1H NMR Profiling of Honey Bee Bodies Revealed Metabolic Differences between Summer and Winter Bees
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Sample Preparation for NMR Analysis
2.3. 1H NMR Analysis
2.4. Data Processing and Statistical Analysis
3. Results
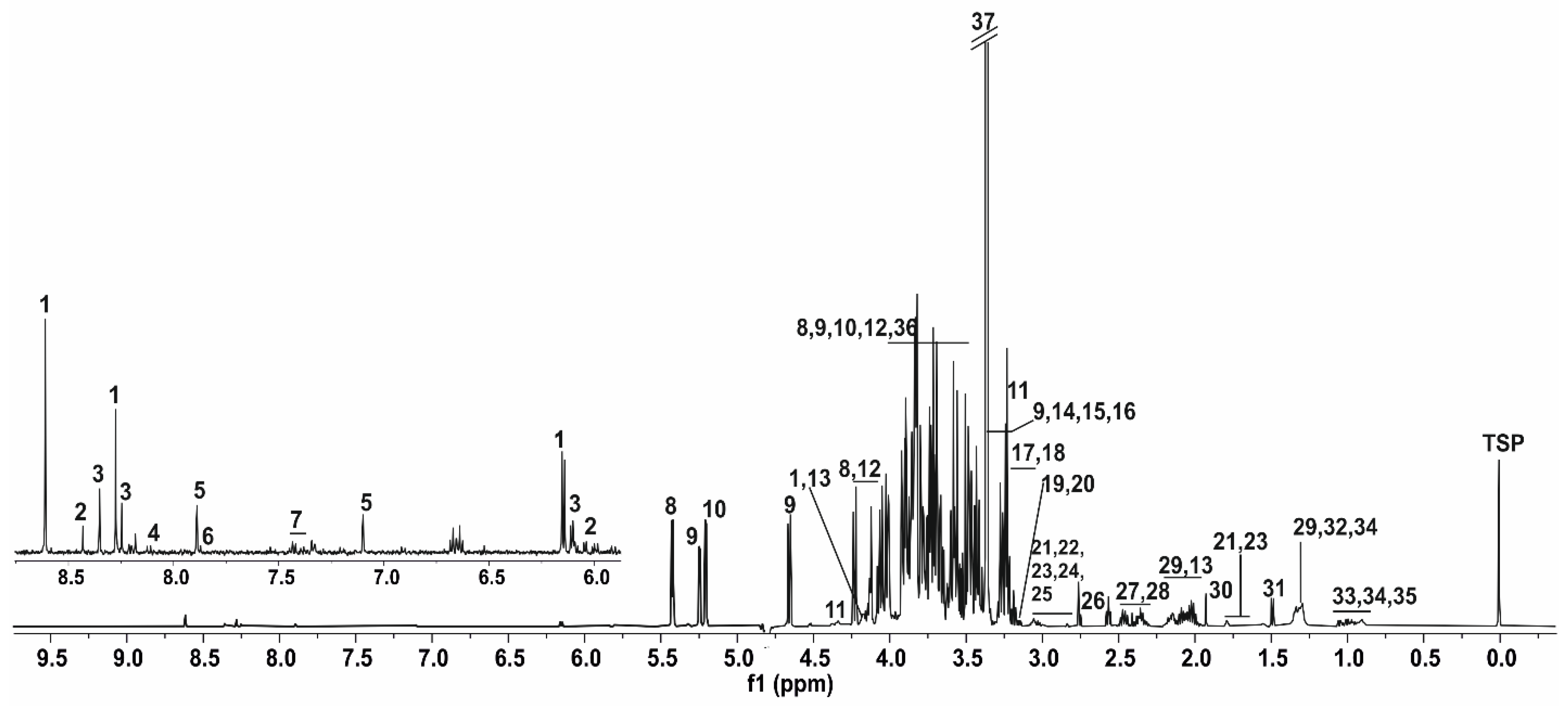
3.1. NMR Spectrum of Honey Bee’s Body
3.2. Multivariate Analysis Explaining the Discrimination between Summer (June) and Winter Bees (Late August)
3.3. Univariate Analysis Confirmed Statistically Significant Metabolites
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Potts, S.G.; Biesmeijer, J.C.; Kremen, C.; Neumann, P.; Schweiger, O.; Kunin, W.E. Global pollinator declines: Trends, impacts and drivers. Trends Ecol. Evol. 2010, 25, 345–353. [Google Scholar] [CrossRef]
- Brodschneider, R.; Gray, A.; Adjlane, N.; Ballis, A.; Brusbardis, V.; Charrière, J.D.; Chlebo, R.; Coffey, M.F.; Dahle, B.; de Graaf, D.C.; et al. Multi-country loss rates of honey bee colonies during winter 2016/2017 from the COLOSS survey. J. Apic. Res. 2018, 57, 452–457. [Google Scholar] [CrossRef] [Green Version]
- Vanengelsdorp, D.; Caron, D.; Hayes, J.; Underwood, R.; Henson, M.; Rennich, K.; Spleen, A.; Andree, M.; Snyder, R.; Lee, K.; et al. A national survey of managed honey bee 2010–11 winter colony losses in the USA: Results from the Bee Informed Partnership. J. Apic. Res. 2012, 51, 115–124. [Google Scholar] [CrossRef] [Green Version]
- Gray, A.; Brodschneider, R.; Adjlane, N.; Ballis, A.; Brusbardis, V.; Charrière, J.-D.; Chlebo, R.; Coffey, M.F.; Cornelissen, B.; da Costa, C.A.; et al. Loss rates of honey bee colonies during winter 2017/18 in 36 countries participating in the COLOSS survey including effects of forage sources. J. Apic. Res. 2019, 58, 479–485. [Google Scholar] [CrossRef] [Green Version]
- Steinhauer, N.A.; Rennich, K.; Wilson, M.E.; Caron, D.M.; Lengerich, E.J.; Pettis, J.S.; Rose, R.; Skinner, J.A.; Tarpy, D.R.; Wilkes, J.T.; et al. A national survey of managed honey bee 2012–2013 annual colony losses in the USA: Results from the Bee Informed Partnership. J. Apic. Res. 2014, 53, 1–18. [Google Scholar] [CrossRef]
- Van Engelsdorp, D.; Evans, J.D.; Saegerman, C.; Mullin, C.; Haubruge, E.; Nguyen, B.K.; Frazier, M.; Frazier, J.; Cox-Foster, D.; Chen, Y.; et al. Colony collapse disorder: A descriptive study. PLoS ONE 2009, 4, e6481. [Google Scholar] [CrossRef]
- Van Dooremalen, C.; Gerritsen, L.; Cornelissen, B.; van der Steen, J.J.M.; van Langevelde, F.; Blacquière, T. Winter survival of individual honey bees and honey bee colonies depends on level of varroa destructor infestation. PLoS ONE 2012, 7, e36285. [Google Scholar] [CrossRef] [Green Version]
- Higes, M.; Martín-Hernández, R.; Botías, C.; Bailón, E.G.; González-Porto, A.V.; Barrios, L.; Del Nozal, M.J.; Bernal, J.L.; Jiménez, J.J.; Palencia, P.G.; et al. How natural infection by Nosema ceranae causes honeybee colony collapse. Environ. Microbiol. 2008, 10, 2659–2669. [Google Scholar] [CrossRef] [PubMed]
- Highfield, A.C.; El Nagar, A.; Mackinder, L.C.M.; Noël, L.M.L.J.; Hall, M.J.; Martin, S.J.; Schroeder, D.C. Deformed wing virus implicated in overwintering honeybee colony losses. Appl. Environ. Microbiol. 2009, 75, 7212–7220. [Google Scholar] [CrossRef] [Green Version]
- Simon-Delso, N.; Martin, G.S.; Bruneau, E.; Minsart, L.A.; Mouret, C.; Hautier, L. Honeybee colony disorder in crop areas: The role of pesticides and viruses. PLoS ONE 2014, 9, e103073. [Google Scholar] [CrossRef]
- Requier, F.; Odoux, J.F.; Henry, M.; Bretagnolle, V. The carry-over effects of pollen shortage decrease the survival of honeybee colonies in farmlands. J. Appl. Ecol. 2017, 54, 1161–1170. [Google Scholar] [CrossRef]
- Seitz, N.; Traynor, K.S.; Steinhauer, N.; Rennich, K.; Wilson, M.E.; Ellis, J.D.; Rose, R.; Tarpy, D.R.; Sagili, R.R.; Caron, D.M.; et al. A national survey of managed honey bee 2014–2015 annual colony losses in the USA Nicola. J. Apic. Res. 2015, 54, 292–304. [Google Scholar] [CrossRef]
- Le Conte, Y.; Navajas, M. Climate change: Impact on honey bee populations and diseases. Rev. Sci. Tech. 2008, 27, 485–497, 499–510. [Google Scholar] [CrossRef]
- Page, R.E.; Peng, C.Y.S. Aging and development in social insects with emphasis on the honey bee, Apis mellifera L. Exp. Gerontol. 2001, 36, 695–711. [Google Scholar] [CrossRef]
- Mattila, H.R.; Harris, J.L.; Otis, G.W. Timing of production of winter bees in honey bee (Apis mellifera) colonies. Insectes Soc. 2001, 48, 88–93. [Google Scholar] [CrossRef]
- Fluri, P.; Lüscher, M.; Wille, H.; Gerig, L. Changes in weight of the pharyngeal gland and haemolymph titres of juvenile hormone, protein and vitellogenin in worker honey bees. J. Insect Physiol. 1982, 28, 61–68. [Google Scholar] [CrossRef]
- Seeley, T.D. The Lives of Bees; Princeton University Press: Princeton, NJ, USA, 2019; ISBN 978-0-691-16676-6. [Google Scholar]
- Omholt, S.W.; Amdam, G.V. Epigenetic regulation of aging in honeybee workers. Sci. Aging Knowl. Environ. 2004, 2004, pe28. [Google Scholar] [CrossRef]
- Corona, M.; Velarde, R.A.; Remolina, S.; Moran-lauter, A.; Wang, Y.; Hughes, K.A.; Robinson, G.E. Vitellogenin, juvenile hormone, insulin signaling, and queen honey bee longevity. Pnas 2007, 104, 7128–7133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harwood, G.P.; Ihle, K.E.; Salmela, H.; Amdam, G.V. Regulation of Honeybee Worker (Apis mellifera) Life Histories by Vitellogenin, 3rd ed.; Elsevier: Amsterdam, The Netherlands, 2017; Volume 2, ISBN 9780128036082. [Google Scholar]
- Kunert, K.; Crailsheim, K. Seasonal changes in carbohydrate, lipid and protein content in emerging worker honeybees and their mortality. J. Apic. Res. 1988, 27, 13–21. [Google Scholar] [CrossRef]
- Kunc, M.; Dobeš, P.; Hurychová, J.; Vojtek, L.; Poiani, S.B.; Danihlík, J.; Havlík, J.; Titěra, D.; Hyršl, P. The year of the honey bee (Apis mellifera L.) with respect to its physiology and immunity: A search for biochemical markers of longevity. Insects 2019, 10, 244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kešnerová, L.; Emery, O.; Troilo, M.; Liberti, J.; Erkosar, B.; Engel, P. Gut microbiota structure differs between honeybees in winter and summer. ISME J. 2020, 14, 801–814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seehuus, S.C.; Krekling, T.; Amdam, G.V. Cellular senescence in honey bee brain is largely independent of chronological age. Exp. Gerontol. 2006, 41, 1117–1125. [Google Scholar] [CrossRef] [Green Version]
- Behrends, A.; Scheiner, R. Learning at old age: A study on winter bees. Front. Behav. Neurosci. 2010, 4, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, C.H.; Ivanisevic, J.; Siuzdak, G. Metabolomics: Beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 2016, 17, 451–459. [Google Scholar] [CrossRef] [Green Version]
- Fan, T.W.M.; Lane, A.N. Applications of NMR spectroscopy to systems biochemistry. Prog. Nucl. Magn. Reson. Spectrosc. 2016, 92–93, 18–53. [Google Scholar] [CrossRef] [Green Version]
- Cox, J.E.; Thummel, C.S.; Tennessen, J.M. Metabolomic studies in Drosophila. Genetics 2017, 206, 1169–1185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lenz, E.M.; Hägele, B.F.; Wilson, I.D.; Simpson, S.J. High resolution 1H NMR spectroscopic studies of the composition of the haemolymph of crowd- and solitary-reared nymphs of the desert locust, Schistocerca gregaria. Insect Biochem. Mol. Biol. 2001, 32, 51–56. [Google Scholar] [CrossRef]
- Kapranas, A.; Snart, C.J.P.; Williams, H.; Hardy, I.C.W.; Barrett, D.A. Metabolomics of aging assessed in individual parasitoid wasps. Sci. Rep. 2016, 6, 34848. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Carolan, J.C.; Hao, F.; Nicholson, J.K.; Wilkinson, T.L.; Douglas, A.E. Integrated metabonomic-proteomic analysis of an insect-bacterial symbiotic system. J. Proteome Res. 2010, 9, 1257–1267. [Google Scholar] [CrossRef] [Green Version]
- Jousse, C.; Dalle, C.; Abila, A.; Traikia, M.; Diogon, M.; Lyan, B.; El Alaoui, H.; Vidau, C.; Delbac, F. A combined LC-MS and NMR approach to reveal metabolic changes in the hemolymph of honeybees infected by the gut parasite Nosema ceranae. J. Invertebr. Pathol. 2020, 176, 1–8. [Google Scholar] [CrossRef]
- Dostálková, S.; Dobeš, P.; Kunc, M.; Hurychová, J.; Škrabišová, M.; Petřivalský, M.; Titěra, D.; Havlík, J.; Hyršl, P.; Danihlík, J. Winter honeybee (Apis mellifera) populations show greater potential to induce immune responses than summer populations after immune stimuli. J. Exp. Biol. 2021, 224. [Google Scholar] [CrossRef] [PubMed]
- Williams, G.R.; Alaux, C.; Costa, C.; Csáki, T.; Doublet, V.; Eisenhardt, D.; Fries, I.; Kuhn, R.; McMahon, D.P.; Medrzycki, P.; et al. Standard methods for maintaining adult Apis mellifera in cages under in vitro laboratory conditions. J. Apic. Res. 2013, 52, 1–36. [Google Scholar] [CrossRef] [Green Version]
- Pang, Z.; Chong, J.; Li, S.; Xia, J. Metaboanalystr 3.0: Toward an optimized workflow for global metabolomics. Metabolites 2020, 10, 186. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.; Wishart, D.S.; Xia, J. Using MetaboAnalyst 4.0 for Comprehensive and Integrative Metabolomics Data Analysis. Curr. Protoc. Bioinforma. 2019, 68, 1–128. [Google Scholar] [CrossRef]
- Lê, S.; Josse, J.; Husson, F. FactoMineR: An R Package for Multivariate Analysis. J. Stat. Softw. 2008, 25, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. [Google Scholar]
- Blatt, J.; Roces, F. Haemolymph sugar levels in foraging honeybees (Apis mellifera carnica): Dependence on metabolic rate and in vivo measurement of maximal rates of trehalose synthesis. J. Exp. Biol. 2001, 204, 2709–2716. [Google Scholar] [CrossRef]
- Suarez, R.K.; Darveau, C.A.; Welch, K.C.; O’Brien, D.M.; Roubik, D.W.; Hochachka, P.W. Energy metabolism in orchid bee flight muscles: Carbohydrate fuels all. J. Exp. Biol. 2005, 208, 3573–3579. [Google Scholar] [CrossRef] [Green Version]
- Arrese, E.L.; Soulages, J.L. Insect Fat Body: Energy, Metabolism, and Regulation. Annu. Rev. Entomol. 2010, 55, 207–225. [Google Scholar] [CrossRef] [Green Version]
- Hollis Woodard, S.; Duennes, M.A.; Watrous, K.M.; Jha, S. Diet and nutritional status during early adult life have immediate and persistent effects on queen bumble bees. Conserv. Physiol. 2019, 7, coz048. [Google Scholar] [CrossRef]
- Thompson, S.N. Trehalose—The Insect “Blood” Sugar. Adv. Insect Phys. 2003, 31, 205–285. [Google Scholar] [CrossRef]
- Koštál, V.; Zahradníčková, H.; Šimek, P.; Zelený, J. Multiple component system of sugars and polyols in the overwintering spruce bark beetle, Ips typographus. J. Insect Physiol. 2007, 53, 580–586. [Google Scholar] [CrossRef] [PubMed]
- Overgaard, J.; Malmendal, A.; Sørensen, J.G.; Bundy, J.G.; Loeschcke, V.; Nielsen, N.C.; Holmstrup, M. Metabolomic profiling of rapid cold hardening and cold shock in Drosophila melanogaster. J. Insect Physiol. 2007, 53, 1218–1232. [Google Scholar] [CrossRef]
- Watanabe, M.; Kikawada, T.; Minagawa, N.; Yukuhiro, F.; Okuda, T. Mechanism allowing an insect to survive complete dehydration and extreme temperatures. J. Exp. Biol. 2002, 205, 2799–2802. [Google Scholar] [CrossRef] [PubMed]
- Thompson, K.M.; Huber, D.P.W.; Murray, B.W. Autumn shifts in cold tolerance metabolites in overwintering adult mountain pine beetles. PLoS ONE 2020, 15, e0227203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, M.; Wang, H.; Liu, Z.; Wang, Y.; Zhang, W.; Xu, B. Changes in cold tolerance during the overwintering period in Apis mellifera ligustica. J. Apic. Res. 2019, 58, 702–709. [Google Scholar] [CrossRef]
- Crailsheim, K. Dependence of protein metabolism on age and season in the honeybee (Apis mellifica carnica Pollm). J. Insect Physiol. 1986, 32, 629–634. [Google Scholar] [CrossRef]
- Haydak, M.H. Honey Bee Nutrition. Annu. Rev. Entomol. 1970, 15, 143–156. [Google Scholar] [CrossRef]
- Crailsheim, K. The protein balance of the honey bee worker. Apidologie 1990, 21, 417–429. [Google Scholar] [CrossRef]
- Crailsheim, K.; Leonhard, B. Amino acids in honeybee worker haemolymph. Amino Acids 1997, 13, 141–153. [Google Scholar] [CrossRef]
- Hrassnigg, N.; Leonhard, B.; Crailsheim, K. Free amino acids in the haemolymph of honey bee queens (Apis mellifera L.). Amino Acids 2003, 24, 205–212. [Google Scholar] [CrossRef]
- Storey, K.B.; Storey, J.M. Natural freezing survival in animals. Annu. Rev. Ecol. Syst. 1996, 27, 365–386. [Google Scholar] [CrossRef] [Green Version]
- Koštál, V.; Šimek, P.; Zahradníčková, H.; Cimlová, J.; Štětina, T. Conversion of the chill susceptible fruit fly larva (Drosophila melanogaster) to a freeze tolerant organism. Proc. Natl. Acad. Sci. USA 2012, 109, 3270–3274. [Google Scholar] [CrossRef] [Green Version]
- Koštál, V.; Korbelová, J.; Rozsypal, J.; Zahradníčková, H.; Cimlová, J.; Tomčala, A.; Šimek, P. Long-term cold acclimation extends survival time at 0°C and modifies the metabolomic profiles of the larvae of the fruit fly drosophila melanogaster. PLoS ONE 2011, 6, e25025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, C.M.; McCue, M.D.; Sunny, N.E.; Szejner-Sigal, A.; Morgan, T.J.; Allison, D.B.; Hahn, D.A. Cold adaptation increases rates of nutrient flow and metabolic plasticity during cold exposure in Drosophila melanogaster. Proc. R. Soc. B Biol. Sci. 2016, 283. [Google Scholar] [CrossRef] [Green Version]
- Mattila, H.R.; Otis, G.W. Dwindling pollen resources trigger the transition to broodless populations of long-lived honeybees each autumn. Ecol. Entomol. 2007, 32, 496–505. [Google Scholar] [CrossRef]
- Le Couteur, D.G.; Solon-Biet, S.; Cogger, V.C.; Mitchell, S.J.; Senior, A.; De Cabo, R.; Raubenheimer, D.; Simpson, S.J. The impact of low-protein high-carbohydrate diets on aging and lifespan. Cell. Mol. Life Sci. 2016, 73, 1237–1252. [Google Scholar] [CrossRef] [PubMed]
- Paoli, P.P.; Wakeling, L.A.; Wright, G.A.; Ford, D. The dietary proportion of essential amino acids and Sir2 influence lifespan in the honeybee. Age 2014, 36, 1239–1247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gómez-Moracho, T.; Durand, T.; Pasquaretta, C.; Heeb, P.; Lihoreau, M. Artificial diets modulate infection rates by nosema ceranae in bumblebees. Microorganisms 2021, 9, 158. [Google Scholar] [CrossRef] [PubMed]
- Amdam, G.V.; Simões, Z.L.P.; Hagen, A.; Norberg, K.; Schrøder, K.; Mikkelsen, Ø.; Kirkwood, T.B.L.; Omholt, S.W. Hormonal control of the yolk precursor vitellogenin regulates immune function and longevity in honeybees. Exp. Gerontol. 2004, 39, 767–773. [Google Scholar] [CrossRef]
- Imboden, H.; König, R.; Ott, P.; Lustig, A.; Kämpfer, U.; Lanzrein, B. Characterization of the native vitellogenin and vitellin of the cockroach, Nauphoeta cinerea, and comparison with other species. Insect Biochem. 1987, 17, 353–365. [Google Scholar] [CrossRef]
- Wheeler, D.E.; Kawooya, J.K. Purification and characterization of honey bee vitellogenin. Arch. Insect Biochem. Physiol. 1990, 14, 253–267. [Google Scholar] [CrossRef] [PubMed]
- Moessinger, C.; Klizaite, K.; Steinhagen, A.; Philippou-Massier, J.; Shevchenko, A.; Hoch, M.; Ejsing, C.S.; Thiele, C. Two different pathways of phosphatidylcholine synthesis, the Kennedy Pathway and the Lands Cycle, differentially regulate cellular triacylglycerol storage. BMC Cell Biol. 2014, 15, 43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hardie, D.G. AMP-activated/SNF1 protein kinases: Conserved guardians of cellular energy. Nat. Rev. Mol. Cell Biol. 2007, 8, 774–785. [Google Scholar] [CrossRef] [PubMed]
- Bonkowski, M.S.; Sinclair, D.A. Slowing ageing by design: The rise of NAD+ and sirtuin-activating compounds. Nat. Rev. Mol. Cell Biol. 2016, 17, 679–690. [Google Scholar] [CrossRef]
- Apfeld, J.; O’Connor, G.; McDonagh, T.; DiStefano, P.S.; Curtis, R. The AMP-activated protein kinase AAK-2 links energy levels and insulin-like signals to lifespan in C. elegans. Genes Dev. 2004, 18, 3004–3009. [Google Scholar] [CrossRef] [Green Version]
- Funakoshi, M.; Tsuda, M.; Muramatsu, K.; Hatsuda, H.; Morishita, S.; Aigaki, T. A gain-of-function screen identifies wdb and lkb1 as lifespan-extending genes in Drosophila. Biochem. Biophys. Res. Commun. 2011, 405, 667–672. [Google Scholar] [CrossRef] [PubMed]
- Chuang, Y.L.; Hsu, C.Y. Changes in mitochondrial energy utilization in young and old worker honeybees (Apis mellifera). Age 2013, 35, 1867–1879. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsu, C.Y.; Chuang, Y.L. Changes in energy-regulated molecules in the trophocytes and fat cells of young and old worker honeybees (Apis mellifera). J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2014, 69, 955–964. [Google Scholar] [CrossRef] [Green Version]


| Summber Bees (n = 75) | Winter Bees (n = 45) | Independent t-Test | Benjamini–Hochberg | ||||
|---|---|---|---|---|---|---|---|
| Metabolite | Mean | SD | Mean | SD | DF 1 | p-Value | p-Value |
| AMP | 27.56 | 6.60 | 36.12 | 7.55 | 118 | 0.0019 | <0.0001 |
| Acetate | 11.29 | 8.42 | 7.71 | 2.69 | 118 | <0.0001 | 0.0027 |
| Adenine | 1.29 | 0.82 | 0.46 | 0.25 | 118 | <0.0001 | <0.0001 |
| Adenosine | 1.25 | 0.68 | 2.76 | 1.58 | 118 | 0.0001 | <0.0001 |
| Alanine | 23.38 | 8.90 | 28.56 | 6.72 | 118 [115.6] | <0.0001 | 0.0001 |
| Asparagine | 10.45 | 3.55 | 5.52 | 2.09 | 118 | <0.0001 | <0.0001 |
| Choline | 27.35 | 9.66 | 12.57 | 3.50 | 118 | <0.0001 | <0.0001 |
| Creatine | 2.72 | 0.71 | 3.08 | 0.72 | 118 | 0.0120 | 0.0166 |
| Fructose | 509.4 | 611.6 | 1109.1 | 1008.3 | 118 [84.2] | 0.0001 | 0.0001 |
| Glucose | 655.8 | 442.2 | 915.0 | 715.6 | 118 [68.8] | 0.1664 | 0.1997 |
| Glutamine | 74.25 | 18.29 | 60.93 | 14.42 | 118 | 0.0001 | 0.0002 |
| Glycine | 11.37 | 3.12 | 13.52 | 11.00 | 118 | 0.3608 | 0.4059 |
| Histidine | 7.82 | 1.85 | 7.75 | 1.73 | 118 | 0.8449 | 0.8449 |
| Inosine | 14.73 | 4.76 | 9.17 | 2.36 | 118 | <0.0001 | <0.0001 |
| Isoleucine | 9.82 | 4.41 | 6.06 | 1.77 | 118 [112.8] | <0.0001 | <0.0001 |
| Leucine | 10.35 | 6.02 | 3.54 | 1.31 | 118 | <0.0001 | <0.0001 |
| Lysine | 10.83 | 4.45 | 6.67 | 2.59 | 118 | <0.0001 | <0.0001 |
| Malonate | 6.68 | 0.93 | 6.46 | 0.97 | 118 | 0.2347 | 0.2726 |
| NAD+ | 4.54 | 1.78 | 8.66 | 2.48 | 118 [116.8] | <0.0001 | <0.0001 |
| O-Phosphocholine | 68.38 | 17.75 | 88.01 | 15.99 | 118 [117.8] | <0.0001 | <0.0001 |
| Phenylalanine | 4.39 | 2.53 | 1.31 | 0.55 | 118 | <0.0001 | <0.0001 |
| Proline | 99.24 | 29.01 | 123.47 | 29.05 | 118 | <0.0001 | <0.0001 |
| Putrescine | 7.41 | 2.62 | 7.28 | 1.75 | 118 [116.6] | 0.7555 | 0.7771 |
| Sarcosine | 4.00 | 1.80 | 4.55 | 0.97 | 118 [117.5] | 0.0008 | 0.0012 |
| Suberate | 5.10 | 2.90 | 4.58 | 2.19 | 118 | 0.5916 | 0.7204 |
| Succinate | 3.86 | 1.53 | 5.49 | 2.47 | 118 | 0.6804 | <0.0001 |
| Sucrose | 284.3 | 301.9 | 1690.3 | 2135.0 | 118 | <0.0001 | <0.0001 |
| Taurine | 87.57 | 16.47 | 79.62 | 18.12 | 118 | <0.0001 | 0.0167 |
| Threonine | 10.19 | 4.73 | 6.00 | 2.97 | 118 | 0.0125 | <0.0001 |
| Trehalose | 416.0 | 181.1 | 801.8 | 212.0 | 118 [115.7] | <0.0001 | <0.0001 |
| Trimethylamine N-oxide | 0.86 | 0.63 | 2.97 | 1.54 | 118 | <0.0001 | <0.0001 |
| Unknown | 19.61 | 7.62 | 1.53 | 0.55 | 118 | <0.0001 | <0.0001 |
| Uridine | 2.38 | 0.85 | 2.01 | 0.59 | 118 | <0.0001 | 0.0499 |
| Valine | 8.83 | 3.60 | 3.61 | 1.52 | 118 | 0.0388 | <0.0001 |
| sn-Glycero-3-phosphocholine | 61.28 | 21.28 | 63.42 | 18.68 | 118 | <0.0001 | 0.6454 |
| β-Alanine | 33.81 | 6.80 | 31.56 | 7.31 | 118 | 0.0761 | 0.0945 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, S.; Kalcic, F.; Duarte, I.F.; Titera, D.; Kamler, M.; Mrna, P.; Hyrsl, P.; Danihlik, J.; Dobes, P.; Kunc, M.; et al. 1H NMR Profiling of Honey Bee Bodies Revealed Metabolic Differences between Summer and Winter Bees. Insects 2022, 13, 193. https://doi.org/10.3390/insects13020193
Lee S, Kalcic F, Duarte IF, Titera D, Kamler M, Mrna P, Hyrsl P, Danihlik J, Dobes P, Kunc M, et al. 1H NMR Profiling of Honey Bee Bodies Revealed Metabolic Differences between Summer and Winter Bees. Insects. 2022; 13(2):193. https://doi.org/10.3390/insects13020193
Chicago/Turabian StyleLee, Saetbyeol, Filip Kalcic, Iola F. Duarte, Dalibor Titera, Martin Kamler, Pavel Mrna, Pavel Hyrsl, Jiri Danihlik, Pavel Dobes, Martin Kunc, and et al. 2022. "1H NMR Profiling of Honey Bee Bodies Revealed Metabolic Differences between Summer and Winter Bees" Insects 13, no. 2: 193. https://doi.org/10.3390/insects13020193
APA StyleLee, S., Kalcic, F., Duarte, I. F., Titera, D., Kamler, M., Mrna, P., Hyrsl, P., Danihlik, J., Dobes, P., Kunc, M., Pudlo, A., & Havlik, J. (2022). 1H NMR Profiling of Honey Bee Bodies Revealed Metabolic Differences between Summer and Winter Bees. Insects, 13(2), 193. https://doi.org/10.3390/insects13020193







