Longer mtDNA Fragments Provide a Better Insight into the Genetic Diversity of the Sycamore Lace Bug, Corythucha ciliata (Say, 1832) (Tingidae, Hemiptera), Both in Its Native and Invaded Areas
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
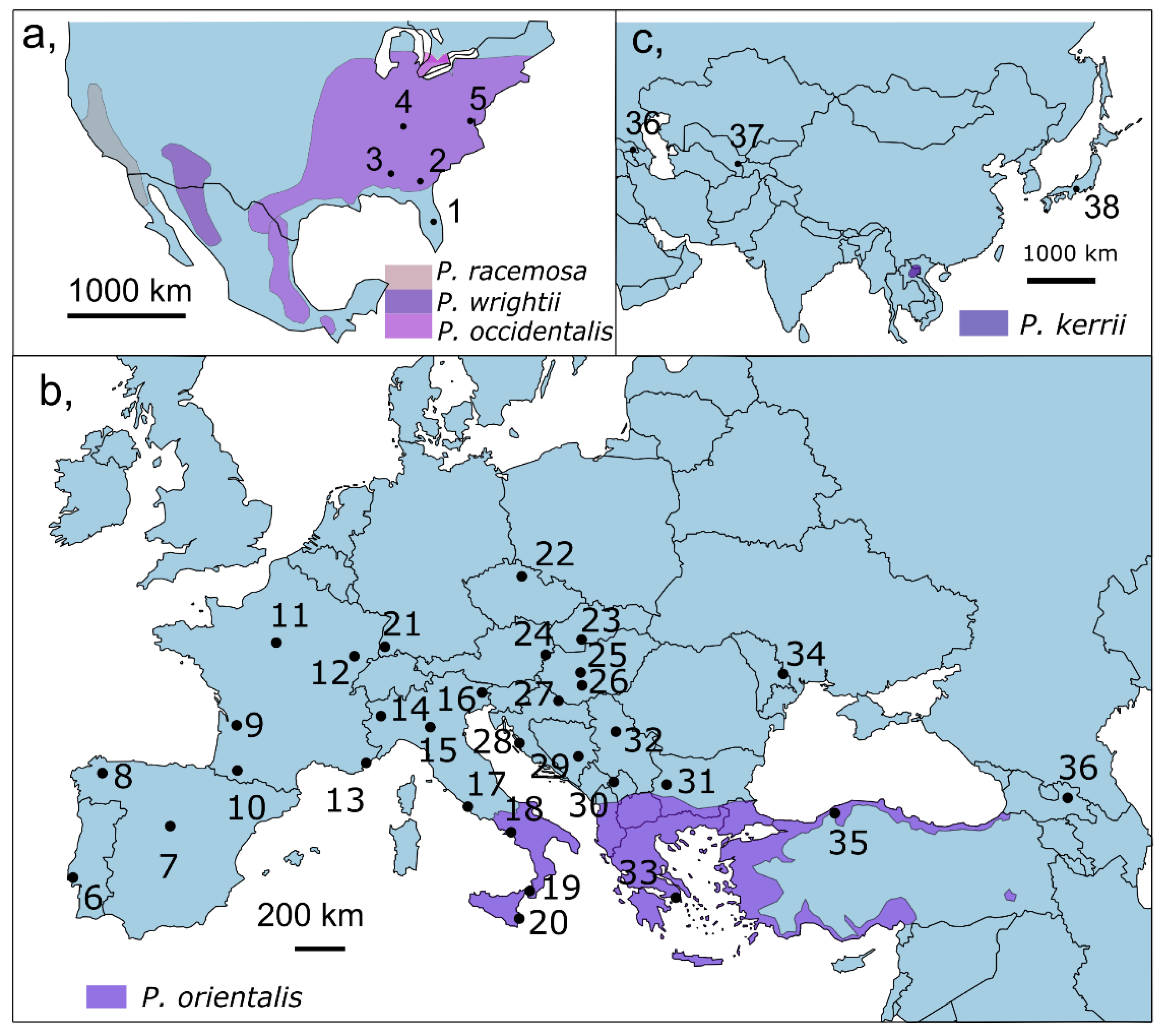
2.1. Sampling and Molecular Methods
2.2. Data Analysis
2.3. Phylogenetic Analyses
3. Results
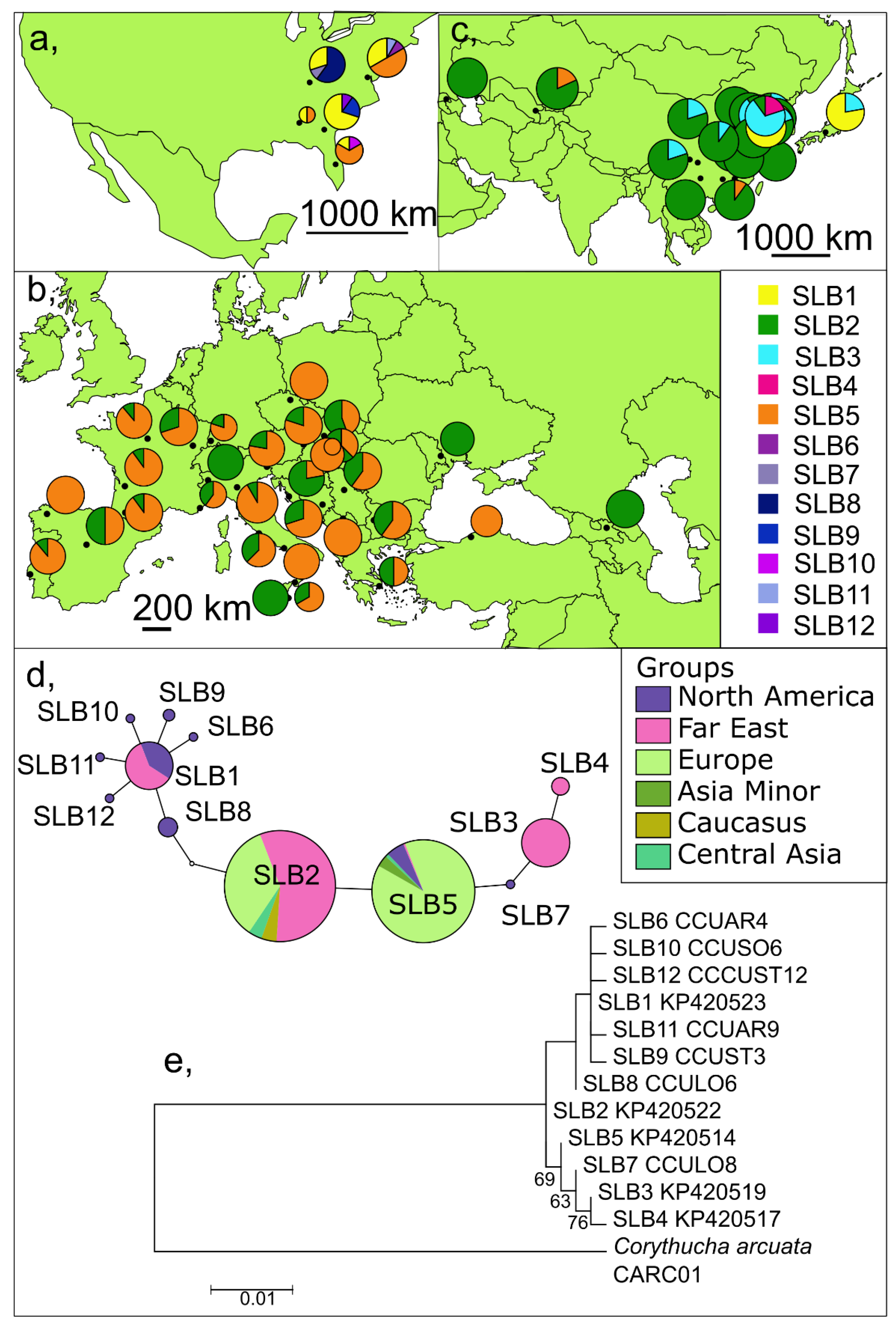
3.1. Long Fragments of the COI Gene
3.1.1. Genetic Diversity and Structure in the Native Range—North America
3.1.2. Genetic Diversity and Structure in the Invaded Range
Europe
Asia
3.2. Short (Barcoding) Fragments of the COI Gene including Already Published Data
3.2.1. Invaded Versus Natural Range
Invaded Area
Native Range
4. Discussion
4.1. Genetic Diversity of SYCAMORE Lace Bug
4.2. Long Versus Short (Barcoding) Fragments of the COI
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Allendorf, F.W.; Luikart, G.; Aitken, S.N. Conservation and the Genetics of Populations, 2nd ed.; Wiley-Blackwell: Hoboken, NJ, USA, 2013. [Google Scholar]
- Bras, A.; Avtzis, D.N.; Kenis, M.; Li, H.; Vétek, G.; Bernard, A.; Colurtin, C.; Rousselet, J.; Roques, A.; Auger-Rozenberg, M.A. A Complex Invasion Story Underlies the Fast Spread of the Invasive Box Tree Moth (Cydalima perspectalis) across Europe. J. Pest Sci. 2019, 92, 1187–1202. [Google Scholar] [CrossRef]
- Kirichenko, N.; Triberti, P.; Ohshima, I.; Haran, H.; Byun, B.-K.; Li, H.; Augustin, S.; Roques, A.; Lopez-Vaamonde, C. From East to West across the Palearctic: Phylogeography of the Invasive Lime Leaf Miner Phyllonorycter issikii (Lepidoptera: Gracillariidae) and Discovery of a Putative New Cryptic Species in East Asia. PLoS ONE 2017, 12, e0171104. [Google Scholar] [CrossRef] [PubMed]
- Carter, M.; Smith, M.; Harrison, R. Genetic Analyses of the Asian Longhorned Beetle (Coleoptera, Cerambycidae, Anoplophora glabripennis), in North America, Europe and Asia. Biol. Invasions 2010, 12, 1165–1182. [Google Scholar] [CrossRef]
- Robinet, C.; Imbert, C.-E.; Rousselet, J.; Sauvard, D.; Garcia, J.; Goussard, F.; Roques, A. Human-Mediated Long-Distance Jumps of the Pine Processionary Moth in Europe. Biol. Invasions 2012, 14, 1557–1569. [Google Scholar] [CrossRef] [Green Version]
- Valade, R.; Kenis, M.; Hernandez-Lopez, A.; Augustin, S.; Mari Mena, N.; Magnoux, E.; Rougerie, R.; Lakatos, F.; Roques, A.; Lopez-Vaamonde, C. Mitochondrial and Microsatellite DNA Markers Reveal a Balkan Origin for the Highly Invasive Horse-Chestnut Leaf Miner Cameraria ohridella (Lepidoptera, Gracillariidae). Mol. Ecol. 2009, 18, 3458–3470. [Google Scholar] [CrossRef]
- Tóth, V.; Lakatos, F. Phylogeographic Pattern of the Plane Leaf Miner, Phyllonorycter platani (STAUDINGER, 1870) (Lepidoptera: Gracillariidae) in Europe. BMC Evol. Biol. 2018, 18, 135. [Google Scholar] [CrossRef]
- Drake, C.J.; Ruhoff, F.A. Lacebugs of the World: A Catalog (Hemiptera: Tingidae); United States National Museum Bulletin: Washington, DC, USA, 1965; Volume 243, pp. 1–634. [Google Scholar]
- D’Aguilar, J.; Pralavorio, R.; Rabasse, J.M.; Mouton, R. Introduction En France Du Tigre Du Platane: Corythucha ciliata (Say) (Het. Tingidae). Bull. Soc. Entomol. Fr. 1977, 82, 1–6. [Google Scholar] [CrossRef]
- Jasinka, J.; Bozsits, G. A Platán Csipkéspoloska (Corythuca ciliata) Fellépése Magyarországon. Növényvédelem 1977, 13, 42–46. [Google Scholar]
- Mutun, S. Corythucha ciliata, a New Platanus Pest in Turkey. Phytoparasitica 2009, 37, 65–66. [Google Scholar] [CrossRef]
- Tzanakakis, M.E. Records of the Sycamore Lace Bug, Corythuca ciliata, (Say), in Greece. Entomol. Hell. 1988, 6, 55–56. [Google Scholar] [CrossRef] [Green Version]
- Grebennikov, K.A.; Mukhanov, S.Y. Corythucha ciliata (Say, 1932) (Hemiptera: Heteroptera: Tingidae): New Alien Species of True Bugs in Uzbekistan Fauna. Russ. J. Biol. Invasions 2019, 10, 126–128. [Google Scholar] [CrossRef]
- Li, F.; Wang, R.; Qu, C.; Fu, N.; Luo, C.; Xu, Y. Sequencing and Characterization of the Invasive Sycamore Lace Bug Corythucha ciliata (Hemiptera: Tingidae) Transcriptome. PLoS ONE 2016, 11, e0160609. [Google Scholar] [CrossRef] [PubMed]
- Picker, M.D.; Griffiths, C.L. Sycamore Tree Lace Bug (Corythucha ciliata Say) (Hemiptera: Tingidae) Reaches Africa. Afr. Entomol. 2015, 23, 247–249. [Google Scholar] [CrossRef]
- Prado, C.E. Presencia En Chile de Corythucha ciliata (Say) (Hemiptera: Heteroptera: Tingidae). Rev. Chil. Entomol. 1990, 18, 53–55. [Google Scholar]
- Rabitsch, W.; Streito, J.C. Corythucha ciliata (Say, 1832)–Sycamore lace bug (Heteroptera: Tingidae). In Alien Terrestrial Arthropds of Europe; Roques, A., Kennis, M., Lees, D., Lopez-Vaamonde, C., Rabitsch, W., Rasplus, J.Y., Roy, D.B., Eds.; Pensoft: Sofia, Bulgaria, 2010; Volume 4, pp. 964–965. [Google Scholar]
- Supatashvili, A.; Goginashvili, N.; Kereselidze, M. Distribution and Some Biological Data of Sycamore Lace Bug–Corythucha ciliata Say (Heteroptera, Tingidae) in Georgia. Ann. Agrar. Sci. 2016, 14, 42–45. [Google Scholar] [CrossRef] [Green Version]
- Cvetkovska-Gjorgievska, A.; Dedov, I.; Hristovski, S.; Langourov, M.; Lazarevska, S.; Prelik, D.; Simov, N. New Records of Allochtonous, Invasive and Pest Invertebrate Species from the Republic of Macedonia. Ecol. Montenegrina 2019, 20, 56–70. [Google Scholar] [CrossRef]
- Yoon, C.; Yang, J.O.; Kang, S.H.; Kim, G.H. Insecticidal Properties of Bistrifluron against Sycamore Lace Bug, Corythucha ciliata (Hemiptera: Tingidae). J. Pestic. Sci. 2008, 33, 44–50. [Google Scholar] [CrossRef] [Green Version]
- Malumphy, C.; Reid, S.; Eyre, D. The Platanus Lace Bug, Corythucha ciliata (Say) (Hemiptera: Tingidae), a Nearctic Pest of Plane Trees, New to Britain. Br. J. Entomol. Nat. Hist. 2007, 20, 233–240. [Google Scholar]
- Grosso-Silva, J.M.; Aguiar, A. Corythucha ciliata (Say, 1832) (Hemiptera, Tingidae), the Nearctic Sycamore Lace Bug, Found in Portugal. Boletín SEA 2007, 40, 366. [Google Scholar]
- Aukema, B.; Hermes, D. Nieuwe en Interessante Nederlandse Wantsen III (Hemiptera: Heteroptera). Ned. Faun. Meded. 2009, 31, 53–87. [Google Scholar]
- Lis, B. Corythucha ciliata (SAY, 1832) (Hemiptera: Heteroptera: Tingidae). Opole Sci. Soc. Nat. J. 2009, 42, 119–122. [Google Scholar]
- Milevoj, L. The Occurence of Some Pests and Diseases on Horse Chestnut, Plane Tree and Indian Bean Tree in Urban Areas of Slovenia. Acta Agric. Slov. 2004, 83, 297–300. [Google Scholar]
- Voigt, K. The First Russian Record of Corythucha ciliata (Say) from Krasnodar (Heteroptera: Tingidae). Zoosystematica Ross. 2001, 10, 76. [Google Scholar]
- Servadei, A. Un Tingide Neartico Comparso in Italia (Corythuca ciliata Say). Bolletino Soc. Entomol. Ital. 1966, 96, 94–96. [Google Scholar]
- Maceljski, M.; Balarin, I. Novi Clan Stetene Entomofaune u Jugoslaviji Corythuca ciliata Say Tingidae, Heteroptera. Zašt. Bilja 1972, 23, 193–206. [Google Scholar]
- Tomić, D.; Mihajlović, L. American Netlike Bug (Corythucha ciliata Say–Heteroptera, Tingidae) New Serious Enemy of Plane Trees in Belgrade. Sumarstvo 1974, 7, 51–54. [Google Scholar]
- Dioli, P. La Presenza in Valtellina Di Alcune Cimici Dannose Alle Piante. Rass. Econ. Prov. Sondrio 1975, 4, 43–46. [Google Scholar]
- Ribes, J. Un Insecte Nordamericä Que Ataca Eis Pätans. Rev. Girona 1980, 93, 299–301. [Google Scholar]
- Mildner, P. Neues Zur Kärntner Arthropodenfauna. Carinthia II 1983, 173, 137–141. [Google Scholar]
- Hopp, I. Die Platanen-Netzwanze Corythucha ciliata (Say) Nun Auch in der Bundesrepublik Deutschland. Entomol. Z. Stuttg. 1984, 94, 60–63. [Google Scholar]
- Tokihiro, G.; Tanaka, K.; Kondo, K. Occurrence of the Sycamore Lace Bug, Corythucha ciliata (Say) (Heteroptera: Tingidae) in Japan. J. Jpn. Soc. Nutr. Food Sci. 2003, 39, 85–87. [Google Scholar]
- Josifov, M. Varchu Pojavata Na Nearkticnija Vid Corythucha ciliata (Say, 1832) (Het., Tingidae) v Balgarija. Acta Zool. Bulg. 1990, 39, 53–55. [Google Scholar]
- Hoffmann, H. Die Platanen-Gitterwanze Corythucha ciliata (Say) Weiter Auf Dem Vormarsch (Hemiptera-Heteroptera: Tingidae). Heteropteron 1996, 2, 19–21. [Google Scholar]
- Hoffmann, H. 50 Jahre Platanengitterwanze Corythucha ciliata (SAY, 1832) in Europa-Ausbreitung des Schädlings in der Paläarktis, Allgemeines und Bibliographie. Heteropteron H 2016, 46, 13–43. [Google Scholar]
- Rabitsch, W. Alien True Bugs of Europe (Insecta: Hemiptera: Heteroptera). Zootaxa 2008, 1827, 1–44. [Google Scholar] [CrossRef] [Green Version]
- Kis, B. Corythucha ciliata (Heteroptera, Tingidae) un Dăunător Forestier Nou Pentru Fauna României [Corythucha ciliata (Heteroptera, Tingidae) a New Pest for the Fauna of Romania]. An. Banat. 1990, 2, 320–321. [Google Scholar]
- Derjanschi, V. Tigrul Platanului Corythucha ciliata Say (Heteroptera, Tingidae)–Specie Nouă Pentru Fauna Republicii Moldova. Bul. Ştiinţific. Rev. Etnogr. Ştiinţele Nat. Şi Muzeol. 2007, 19, 46–53. [Google Scholar]
- Chung, Y.; Kwon, T.; Yeo, W.; Byun, B.; Park, C. Occurrence of the Sycamore Lace Bug, Corythucha ciliata (Say) (Hemiptera: Tingidae) in Korea. Korean J. Appl. Entomol. 1996, 35, 137–139. [Google Scholar]
- Streito, J.-C. Note Sur Quelques Espèces Envahissantes de Tingidae: Corythucha ciliata (Say, 1932), Stephanitis Pyrioides (Scott, 1874) et Stephanitis Takeyai Drake & Maa, 1955 (Hemiptera Tingidae). L’Entomologiste 2006, 62, 31–36. [Google Scholar]
- Gninenko, Y.I. Plane Lace Bug Corythuca ciliata SAY. in North-East Black Sea Costal Area. In Proceedings of the Alien Arthropods in South East Europe–Crossroad of Three Continents Conference, Sofia, Bulgaria, 19–21 September 2007; p. 69. [Google Scholar]
- Schneider, N.; Christian, S. Découverte de Corythucha ciliata (Say, 1832) et de Derephysia SinuatocollisPuton, 1879 Au Luxembourg et Autres Observa-Tions Dignes d’intérêt (Insecta, Hemiptera, Heteroptera). Bull. Soc. Nat. Luxemb. 2013, 114, 105. [Google Scholar]
- Dominiak, B.C.; Gillespie, P.S.; Worsley, P.; Locker, H. Survey for Sycamore Lace Bug ‘Corythucha ciliata’ (Say) (Hemiptera: Tingidae) in New South Wales during 2007. J. Entomol. Soc. New South Wales 2008, 37, 27. [Google Scholar]
- Hrubík, P. Alien Insect Pests on Introduced Woody Plants in Slovakia. Acta Entomol. Serbica 2007, 12, 81–85. [Google Scholar]
- Catalogue of Life. Available online: https://www.catalogueoflife.org/ (accessed on 29 December 2021).
- Halbert, S.E.; Meeker, J.R. Sycamore Lace Bug, Corythucha ciliata (Say) (Insecta: Hemiptera: Tingidae); University of Florida: Gainesville, FL, USA, 2001; pp. 1–4. [Google Scholar]
- Torres-Miller, L. New Records of Lace Bugs from West Virginia (Hemiptera: Tingidae). Insecta Mundi 1989, 3, 10. [Google Scholar]
- Buttram, J.R.; Boyer, W.P.; Hawthorne, R.M.; Jenkins, L.E.; Woodruff, R.E.; Johnson, W.C.; White, C.E.; Peters, L.L.; Harding, W.C., Jr.; Dowdy, A.C.; et al. Summary of Insect Conditions in the United States—1962: Forest and Shade Tree Insects. Coop. Econ. Insect Rep. 1963, 13, 327–356. [Google Scholar]
- McQueen, H.F.; Fullerton, D.G.; Boyer, W.P.; Hawthorne, R.M.; Jenkins, L.E.; Mead, F.W.; Johnson, W.C.; White, C.E.; Peters, L.L.; Harding, W.C., Jr.; et al. Summary of Insect Conditions in the United States-1963: Shade Tree Insects. Coop. Econ. Insect Rep. 1964, 14, 343–352. [Google Scholar]
- McQueen, H.F.; Fullerton, D.G.; Boyer, W.P.; Hawthorne, R.M.; Jenkins, L.E.; Mead, F.W.; Johnson, W.C.; White, C.E.; Peters, L.L.; Harding, W.C., Jr.; et al. Forest, Ornamental and Shade Tree Insects. Coop. Econ. Insect Rep. 1964, 14, 581–585, 693–699. [Google Scholar]
- Leininger, T.D. A Guide to Major Insects, Diseases, Air Pollution Injury, and Chemical Injury of Sycamore; Southern Research Station: Asheville, NC, USA, 1999; Volume 28.
- Coyle, D.R.; Nebeker, T.E.; Hart, E.R.; Mattson, W.J. Biology and Management of Insect Pests in North American Intensively Managed Hardwood Forest Systems. Annu. Rev. Entomol. 2005, 50, 1–29. [Google Scholar] [CrossRef] [Green Version]
- Ripka, G.; Reiderné Saly, K.; Jensen, G.; Rácz, V.; Orosz, A. Díszfák és Díszcserjék Tripsz-, Poloska-És Kabócafaunája a Fővárosban. Növényvédelem 1993, 29, 569–572. [Google Scholar]
- Besedina, E.; Kil, V.; Ismailov, V.; Karpunina, M. Molecular Genetic Analysis and Phenology of the Plane Lace Bug Corythucha ciliata Say (Hemiptera: Tingidae) in Different Parts of Krasnodar Krai. In Proceedings of the XI International Scientific and Practical Conference “Biological Plant Protection is the Basis of Agroecosystems Stabilization”, Krasnodar, Russia, 21–24 September 2020; Volume 21, p. 00011. [Google Scholar]
- Besnard, G.; Tagmount, A.; Baradat, P.; Vigouroux, A.; Berville, A. Molecular Approach of Genetic Affinities between Wild and Ornamental Platanus. Euphytica 2002, 126, 401–412. [Google Scholar] [CrossRef]
- Liu, G.; Bao, M. Adventitious Shoot Regeneration from in Vitro Cultured Leaves of London Plane Tree (Platanus acerifolia Willd.). Plant Cell Rep. 2003, 21, 640–644. [Google Scholar] [CrossRef]
- Warrick, R.B.; Williams, C.F. Checklist of the Cultivated Trees of St. George, Washington County, Utah. Great Basin Nat. 1991, 51, 296–299. [Google Scholar]
- Wu, Z.Y.; Raven, P.H.; Hong, D.Y. Flora of China: Pittosporaceae through Connaraceae; Science Press: Beijing, China; Missouri Botanical Garden Press: St. Louis, MO, USA, 2003; Volume 9. [Google Scholar]
- Ju, R.-T.; Chen, G.-B.; Wang, F.; Li, B. Effects of Heat Shock, Heat Exposure Pattern, and Heat Hardening on Survival of the Sycamore Lace Bug, Corythucha ciliata. Entomol. Exp. Appl. 2011, 141, 168–177. [Google Scholar] [CrossRef]
- Ju, R.-T.; Luo, Q.-Q.; Gao, L.; Yang, J.; Li, B. Identification of HSP70 Gene in Corythucha ciliata and Its Expression Profiles under Laboratory and Field Thermal Conditions. Cell Stress Chaperones 2018, 23, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Ju, R.-T.; Gao, L.; Wei, S.-J.; Li, B. Spring Warming Increases the Abundance of an Invasive Specialist Insect: Links to Phenology and Life History. Sci. Rep. 2017, 7, 1–12. [Google Scholar]
- Pavela, R.; Žabka, M.; Kalinkin, V.; Kotenev, E.; Gerus, A.; Shchenikova, A.; Chermenskaya, T. Systemic Applications of Azadirachtin in the Control of Corythucha ciliata (Say, 1832) (Hemiptera, Tingidae), a Pest of Platanus sp. Plant Prot. Sci. 2013, 49, 27–33. [Google Scholar] [CrossRef] [Green Version]
- Neal, J., Jr.; Schaefer, C. Lace Bugs (Tingidae). In Heteroptera of Economic Importance; Schaefer, C.W., Panizzi, A.R., Eds.; CRC Press: Boca Raton, FL, USA, 2000; pp. 85–134. [Google Scholar]
- Julià, I.; Morton, A.; Roca, M.; Garcia-del-Pino, F. Evaluation of Three Entomopathogenic Nematode Species against Nymphs and Adults of the Sycamore Lace Bug, Corythucha ciliata. BioControl 2020, 65, 623–633. [Google Scholar] [CrossRef]
- Sevim, A.; Demir, I.; Sönmez, E.; Kocacevik, S.; Demirbağ, Z. Evaluation of Entomopathogenic Fungi against the Sycamore Lace Bug, Corythucha ciliata (Say) (Hemiptera: Tingidae). Turk. J. Agric. For. 2013, 37, 595–603. [Google Scholar] [CrossRef]
- Verfaille, T.; Piron, M.; Gutleben, C.; Hecker, C.; Maury-Roberti, A.; Chapin, E.; Clément, A.; Jaloux, B. Program PETAAL: A Biocontrol Strategy of the Sycamore Lace Bug Corythucha ciliata (Say) (Hemiptera: Tingidae) in Urban Areas. In Proceedings of the II International Symposium on Horticulture in Europe 1099, Angers, France, 1–5 July 2012; pp. 375–382. [Google Scholar]
- Yang, W.Y.; Tang, X.T.; Cai, L.; Dong, C.S.; Du, Y.Z. Isolation and Characterization of Nine Microsatellite Loci from the Sycamore Lace Bug Corythucha ciliata (Hemiptera: Tingidae). Fla. Entomol. 2014, 97, 1070–1074. [Google Scholar] [CrossRef]
- Li, F.-Q.; Fu, N.-N.; Qu, C.; Wang, R.; Xu, Y.-H.; Luo, C. Understanding the Mechanisms of Dormancy in an Invasive Alien Sycamore Lace Bug, Corythucha ciliata through Transcript and Metabolite Profiling. Sci. Rep. 2017, 7, 1–11. [Google Scholar]
- Gwiazdowski, R.A.; Foottit, R.G.; Maw, H.E.L.; Hebert, P.D. The Hemiptera (Insecta) of Canada: Constructing a Reference Library of DNA Barcodes. PLoS ONE 2015, 10, e0125635. [Google Scholar] [CrossRef] [Green Version]
- Raupach, M.J.; Hendrich, L.; Küchler, S.M.; Deister, F.; Morinière, J.; Gossner, M.M. Building-up of a DNA Barcode Library for True Bugs (Insecta: Hemiptera: Heteroptera) of Germany Reveals Taxonomic Uncertainties and Surprises. PLoS ONE 2014, 9, e106940. [Google Scholar] [CrossRef] [PubMed]
- Kocher, A.; Guilbert, É.; Lhuillier, É.; Murienne, J. Sequencing of the Mitochondrial Genome of the Avocado Lace Bug Pseudacysta perseae (Heteroptera, Tingidae) Using a Genome Skimming Approach. Comptes Rendus Biol. 2015, 338, 149–160. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.Y.; Tang, X.T.; Ju, R.T.; Zhang, Y.; Du, Y.Z. The Population Genetic Structure of Corythucha ciliata (Say) (Hemiptera: Tingidae) Provides Insights into Its Distribution and Invasiveness. Sci. Rep. 2017, 7, 635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- GenBank Overview. Available online: https://www.ncbi.nlm.nih.gov/genbank/ (accessed on 29 December 2021).
- Tóth, V.; Bender, B.; Tuba, K.; Horváth, B.; Lakatos, F. A Platán Csipkéspoloska (Corythucha ciliata Say, 1832) Populációgenetikai Mintázata. Növényvédelem 2020, 81, 168–175. [Google Scholar]
- Zhang, L.; Cai, W.; Luo, J.; Zhang, S.; Li, W.; Wang, C.; Lv, L.; Cui, J. Population Genetic Structure and Expansion Patterns of the Cotton Pest Adelphocoris fasciaticollis. J. Pest Sci. 2018, 91, 539–550. [Google Scholar] [CrossRef]
- Vujić, A.; Šašić Zorić, L.; Ačanski, J.; Likov, L.; Radenković, S.; Djan, M.; Milić, D.; Šebić, A.; Ranković, M.; Khaghaninia, S. Hide-and-Seek with Hoverflies: Merodon aureus—A Species, a Complex or a Subgroup? Zool. J. Linn. Soc. 2020, 190, 974–1001. [Google Scholar] [CrossRef]
- Naranjo-Díaz, N.; Conn, J.E.; Correa, M.M. Behavior and Population Structure of Anopheles darlingi in Colombia. Infect. Genet. Evol. 2016, 39, 64–73. [Google Scholar] [CrossRef]
- Han, T.; Kim, J.; Yi, D.-A.; Jeong, J.; An, S.L.; Park, I.G.; Park, H. An Integrative Taxonomy on the Locally Endangered Species of the Korean Scarabaeus (Coleoptera, Scarabaeidae). Zootaxa 2016, 4139, 515–526. [Google Scholar] [CrossRef]
- Aly, S.M. Reliability of Long vs. Short COI Markers in Identification of Forensically Important Flies. Croat. Med. J. 2014, 55, 19–26. [Google Scholar] [CrossRef] [Green Version]
- Feng, Y.; Oh, S.-H.; Manos, P.S. Phylogeny and Historical Biogeography of the Genus Platanus as Inferred from Nuclear and Chloroplast DNA. Syst. Bot. 2005, 30, 786–799. [Google Scholar] [CrossRef]
- Dinca, V.; Zakharov, E.V.; Hebert, P.D.N.; Vila, R. Complete DNA Barcode Reference Library for a Country’s Butterfly Fauna Reveals High Performance for Temperate Europe. Proc. R. Soc. B 2011, 278, 347–355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lunt, D.H.; Zhang, D.-X.; Szymura, J.M.; Hewitt, G.M. The Insect COI Gene: Evolutionary Patterns and Conserved Primers for Phylogenetic Studies. Insect Mol. Biol. 1996, 5, 153–165. [Google Scholar] [CrossRef] [PubMed]
- Geospiza Inc. Available online: http://www.geospiza.com/finchtv (accessed on 5 January 2021).
- Thompson, J.D.; Higgins, D.G.; Gibbson, T.J. Clustal W: Improving the Sensitivity of Progressive Multiple Sequence Alignment through Sequence Weighting, Position Specific Gap Penalties and Weight Matrix Choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. JModelTest 2: More Models, New Heuristics and Parallel Computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [Green Version]
- Guindon, S.; Gascuel, O. A Simple, Fast and Accurate Method to Estimate Large Phylogenies by Maximum-Likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef] [Green Version]
- Akaike, H. A New Look at the Statistical Model Identification. IEEE Trans. Autom. Control 1974, 19, 716–723. [Google Scholar] [CrossRef]
- Nei, M. Molecular Evolutionary Genetics; Columbia University Press: New York, NY, USA, 1987. [Google Scholar]
- Ewens, W.J. The Sampling Theory of Selectively Neutral Alleles. Theor. Popul. Biol. 1972, 3, 87–112. [Google Scholar] [CrossRef]
- Zouros, E. Mutation Rates, Population Sizes and Amounts of Electrophoretic Variation of Enzyme Loci in Natural Populations. Genetics 1979, 92, 623–646. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E.L. Arlequin Suite Ver 3.5: A New Series of Programs to Perform Population Genetics Analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A Software for Comprehensive Analysis of DNA Polymorphism Data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramos-Onsins, S.E.; Rozas, J. Statistical Properties of New Neutrality Tests Against Population Growth. Mol. Biol. Evol. 2002, 19, 2092–2100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dupanloup, I.; Schneider, S.; Excoffier, L. A Simulated Annealing Approach to Define the Genetic Structure of Populations. Mol. Ecol. 2002, 11, 2571–2581. [Google Scholar] [CrossRef] [PubMed]
- Chiari, Y.; van der Meijden, A.; Mucedda, M.; Lourenço, J.M.; Hochkirch, A.; Veith, M. Phylogeography of Sardinian Cave Salamanders (Genus Hydromantes) Is Mainly Determined by Geomorphology. PLoS ONE 2012, 7, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weir, B.S.; Cockerham, C.C. Estimating F-Statistics for the Analysis of Population Structure. Evolution 1984, 38, 1358–1370. [Google Scholar] [PubMed]
- Excoffier, L.; Smouse, P.; Quattro, J. Analysis of Molecular Variance Inferred from Metric Distances among DNA Haplotypes: Application to Human Mitochondrial DNA Restriction Data. Genetics 1992, 131, 479–491. [Google Scholar] [CrossRef] [PubMed]
- Weir, B.S. Genetic Data Analysis II: Methods for Discrete Population Genetic Data; Sinauer Associates, Inc.: Sunderland, MA, USA, 1996. [Google Scholar]
- Templeton, A.R.; Crandall, K.A.; Sing, C.F. A Cladistic Analysis of Phenotypic Associations with Haplotypes Inferred from Restriction Endonuclease Mapping and DNA Sequence Data. III. Cladogram Estimation. Genetics 1992, 132, 619–633. [Google Scholar] [CrossRef]
- Clement, M.; Posada, D.; Crandall, K.A. TCS: A Computer Program to Estimate Gene Genealogies. Mol. Ecol. 2000, 9, 1657–1660. [Google Scholar] [CrossRef] [Green Version]
- Múrias dos Santos, A.; Cabezas, M.P.; Tavares, A.I.; Xavier, R.; Branco, M. TcsBU: A Tool to Extend TCS Network Layout and Visualization. Bioinformatics 2016, 32, 627–628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- QGIS Geographic Information System. Open-Source Geospatial Foundation Project. Available online: http://qgis.osgeo.org (accessed on 9 July 2021).
- Inkscape. Available online: https://inkscape.org/release/inkscape-1.0.2/ (accessed on 15 January 2021).
- Rogers, A.R.; Harpending, H. Population Growth Makes Waves in the Distribution of Pairwise Genetic Differences. Mol. Biol. Evol. 1992, 9, 552–569. [Google Scholar]
- Slatkin, M.; Hudson, R.R. Pairwise Comparisons of Mitochondrial DNA Sequences in Stable and Exponentially Growing Populations. Genetics 1991, 129, 555–562. [Google Scholar] [CrossRef]
- Jung, S.; Duwal, R.K.; Lee, S. COI Barcoding of True Bugs (Insecta, Heteroptera). Mol. Ecol. Resour. 2011, 11, 266–270. [Google Scholar] [CrossRef] [PubMed]
- Park, D.S.; Foottit, R.; Maw, E.; Hebert, P.D.N. Barcoding Bugs: DNA-Based Identification of the True Bugs (Insecta: Hemiptera: Heteroptera). PLoS ONE 2011, 6, e18749. [Google Scholar] [CrossRef] [PubMed]
- Rugman-Jones, P.F.; Hoddle, M.S.; Phillips, P.A.; Jeong, G.; Stouthamer, R. Strong Genetic Structure among Populations of the Invasive Avocado Pest Pseudacysta perseae (Heidemann) (Hemiptera: Tingidae) Reveals the Source of Introduced Populations. Biol. Invasions 2012, 14, 1079–1100. [Google Scholar] [CrossRef]
- Grant, W.A.S.; Bowen, B.W. Shallow Population Histories in Deep Evolutionary Lineages of Marine Fishes: Insights from Sardines and Anchovies and Lessons for Conservation. J. Hered. 1998, 89, 415–426. [Google Scholar] [CrossRef]
- Dlugosch, K.M.; Anderson, S.R.; Braasch, J.; Cang, F.A.; Gillette, H.D. The Devil Is in the Details: Genetic Variation in Introduced Populations and Its Contributions to Invasion. Mol. Ecol. 2015, 24, 2095–2111. [Google Scholar] [CrossRef] [PubMed]
- Roe, A.D.; Sperling, F.A. Patterns of Evolution of Mitochondrial Cytochrome c Oxidase I and II DNA and Implications for DNA Barcoding. Mol. Phylogenetics Evol. 2007, 44, 325–345. [Google Scholar] [CrossRef]
- Maggioni, D.; Assandri, G.; Ramazzotti, F.; Magnani, D.; Pellegrino, I.; Valsecchi, E.; Galimberti, A. Differential Genetic Variability at Two MtDNA COI Regions Does Not Imply Mismatches in Odonata Molecular Identification Performances. Eur. Zool. J. 2021, 88, 425–435. [Google Scholar] [CrossRef]

| Group | n | No. | Nex | S | ts/tv | h ± SD | π (%) ± SD |
|---|---|---|---|---|---|---|---|
| Invaded | 287 | 7 | 4 | 16 | 14/2 | 0.6717 ± 0.0124 | 0.1196 ± 0.0785 |
| E | 250 | 5 | 3 | 6 | 6/0 | 0.6542 ± 0.0123 | 0.0937 ± 0.0655 |
| A | 37 | 4 | 1 | 12 | 10/2 | 0.6562 ± 0.0555 | 0.2401 ± 0.1406 |
| Native/NA | 40 | 12 | 10 | 21 | 20/1 | 0.8462 ± 0.0378 | 0.3630 ± 0.2005 |
| Total | 327 | 17 | n.r. | 26 | 25/2 | 0.7320 ± 0.0136 | 0.1945 ± 0.1152 |
| Groups | Source of Variation | var% | Fixation Indices |
|---|---|---|---|
| Natural | Among groups | Va = 44.64 | FCT = 0.446 *** |
| Invaded | Among populations within groups | Vb = 31.08 | FSC = 0.561 *** |
| Within populations | Vc = 24.28 | FST = 0.757 *** |
| Group | n | No. | Nex | S | ts/tv | h ± SD | π (%) ± SD |
|---|---|---|---|---|---|---|---|
| Invaded | 477 | 5 | 3 | 7 | 6/1 | 0.6232 ± 0.0125 | 0.2120 ± 0.1517 |
| E | 250 | 2 | 0 | 1 | 1/0 | 0.4310 ± 0.0222 | 0.0789 ± 0.0791 |
| A | 227 | 7 | 2 | 7 | 6/1 | 0.5368 ± 0.0331 | 0.2996 ± 0.1964 |
| Native/NA | 40 | 9 | 7 | 10 | 10/1 | 0.7551 ± 0.0456 | 0.3933 ± 0.2479 |
| Total | 517 | 12 | n.r. | 11 | 11/1 | 0.6600 ± 0.0122 | 0.2552 ± 0.1737 |
| Groups | Source of Variation | var% | Fixation Indices |
|---|---|---|---|
| Natural | Among groups | Va = 57.47 | FCT = 0.575 *** |
| Invaded | Among populations within groups | Vb = 15.97 | FSC = 0.376 *** |
| Within populations | Vc = 26.56 | FST = 0.734 *** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lakatos, F.; Tuba, K.; Bender, B.; Kajimura, H.; Tóth, V. Longer mtDNA Fragments Provide a Better Insight into the Genetic Diversity of the Sycamore Lace Bug, Corythucha ciliata (Say, 1832) (Tingidae, Hemiptera), Both in Its Native and Invaded Areas. Insects 2022, 13, 123. https://doi.org/10.3390/insects13020123
Lakatos F, Tuba K, Bender B, Kajimura H, Tóth V. Longer mtDNA Fragments Provide a Better Insight into the Genetic Diversity of the Sycamore Lace Bug, Corythucha ciliata (Say, 1832) (Tingidae, Hemiptera), Both in Its Native and Invaded Areas. Insects. 2022; 13(2):123. https://doi.org/10.3390/insects13020123
Chicago/Turabian StyleLakatos, Ferenc, Katalin Tuba, Boglárka Bender, Hisashi Kajimura, and Viktória Tóth. 2022. "Longer mtDNA Fragments Provide a Better Insight into the Genetic Diversity of the Sycamore Lace Bug, Corythucha ciliata (Say, 1832) (Tingidae, Hemiptera), Both in Its Native and Invaded Areas" Insects 13, no. 2: 123. https://doi.org/10.3390/insects13020123
APA StyleLakatos, F., Tuba, K., Bender, B., Kajimura, H., & Tóth, V. (2022). Longer mtDNA Fragments Provide a Better Insight into the Genetic Diversity of the Sycamore Lace Bug, Corythucha ciliata (Say, 1832) (Tingidae, Hemiptera), Both in Its Native and Invaded Areas. Insects, 13(2), 123. https://doi.org/10.3390/insects13020123







