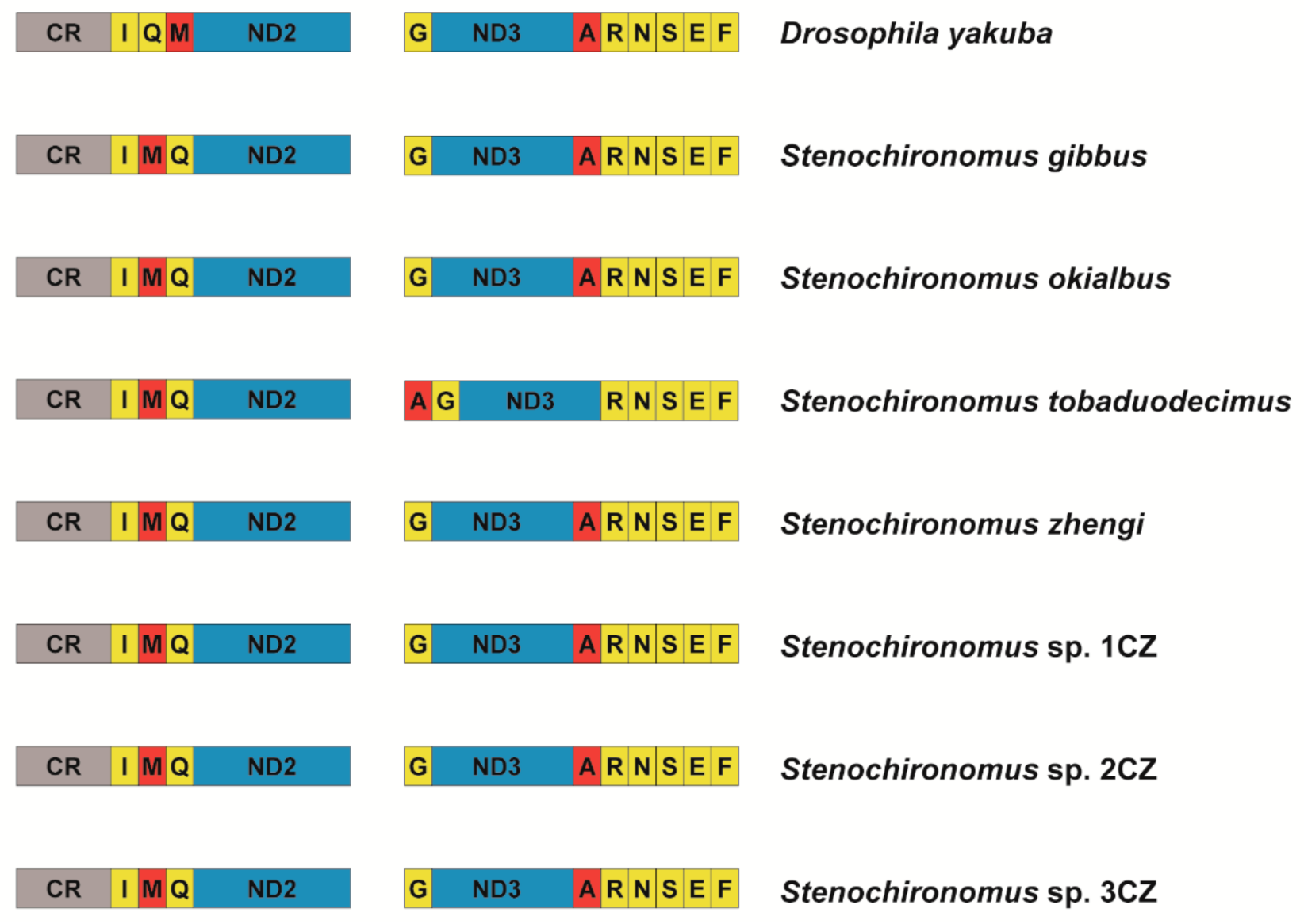
The gene arrangement of mitogenomes is conservative in most groups of Diptera [30,54,55,56]. New gene orders have only been reported in a few taxa, for example: The trnI gene inverted and transposed from the position between the control region and the ND2 gene to the block of tRNA genes between ND3 and ND5 in two gall midges [28], and the midge Arachnocampa flava has an inversion of the trnE gene [30]. Prior to this study, no examples of gene rearrangement were reported from mitogenomes of non-biting midge species. The gene order rearranges from trnI-trnQ-trnM to trnI-trnM-trnQ in all the seven Stenochironomus mitogenomes and the trnA gene moves to upstream, forming a new gene cluster: trnA-trnG-ND3-trnR in Stenochironomus tobaduodecimus (Figure 4), which is the first instance of mitochondrial gene rearrangement discovered in Chironomidae. Previous studies have shown that gene rearrangement can act as a synapomorphy and be shared within different taxonomic levels [1,11,57,58]. In this study, the gene rearrangement (trnI-trnM-trnQ) is discovered in all the seven Stenochironomus mitogenomes, which might act as a synapomorphy of the genus, supporting the monophyletic of the Stenochironomus species.
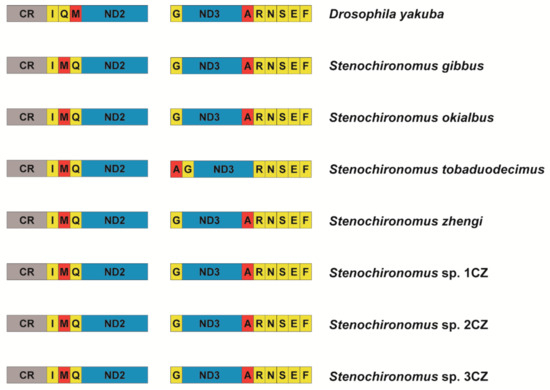
Figure 4.
Gene rearrangement of Stenochironomus mitogenomes. The names of PCGs are indicated by standard abbreviations and the names of tRNAs are represented by a single letter abbreviation.
In the insect mitogenome, the gene cluster trnI-trnM-trnQ is one of the regions with the highest probability of gene rearrangement. Many gene rearrangement events related to this region have been reported, including gene duplication [2,20,59], and gene translocation [21,60]. The derived gene order trnI-trnM-trnQ, same as in this study have been observed in 14 species belonging to 5 different orders in previous studies [2]. The gene cluster trnI-trnM-trnQ is adjacent to the control region of mitogenome, and transcription of the entire mitogenome starts from this region. The probability of rearrangement may be higher at the beginning of transcription [1,2,11]. The six-tRNA gene cluster trnA-trnR-trnN-trnS-trnE-trnF is also a gene rearrangement the hotspot region. Cases of gene rearrangement in this region have been reported in many different orders of insects [1,2].
3.3. Phylogenetic Relationships
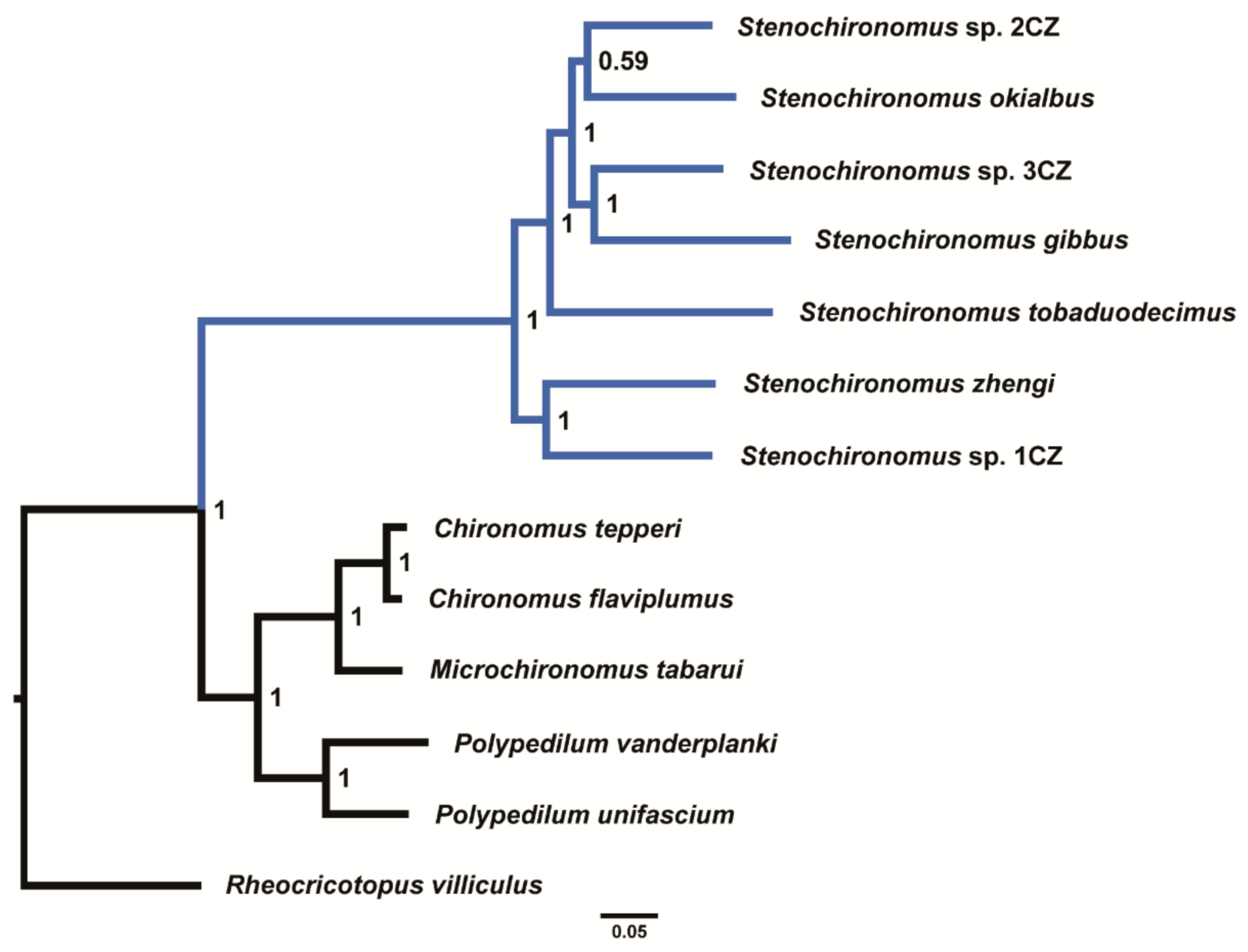
The phylogenetic analyses were performed based on the concatenated nucleotide sequences of 13 PCGs. All phylogenetic trees based on different datasets show that the seven Stenochironomus species form a monophyletic group with strong support (Figure 5 and Supplementary Figures S2–S4). Our study reveals that the mitogenomes of Stenochironomus are useful for phylogenetic inference. Although mitogenomes have poor phylogenetic signals at the subfamily level of Chironomidae [33], mitogenomes are still useful for phylogeny at the genus level within Chironomidae according to the present and recent studies [32].
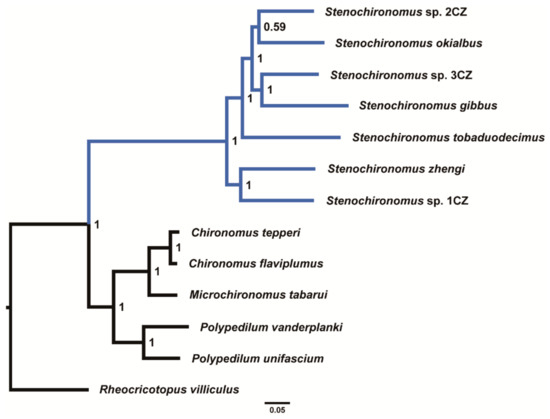
Figure 5.
Phylogenetic tree of Chironominae based on dataset AA. Numbers at the nodes are BI.
4. Conclusions
In this study, seven new mitogenomes of the genus Stenochironomus were sequenced and analyzed. Coupled with published data, phylogenetic analyses were performed within Chironominae. The present study showed that mitogenomes of Stenochironomus show a higher A and T bias than other chironomid species. A synapomorphic gene rearrangement that the gene order rearranges from trnI-trnQ-trnM to trnI-trnM-trnQ was identified within Stenochironomus, which is the first instance of mitochondrial gene rearrangement discovered in Chironomidae. The monophyly of the genus Stenochironomus was strongly supported by mitogenomes.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/insects13020115/s1, Supplementary Table S1: The best model for each partition of the three datasets. Supplementary Table S2: Start and stop codons of PCGs among the seven Stenochironomus mitogenomes. Supplementary Table S3: Total number of codons of the seven Stenochironomus mitogenomes. Supplementary Figure S1: Patterns of codon usage of the seven Stenochironomus mitogenomes. The X-axis shows the codon families and the Y-axis shows the total codons. Supplementary Figure S2: Phylogenetic relationships of Chironominae based on AA. Numbers at the nodes are ML bootstrap values. Supplementary Figure S3: Phylogenetic relationships of Chironominae based on PCG12. Numbers at the nodes are BI posterior probabilities and ML bootstrap values. Supplementary Figure S4: Phylogenetic relationships of Chironominae based on PCG123. Numbers at the nodes are BI posterior probabilities and ML bootstrap values.
Author Contributions
X.-L.L. and C.-G.Z. conceived the research. X.-L.L. collected samples. Y.-M.Z. performed laboratory work. Y.W. and C.-G.Z. conducted analyses. X.-L.L. and C.-G.Z. drafted the manuscript. Z.L., W.-J.B. and X.-H.W. revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China, grant number 31900344, 41502021, China Postdoctoral Science Foundation Grant, grant number 2018M640227, and Technological Innovation Talent Training Project of the Ministry of Natural Resources of China, grant number 12110600000018003910.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The following information was supplied regarding the availability of DNA sequences: The new mitogenomes of Stenochironomus gibbus, Stenochironomus okialbus, Stenochironomus sp. 1CZ, Stenochironomus sp. 2CZ, Stenochironomus sp. 3CZ, Stenochironomus tobaduodecimus, Stenochironomus zhengi are deposited in GenBank of NCBI under accession numbers OL742440, OL753645, OL753646, OL742441, OL753647, OL753648 and OL753649, respectively.
Acknowledgments
We would like to thank three anonymous reviewers for their suggestions and comments. We sincerely thank Xin Duan for giving assistance in software analysis.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Cameron, S.L. Insect mitochondrial genomics: Implications for evolution and phylogeny. Annu. Rev. Entomol. 2014, 59, 95–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moreno-Carmona, M.; Cameron, S.L.; Quiroga, C.F.P. How are the mitochondrial genomes reorganized in Hexapoda? Differential evolution and the first report of convergences within Hexapoda. Gene 2021, 791, 145719. [Google Scholar] [CrossRef] [PubMed]
- Brown, W.M. The mitochondrial genome of animals. In Molecular Evolutionary Genetics; Plenum Press: New York, NY, USA, 1985; pp. 95–130. [Google Scholar]
- Boore, J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, W.M.; George, M.; Wilson, A.C. Rapid evolution of animal mitochondrial DNA. Proc. Natl. Acad. Sci. USA 1979, 76, 1967–1971. [Google Scholar] [CrossRef] [Green Version]
- Curole, J.P.; Kocher, T.D. Mitogenomics: Digging deeper with complete mitochondrial genomes. Trends Ecol. Evol. 1999, 14, 394–398. [Google Scholar] [CrossRef]
- Du, Z.; Hasegawa, H.; Cooley, J.R.; Simon, C.; Yoshimura, J.; Cai, W.; Sota, T.; Li, H. Mitochondrial genomics reveals shared phylogeographic patterns and demographic history among three periodical cicada species groups. Mol. Biol. Evol. 2019, 36, 1187–1200. [Google Scholar] [CrossRef]
- Kieran, T.J. Mitochondrial, metagenomic, and phylogenetic analysis of the ground beetle Harpalus pensylvanicus (Coleoptera: Carabidae). Gene 2020, 740, 144540. [Google Scholar] [CrossRef]
- Manchola, O.F.S.; Virrueta Herrera, S.; D’Alessio, L.M.; Yoshizawa, K.; Garcia Aldrete, A.N.; Johnson, K.P. Mitochondrial genomes within bark lice (Insecta: Psocodea: Psocomorpha) reveal novel gene rearrangements containing phylogenetic signal. Syst. Entomol. 2021, 46, 938–951. [Google Scholar] [CrossRef]
- Rokas, A.; Holland, P.W. Rare genomic changes as a tool for phylogenetics. Trends Ecol. Evol. 2000, 15, 454–459. [Google Scholar] [CrossRef]
- Zhang, J.; Kan, X.; Miao, G.; Hu, S.; Sun, Q.; Tian, W. qMGR: A new approach for quantifying mitochondrial genome rearrangement. Mitochondrion 2020, 52, 20–23. [Google Scholar] [CrossRef]
- Zheng, B.-Y.; Cao, L.-J.; Tang, P.; van Achterberg, K.; Hoffmann, A.A.; Chen, H.-Y.; Chen, X.-X.; Wei, S.-J. Gene arrangement and sequence of mitochondrial genomes yield insights into the phylogeny and evolution of bees and sphecid wasps (Hymenoptera: Apoidea). Mol. Phylogenet. Evol. 2018, 124, 1–9. [Google Scholar] [CrossRef]
- Sweet, A.D.; Johnson, K.P.; Cao, Y.; de Moya, R.S.; Skinner, R.K.; Tan, M.; Herrera, S.V.; Cameron, S.L. Structure, gene order, and nucleotide composition of mitochondrial genomes in parasitic lice from Amblycera. Gene 2021, 768, 145312. [Google Scholar] [CrossRef]
- Dietrich, C.; Brune, A. The complete mitogenomes of six higher termite species reconstructed from metagenomic datasets (Cornitermes sp., Cubitermes ugandensis, Microcerotermes parvus, Nasutitermes corniger, Neocapritermes taracua, and Termes hospes). Mitochondrial DNA Part. A 2016, 27, 3903–3904. [Google Scholar] [CrossRef]
- Zhang, J.; Zhou, C.; Gai, Y.; Song, D.; Zhou, K. The complete mitochondrial genome of Parafronurus youi (Insecta: Ephemeroptera) and phylogenetic position of the Ephemeroptera. Gene 2008, 424, 18–24. [Google Scholar] [CrossRef]
- Zhang, W.; Li, R.; Zhou, C. Complete mitochondrial genomes of Epeorus carinatus and E. dayongensis (Ephemeroptera: Heptageniidae): Genomic comparison and phylogenetic inference. Gene 2021, 777, 145467. [Google Scholar] [CrossRef]
- Shi, A.; Li, H.; Bai, X.; Dai, X.; Chang, J.; Guilbert, E.; Cai, W. The complete mitochondrial genome of the flat bug Aradacanthia heissi (Hemiptera: Aradidae). Zootaxa 2012, 3238, 23–38. [Google Scholar] [CrossRef]
- Li, H.; Leavengood Jr, J.M.; Chapman, E.G.; Burkhardt, D.; Song, F.; Jiang, P.; Liu, J.; Zhou, X.; Cai, W. Mitochondrial phylogenomics of Hemiptera reveals adaptive innovations driving the diversification of true bugs. Proc. R. Soc. B Biol. Sci. 2017, 284, 20171223. [Google Scholar] [CrossRef]
- Cheng, Y.; Yan, Y.; Wei, M.; Niu, G. Characterization of mitochondrial genomes of three new species: Leptocimbex praiaformis, L. clavicornis, and L. yanniae (Hymenoptera: Cimbicidae). Entomol. Res. 2021, 51, 287–304. [Google Scholar] [CrossRef]
- Hu, J.; Zhang, D.; Hao, J.; Huang, D.; Cameron, S.; Zhu, C. The complete mitochondrial genome of the yellow coaster, Acraea issoria (Lepidoptera: Nymphalidae: Heliconiinae: Acraeini): Sequence, gene organization and a unique tRNA translocation event. Mol. Biol. Rep. 2010, 37, 3431–3438. [Google Scholar] [CrossRef]
- Ye, F.; Lan, X.-E.; Zhu, W.-B.; You, P. Mitochondrial genomes of praying mantises (Dictyoptera, Mantodea): Rearrangement, duplication, and reassignment of tRNA genes. Sci. Rep. 2016, 6, 25634. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.-P.; Cai, Y.-Y.; Yu, D.-N.; Storey, K.B.; Zhang, J.-Y. Gene characteristics of the complete mitochondrial genomes of Paratoxodera polyacantha and Toxodera hauseri (Mantodea: Toxoderidae). Peer J. 2018, 6, e4595. [Google Scholar] [CrossRef] [Green Version]
- Fenn, J.D.; Song, H.; Cameron, S.L.; Whiting, M.F. A preliminary mitochondrial genome phylogeny of Orthoptera (Insecta) and approaches to maximizing phylogenetic signal found within mitochondrial genome data. Mol. Phylogenet. Evol. 2008, 49, 59–68. [Google Scholar] [CrossRef]
- Leavitt, J.R.; Hiatt, K.D.; Whiting, M.F.; Song, H. Searching for the optimal data partitioning strategy in mitochondrial phylogenomics: A phylogeny of Acridoidea (Insecta: Orthoptera: Caelifera) as a case study. Mol. Phylogenet. Evol. 2013, 67, 494–508. [Google Scholar] [CrossRef]
- Cameron, S.L.; Johnson, K.P.; Whiting, M.F. The mitochondrial genome of the screamer louse Bothriometopus (Phthiraptera: Ischnocera): Effects of extensive gene rearrangements on the evolution of the genome. J. Mol. Evol. 2007, 65, 589–604. [Google Scholar] [CrossRef]
- Dickey, A.M.; Kumar, V.; Morgan, J.K.; Jara-Cavieres, A.; Shatters, R.G.; McKenzie, C.L.; Osborne, L.S. A novel mitochondrial genome architecture in thrips (Insecta: Thysanoptera): Extreme size asymmetry among chromosomes and possible recent control region duplication. BMC Genom. 2015, 16, 439. [Google Scholar] [CrossRef] [Green Version]
- Nelson, L.A.; Lambkin, C.L.; Batterham, P.; Wallman, J.F.; Dowton, M.; Whiting, M.F.; Yeates, D.K.; Cameron, S.L. Beyond barcoding: A mitochondrial genomics approach to molecular phylogenetics and diagnostics of blowflies (Diptera: Calliphoridae). Gene 2012, 511, 131–142. [Google Scholar] [CrossRef] [Green Version]
- Beckenbach, A.T.; Joy, J.B. Evolution of the mitochondrial genomes of gall midges (Diptera: Cecidomyiidae): Rearrangement and severe truncation of tRNA genes. Genome Biol. Evol. 2009, 1, 278–287. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Huang, J.; Wu, H. Mitogenomes provide insights into the phylogeny of Mycetophilidae (Diptera: Sciaroidea). Gene 2021, 783, 145564. [Google Scholar] [CrossRef]
- Beckenbach, A.T. Mitochondrial genome sequences of Nematocera (lower Diptera): Evidence of rearrangement following a complete genome duplication in a winter crane fly. Genome Biol. Evol. 2012, 4, 89–101. [Google Scholar] [CrossRef] [Green Version]
- Deviatiiarov, R.; Kikawada, T.; Gusev, O. The complete mitochondrial genome of an anhydrobiotic midge Polypedilum vanderplanki (Chironomidae, Diptera). Mitochondrial DNA Part. A 2017, 28, 218–220. [Google Scholar] [CrossRef]
- Lin, X.L.; Zhao, Y.M.; Yan, L.P.; Liu, W.B.; Bu, W.J.; Wang, X.H.; Zheng, C.G. Mitogenomes provide new insights into the evolutionary history of Prodiamesinae (Diptera: Chironomidae). Zool. Scr. 2022, 51, 119–132. [Google Scholar] [CrossRef]
- Zheng, C.-G.; Zhu, X.-X.; Yan, L.-P.; Yao, Y.; Bu, W.-J.; Wang, X.-H.; Lin, X.-L. First complete mitogenomes of Diamesinae, Orthocladiinae, Prodiamesinae, Tanypodinae (Diptera: Chironomidae) and their implication in phylogenetics. Peer J. 2021, 9, e11294. [Google Scholar] [CrossRef]
- Lei, T.; Song, C.; Zhu, X.-D.; Xu, B.-Y.; Qi, X. The complete mitochondrial genome of a non-biting midge Polypedilum unifascium (Tokunaga, 1938) (Diptera: Chironomidae). Mitochondrial DNA Part. B 2021, 6, 2212–2213. [Google Scholar] [CrossRef]
- Kong, F.-Q.; Zhao, Y.-C.; Chen, J.-L.; Lin, X.-L. First report of the complete mitogenome of Microchironomus tabarui Sasa, 1987 (Diptera, Chironomidae) from Hebei Province, China. Mitochondrial DNA Part. B 2021, 6, 2845–2846. [Google Scholar] [CrossRef]
- Hiki, K.; Oka, K.; Nakajima, N.; Yamamoto, H.; Yamagishi, T.; Sugaya, Y. The complete mitochondrial genome of the non-biting midge Chironomus yoshimatsui (Diptera: Chironomidae). Mitochondrial DNA Part. B 2021, 6, 2995–2996. [Google Scholar] [CrossRef]
- Jiang, Y.-W.; Zhao, Y.-M.; Lin, X.-L. First report of the complete mitogenome of Tanypus punctipennis Meigen, 1818 (Diptera, Chironomidae) from Hebei Province, China. Mitochondrial DNA Part. B 2022, 7, 215–216. [Google Scholar] [CrossRef]
- Cranston, P.S.; Dillon, M.E.; Pinder, L.C.V.; Reiss, F. The Adult Males of Chironominae (Diptera, Chironomidae) of the Holarctic Region—Keys and Diagnoses. In Chironomidae of the Holarctic region. Keys and diagnoses. Part 3—Adult Males; Wiederholm, T., Ed.; Entomologica Scandinavica: Lund, Sweden, 1989; pp. 353–502, Supplement 34. [Google Scholar]
- Wantzen, K.M.; Wagner, R. Detritus processing by invertebrate shredders: A neotropical–temperate comparison. J. North. Am. Benthol. Soc. 2006, 25, 216–232. [Google Scholar] [CrossRef]
- Valente-Neto, F.; Koroiva, R.; Fonseca-Gessner, A.A.; de Oliveira Roque, F. The effect of riparian deforestation on macroinvertebrates associated with submerged woody debris. Aquat. Ecol. 2015, 49, 115–125. [Google Scholar] [CrossRef]
- Martins, I.; Castro, D.M.; Macedo, D.R.; Hughes, R.M.; Callisto, M. Anthropogenic impacts influence the functional traits of Chironomidae (Diptera) assemblages in a neotropical savanna river basin. Aquat. Ecol. 2021, 55, 1081–1095. [Google Scholar] [CrossRef]
- Park, K.; Kim, W.-S.; Park, J.-W.; Kwak, I.-S. Complete mitochondrial genome of Chironomus flaviplumus (Diptera: Chironomidae) collected in Korea. Mitochondrial DNA Part. B 2021, 6, 2843–2844. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, Y.; Leung, H.C.; Yiu, S.-M.; Chin, F.Y. IDBA-UD: A de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 2012, 28, 1420–1428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Grant, J.R.; Stothard, P. The CGView Server: A comparative genomics tool for circular genomes. Nucleic Acids Res. 2008, 36, 181–184. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Vaidya, G.; Lohman, D.J.; Meier, R. SequenceMatrix: Concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 2011, 27, 171–180. [Google Scholar] [CrossRef]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [CrossRef] [Green Version]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [Green Version]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, L.-T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- de Oliveira Aragão, A.; Neto, J.P.N.; Cruz, A.C.R.; Casseb, S.M.M.; Cardoso, J.F.; da Silva, S.P.; Ishikawa, E.A.Y. Description and phylogeny of the mitochondrial genome of Sabethes chloropterus, Sabethes glaucodaemon and Sabethes belisarioi (Diptera: Culicidae). Genomics 2019, 111, 607–611. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-Y.; Yan, L.-P.; Pape, T.; Gao, Y.-Y.; Zhang, D. Evolutionary insights into bot flies (Insecta: Diptera: Oestridae) from comparative analysis of the mitochondrial genomes. Int. J. Biol. Macromol. 2020, 149, 371–380. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, C.; Alves, J.M.; Foster, P.G.; Suesdek, L.; Sallum, M.A.M. Phylogeny and temporal diversification of mosquitoes (Diptera: Culicidae) with an emphasis on the Neotropical fauna. Syst. Entomol. 2021, 46, 798–811. [Google Scholar] [CrossRef]
- Thao, M.L.; Baumann, L.; Baumann, P. Organization of the mitochondrial genomes of whiteflies, aphids, and psyllids (Hemiptera, Sternorrhyncha). BMC Evol. Biol. 2004, 4, 25. [Google Scholar] [CrossRef] [Green Version]
- Tan, H.-W.; Liu, G.-H.; Dong, X.; Lin, R.-Q.; Song, H.-Q.; Huang, S.-Y.; Yuan, Z.-G.; Zhao, G.-H.; Zhu, X.-Q. The complete mitochondrial genome of the Asiatic cavity-nesting honeybee Apis cerana (Hymenoptera: Apidae). PLoS ONE 2011, 6, e23008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, P.; Li, H.; Song, F.; Cai, Y.; Wang, J.; Liu, J.; Cai, W. Duplication and remolding of tRNA genes in the mitochondrial genome of Reduvius tenebrosus (Hemiptera: Reduviidae). Int. J. Mol. Sci. 2016, 17, 951. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Li, H.; Song, F.; Zhao, Y.; Wilson, J.J.; Cai, W. Higher-level phylogeny and evolutionary history of Pentatomomorpha (Hemiptera: Heteroptera) inferred from mitochondrial genome sequences. Syst. Entomol. 2019, 44, 810–819. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).