Multiplex PCR Assay for the Identification of Four Species of the Anopheles Leucosphyrus Sub-Group in Malaysia
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
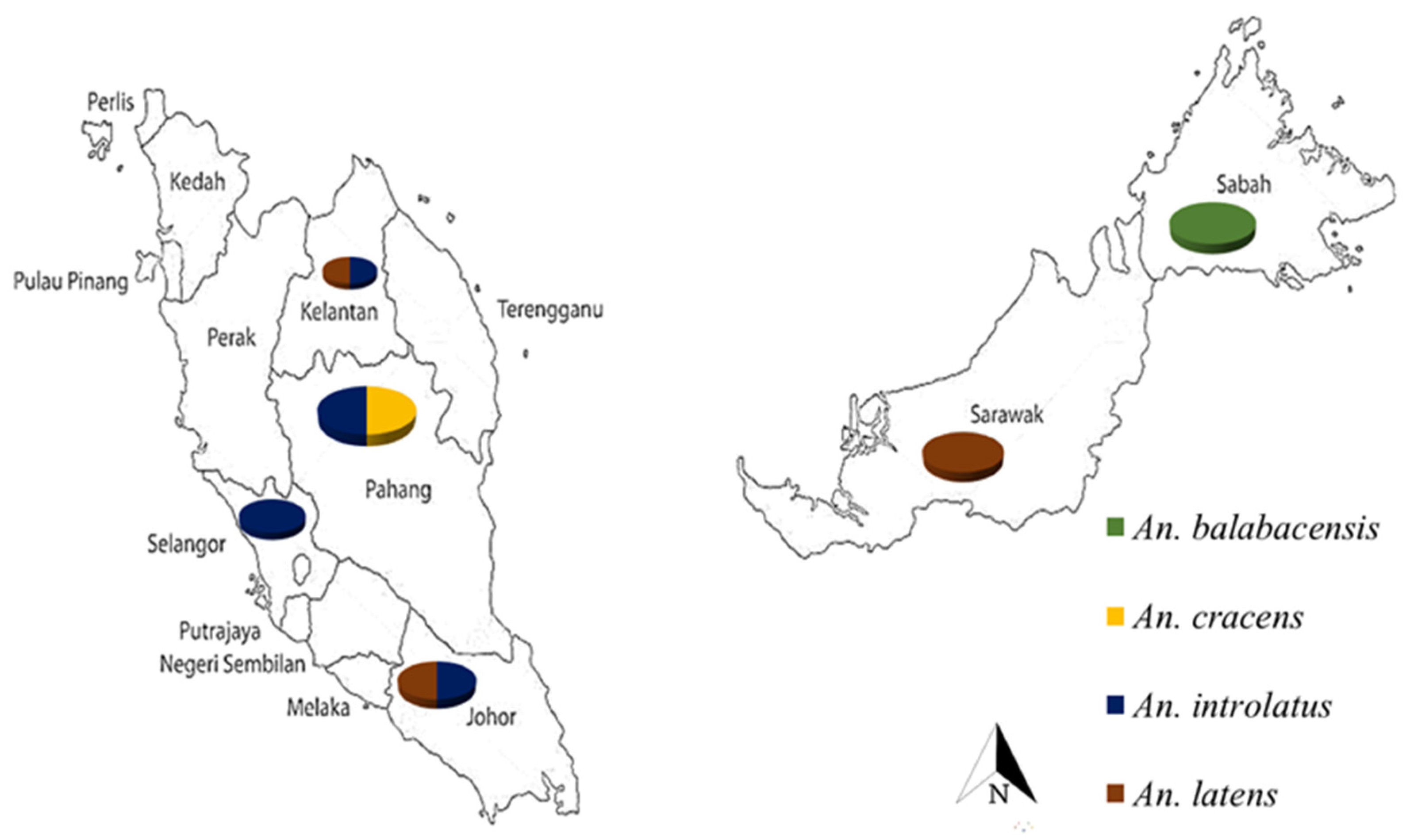
2.1. Sample Collection
2.2. DNA Extraction and Molecular Identification of Field Caught Anopheles
2.3. Sequence Analysis
2.4. Primer Design
2.5. Multiplex PCR Assay for Four Anopheles Species
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. World Malaria Report 2020: 20 Years of Global Progress and Challenges. Available online: https://cdn.who.int/media/docs/default-source/malaria/world-malaria-reports/9789240015791-double-page-view.pdf?sfvrsn=2c24349d10 (accessed on 6 August 2021).
- World Health Organization. WHO Certifies Sri Lanka Malaria-Free. Available online: https://www.who.int/southeastasia/news/detail/05-09-2016-who-certifies-sri-lanka-malaria-free (accessed on 6 August 2021).
- World Health Organization. From 30 Million Cases to Zero: China Is Certified Malaria-Free by WHO. Available online: https://www.who.int/news/item/30-06-2021-from-30-million-cases-to-zero-china-is-certified-malaria-free-by-who (accessed on 6 August 2021).
- World Health Organization. Update on the E-2020 Initiative of 21 Malaria-Eliminating Countries: Report and Country Briefs. Available online: https://apps.who.int/iris/bitstream/handle/10665/273633/WHO-CDS-GMP-2018.13-eng.pdf (accessed on 6 August 2021).
- Jeyaprakasam, N.K.; Liew, J.W.K.; Low, V.L.; Wan-Sulaiman, W.-Y.; Vythilingam, I. Plasmodium knowlesi infecting humans in Southeast Asia: What’s next? PLoS Negl. Trop. Dis. 2020, 14, e0008900. [Google Scholar] [CrossRef] [PubMed]
- Ta, T.H.; Hisam, S.; Lanza, M.; Jiram, A.I.; Ismail, N.; Rubio, J.M. First case of a naturally acquired human infection with Plasmodium cynomolgi. Malar. J. 2014, 13, 68. [Google Scholar] [CrossRef] [PubMed]
- Grignard, L.; Shah, S.; Chua, T.H.; William, T.; Drakeley, C.J.; Fornace, K.M. Natural human infections with Plasmodium cynomolgi and other malaria species in an elimination setting in Sabah, Malaysia. J. Infect. Dis. 2019, 220, 1946–1949. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.; Kadir, K.A.; Hu, T.H.; Raja, T.N.; Mohamad, D.S.; Lin, L.W.; Hii, K.C. Naturally acquired human infections with the simian malaria parasite, Plasmodium cynomolgi, in Sarawak, Malaysian Borneo. Int. J. Infect. Dis. 2018, 73, 68. [Google Scholar] [CrossRef]
- Imwong, M.; Madmanee, W.; Suwannasin, K.; Kunasol, C.; Peto, T.; Tripura, R.; Von Seidlein, L.; Nguon, C.; Davoeung, C.; Day, N.P.J.; et al. Asymptomatic natural human infections with the simian malaria parasites Plasmodium cynomolgi and Plasmodium knowlesi. J. Infect. Dis. 2019, 219, 695–702. [Google Scholar] [CrossRef]
- Hartmeyer, G.N.; Stensvold, C.R.; Fabricius, T.; Marmolin, E.S.; Hoegh, S.V.; Nielsen, H.V.; Kemp, M.; Vestergaard, L.S. Plasmodium cynomolgi as cause of malaria in tourist to Southeast Asia. Emerg. Infect. Dis. 2019, 25, 1936–1939. [Google Scholar] [CrossRef]
- Yap, N.J.; Hossain, H.; Nada-Raja, T.; Ngui, R.; Muslim, A.; Hoh, B.-P.; Khaw, L.T.; Kadir, K.A.; Divis, P.C.S.; Vythilingam, I.; et al. Natural human infections with Plasmodium cynomolgi, P. inui, and 4 other simian malaria parasites, Malaysia. Emerg. Infect. Dis. 2021, 27, 2187–2191. [Google Scholar] [CrossRef]
- Liew, J.W.; Bukhari, F.D.M.; Jeyaprakasam, N.K.; Phang, W.K.; Vythilingam, I.; Lau, Y.L. Natural Plasmodium inui infections in humans and Anopheles cracens Mosquito, Malaysia. Emerg. Infect. Dis. 2021, 27, 2700–2703. [Google Scholar] [CrossRef]
- Ministry of Health. Press Statement Zoonotic Malaria and the Prevention Program in Malaysia 2018. Available online: https://www.moh.gov.my/index.php/database_stores/attach_download/337/1087 (accessed on 7 August 2021).
- Warren, M.; Wharton, R.H. The vectors of simian malaria: Identity, biology, and geographical distribution. J. Parasitol. 1963, 49, 892–904. [Google Scholar] [CrossRef]
- Peyton, E.L. A new classification for the Leucosphyrus Group of Anopheles (Cellia). Mosq. Syst. 1989, 21, 197–205. [Google Scholar]
- Sallum, M.; Peyton, E.L.; Wilkerson, R. Six new species of the Anopheles leucosphyrus Group, reinterpretation of An. elegans and vector implications. Med. Vet. Entomol. 2005, 19, 158–199. [Google Scholar] [CrossRef] [PubMed]
- Manguin, S.; Garros, C.; Dusfour, I.; Harbach, R.; Coosemans, M. Bionomics, taxonomy, and distribution of the major malaria vector taxa of Anopheles subgenus Cellia in Southeast Asia: An updated review. Infect. Genet. Evol. 2008, 8, 489–503. [Google Scholar] [CrossRef]
- Sallum, M.A.M.; Peyton, E.L.; Harrison, B.A.; Wilkerson, R.C. Revision of the Leucosphyrus Group of Anopheles (Cellia) (Diptera, Culicidae). Rev. Bras. Entomol. 2005, 49 (Suppl. 1), 1–152. [Google Scholar] [CrossRef]
- Vythilingam, I.; Lim, Y.A.L.; Venugopalan, B.; Ngui, R.; Leong, C.S.; Wong, M.L.; Khaw, L.; Goh, X.; Yap, N.; Sulaiman, W.Y.W.; et al. Plasmodium knowlesi malaria an emerging public health problem in Hulu Selangor, Selangor, Malaysia (2009–2013): Epidemiologic and entomologic analysis. Parasites Vectors 2014, 7, 436. [Google Scholar] [CrossRef] [PubMed]
- Vythilingam, I.; Tan, C.H.; Asmad, M.; Chan, S.T.; Lee, K.S.; Singh, B. Natural transmission of Plasmodium knowlesi to humans by Anopheles latens in Sarawak, Malaysia. Trans. R. Soc. Trop. Med. Hyg. 2006, 100, 1087–1088. [Google Scholar] [CrossRef]
- Wong, M.L.; Chua, T.H.; Leong, C.S.; Khaw, L.T.; Fornace, K.; Wan-Sulaiman, W.-Y.; William, T.; Drakeley, C.; Ferguson, H.M.; Vythilingam, I. Seasonal and spatial dynamics of the primary vector of Plasmodium knowlesi within a major transmission focus in Sabah, Malaysia. PLoS Negl. Trop. Dis. 2015, 9, e0004135. [Google Scholar] [CrossRef]
- Vythilingam, I.; NoorAzian, Y.M.; Huat, T.C.; Jiram, A.I.; Yusri, Y.M.; Azahari, A.H.; NorParina, I.; NoorRain, A.; LokmanHakim, S. Plasmodium knowlesi in humans, macaques and mosquitoes in peninsular Malaysia. Parasit Vectors 2008, 1, 26. [Google Scholar] [CrossRef]
- Wharton, R.H.; Eyles, D.E. Anopheles hackeri, a vector of Plasmodium knowlesi in Malaya. Science 1961, 134, 279–280. [Google Scholar] [CrossRef]
- Van Bortel, W.; Trung, H.D.; Manh, N.D.; Roelants, P.; Verle, P.; Coosemans, M. Identification of two species within the Anopheles minimus complex in northern Vietnam and their behavioural divergences. Trop. Med. Int. Health 1999, 4, 257–265. [Google Scholar] [CrossRef]
- Green, C.A.; Gass, R.F.; Munstermann, L.E.; Baimai, V. Population-genetic evidence for two species in Anopheles minimus in Thailand. Med. Vet. Entomol. 1990, 4, 25–34. [Google Scholar] [CrossRef]
- Van Bortel, W.; Trung, H.D.; Roelants, P.; Harbach, R.E.; Backeljau, T.; Coosemans, M. Molecular identification of Anopheles minimus s.l. beyond distinguishing the members of the species complex. Insect Mol. Biol. 2000, 9, 335–340. [Google Scholar] [CrossRef] [PubMed]
- Tananchai, C.; Tisgratog, R.; Juntarajumnong, W.; Grieco, J.P.; Manguin, S.; Prabaripai, A.; Chareonviriyaphap, T. Species diversity and biting activity of Anopheles dirus and Anopheles baimaii (Diptera: Culicidae) in a malaria prone area of western Thailand. Parasites Vectors 2012, 5, 211. [Google Scholar] [CrossRef] [PubMed]
- Phuc, H.K.; Ball, A.J.; Son, L.; Hanh, N.V.; Tu, N.D.; Lien, N.G.; Verardi, A.; Townson, H. Multiplex PCR assay for malaria vector Anopheles minimus and four related species in the Myzomyia Series from Southeast Asia. Med. Vet. Entomol. 2003, 17, 423–428. [Google Scholar] [CrossRef] [PubMed]
- Choochote, W.; Saeung, A. Anopheles Mosquitoes-New Insights into Malaria Vectors; IntechOpen: London, UK, 2013; pp. 57–79. [Google Scholar]
- De Ang, J.X.; Yaman, K.; Kadir, K.A.; Matusop, A.; Singh, B. New vectors that are early feeders for Plasmodium knowlesi and other simian malaria parasites in Sarawak, Malaysian Borneo. Sci. Rep. 2021, 11, 7739. [Google Scholar] [CrossRef] [PubMed]
- Paredes-Esquivel, C.; Donnelly, M.J.; Harbach, R.E.; Townson, H. A molecular phylogeny of mosquitoes in the Anopheles barbirostris Subgroup reveals cryptic species: Implications for identification of disease vectors. Mol. Phylogenet. Evol. 2009, 50, 141–151. [Google Scholar] [CrossRef] [PubMed]
- Sungvornyothin, S.; Garros, C.; Chareonviriyaphap, T.; Manguin, S. How reliable is the humeral pale spot for identification of cryptic species of the Minimus Complex? J. Am. Mosq. Control Assoc. 2006, 22, 185–191. [Google Scholar] [CrossRef]
- Dusfour, I.; Blondeau, J.; Harbach, R.E.; Vythilingham, I.; Baimai, V.; Trung, H.D.; Sochanta, T.; Bangs, M.J.; Manguin, S. Polymerase chain reaction identification of three members of the Anopheles sundaicus (Diptera: Culicidae) complex, malaria vectors in Southeast Asia. J. Med. Entomol. 2007, 44, 723–731. [Google Scholar] [CrossRef]
- Baimai, V.; Kijchalao, U.; Sawadwongporn, P.; Green, C.A. Geographic distribution and biting behaviour of four species of the Anopheles dirus complex (Diptera: Culicidae) in Thailand. Southeast Asian J. Trop. Med. Public Health 1988, 19, 151–161. [Google Scholar]
- Baimai, V. Population cytogenetics of the malaria vector Anopheles Leucosphyrus Group. Southeast Asian J. Trop. Med. Public Health 1988, 19, 667–680. [Google Scholar]
- Baimai, V.; Harbach, R.; Kijchalao, U. Cytogenetic evidence for a fifth species within the taxon Anopheles dims in Thailand. J. Am. Mosq. Control Assoc. 1988, 4, 333–338. [Google Scholar]
- Baimai, V.; Poopittayasataporn, A.; Kijchalao, U. Cytological differences and chromosomal rearrangements in four members of the Anopheles dirus complex (Diptera: Culicidae). Genome 1988, 30, 372–379. [Google Scholar] [CrossRef] [PubMed]
- Baimai, V.; Thu, M.M.; Paing, M.; Maheswary, N.P. Distribution and chromosomal polymorphism of the malaria vector Anopheles dirus species D. Southeast Asian J. Trop. Med. Public Health 1988, 19, 661–665. [Google Scholar] [PubMed]
- Poopittayasataporn, A.; Baimai, V. Polytene chromosome relationships of five species of the Anopheles dirus complex in Thailand. Genome 1995, 38, 426–434. [Google Scholar] [CrossRef] [PubMed]
- Sawadipanich, Y.; Baimai, V.; Harrison, B.A. Anopheles dirus species E: Chromosomal and crossing evidence for another member of the dirus complex. J. Am. Mosq. Control Assoc. 1990, 6, 477–481. [Google Scholar] [PubMed]
- Green, C.A.; Munstermann, L.E.; Tan, S.; Panyim, S.; Baimai, V. Population genetic evidence for species A, B, C and D of the Anopheles dirus complex in Thailand and enzyme electromorphs for their identification. Med. Vet. Entomol. 1992, 6, 29–36. [Google Scholar] [CrossRef]
- Panyim, S.; Yasothornsrikul, S.; Tungpradubkul, S.; Baimai, V.; Rosenberg, R.; Andre, R.G.; Green, C.A. Identification of isomorphic malaria vectors using a DNA probe. Am. J. Trop. Med. Hyg. 1988, 38, 47–49. [Google Scholar] [CrossRef]
- Audtho, M.; Tassanakajon, A.; Boonsaeng, V.; Tpiankijagum, S.; Panyim, S. Simple nonradioactive DNA hybridization method for identification of sibling species of Anopheles dirus (Diptera: Culicidae) complex. J. Med. Entomol. 1995, 32, 107–111. [Google Scholar] [CrossRef]
- Manguin, S.; Kengne, P.; Sonnier, L.; Harbach, R.E.; Baimai, V.; Trung, H.D.; Coosemans, M. SCAR markers and multiplex PCR-based identification of isomorphic species in the Anopheles dirus complex in Southeast Asia. Med. Vet. Entomol. 2002, 16, 46–54. [Google Scholar] [CrossRef]
- Walton, C.; Handley, J.M.; Kuvangkadilok, C.; Collins, F.H.; Harbach, R.E.; Baimai, V.; Butlin, R. Identification of five species of the Anopheles dirus complex from Thailand, using allele-specific polymerase chain reaction. Med. Vet. Entomol. 1999, 13, 24–32. [Google Scholar] [CrossRef]
- Pramasivan, S.; Ngui, R.; Jeyaprakasam, N.K.; Liew, J.W.K.; Low, V.L.; Hassan, N.M.; Sulaiman, W.Y.W.; Jaraee, R.; Rahman, R.A.; Jelip, J.; et al. Spatial distribution of Plasmodium knowlesi cases and their vectors in Johor, Malaysia: In the light of human malaria elimination. Malar. J. 2021, 20, 426. [Google Scholar] [CrossRef]
- Yusof, R.; Lau, Y.L.; Mahmud, R.; Fong, M.Y.; Jelip, J.; Ngian, H.U.; Mustakim, S.; Hussin, H.M.; Marzuki, N.; Mohd Ali, M. High proportion of knowlesi malaria in recent malaria cases in Malaysia. Malar. J. 2014, 13, 168. [Google Scholar] [CrossRef] [PubMed]
- Vythilingam, I.; Foo, L.C.; Chiang, G.L.; Chan, S.T.; Eng, K.L.; Mahadevan, S.; Mak, J.W.; Singh, K.I. The impact of permethrin impregnated bednets on the malaria vector Anopheles maculatus (Diptera: Culicidae) in aboriginal villages of Pos Betau Pahang, Malaysia. Southeast Asian J. Trop. Med. Public Health 1995, 26, 354–358. [Google Scholar] [PubMed]
- Jeyaprakasam, N.K.; Pramasivan, S.; Liew, J.W.K.; Van Low, L.; Wan-Sulaiman, W.-Y.; Ngui, R.; Jelip, J.; Vythilingam, I. Evaluation of Mosquito Magnet and other collection tools for Anopheles mosquito vectors of simian malaria. Parasites Vectors 2021, 14, 184. [Google Scholar] [CrossRef] [PubMed]
- Reid, J.A. Anopheline Mosquitoes of Malaya and Borneo; Government of Malaysia: Kuala Lumpur, Malaysia, 1968.
- Garros, C.; Koekemoer, L.L.; Coetzee, M.; Coosemans, M.; Manguin, S. A single multiplex assay to identify major malaria vectors within the African Anopheles funestus and the Oriental An. minimus groups. Am. J. Trop. Med. Hyg. 2004, 70, 583–590. [Google Scholar] [CrossRef]
- Hempolchom, C.; Otsuka, Y.; Baimai, V.; Thongsahuan, S.; Saeung, A.; Taai, K.; Srisuka, W.; Somboon, P.; Choochote, W. Development of a multiplex PCR assay for the identification of eight species members of the Thai Hyrcanus Group (Diptera: Culicidae). Appl. Entomol. Zool. 2013, 48, 469–476. [Google Scholar] [CrossRef]
- Walton, C.; Somboon, P.; O’Loughlin, S.; Zhang, S.; Harbach, R.; Linton, Y.-M.; Chen, B.; Nolan, K.; Duong, S.; Fong, M.-Y.; et al. Genetic diversity and molecular identification of mosquito species in the Anopheles maculatus group using the ITS2 region of rDNA. Infect. Genet. Evol. 2007, 7, 93–102. [Google Scholar] [CrossRef]
- Manonmani, A.; Townson, H.; Adeniran, T.; Jambulingam, P.; Sahu, S.; Vijayakumar, T. rDNA-ITS2 polymerase chain reaction assay for the sibling species of Anopheles fluviatilis. Acta Trop. 2001, 78, 3–9. [Google Scholar] [CrossRef]
- Taai, K.; Harbach, R.E. Systematics of the Anopheles barbirostris species complex (Diptera: Culicidae: Anophelinae) in Thailand. Zool. J. Linn. Soc. 2015, 174, 244–264. [Google Scholar] [CrossRef]
- Dahan-Moss, Y.; Hendershot, A.; Dhoogra, M.; Julius, H.; Zawada, J.; Kaiser, M.; Lobo, N.F.; Brooke, B.D.; Koekemoer, L.L. Member species of the Anopheles gambiae complex can be misidentified as Anopheles leesoni. Malar. J. 2020, 19, 89. [Google Scholar] [CrossRef]
- Bang, W.J.; Kim, H.C.; Ryu, J.; Lee, H.S.; Lee, S.Y.; Kim, M.S.; Chong, S.T.; Klein, T.A.; Choi, K.S. Multiplex PCR assay for the identification of eight Anopheles species belonging to the Hyrcanus, Barbirostris and Lindesayi groups. Malar. J. 2021, 20, 287. [Google Scholar] [CrossRef]
- Beebe, N.; Cooper, R. Systematics of malaria vectors with particular reference to the Anopheles punctulatus group. Int. J. Parasitol. 2000, 30, 1–17. [Google Scholar] [CrossRef]
- Sallum, M.A.M.; Foster, P.G.; Li, C.; Sithiprasasna, R.; Wilkerson, R.C. Phylogeny of the Leucosphyrus Group of Anopheles (Cellia) (Diptera: Culicidae) Based on Mitochondrial Gene Sequences. Ann. Entomol. Soc. Am. 2007, 100, 27–35. [Google Scholar] [CrossRef]
- Keller, D.; Holderegger, R.; van Strien, M.J.; Bolliger, J. How to make landscape genetics beneficial for conservation management. Conserv. Genet. 2015, 16, 503–512. [Google Scholar] [CrossRef]
- Marrelli, M.T.; Sallum, M.A.M.; Marinotti, O. The second internal transcribed spacer of nuclear ribosomal DNA as a tool for Latin American anopheline taxonomy. Memórias Do Inst. Oswaldo Cruz 2006, 101, 817–832. [Google Scholar] [CrossRef] [PubMed]
- Wesson, D.M.; Porter, C.H.; Collins, F.H. Sequence and secondary structure comparisons of ITS rDNA in mosquitoes (Diptera: Culicidae). Mol. Phylogenet. Evol. 1992, 1, 253–269. [Google Scholar] [CrossRef]
- Widiarti, W.; Garjito, T.; Widyastuti, U. Diversitas Genetik Anopheles balabacensis, Baisas di Berbagai Daerah Indonesia Berdasarkan Sekuen Gen ITS 2 DNA Ribosom. Indones. Bull. Health Res. 2016, 44, 20141. [Google Scholar] [CrossRef]
- Erlank, E.; Koekemoer, L.L.; Coetzee, M. The importance of morphological identification of African anopheline mosquitoes (Diptera: Culicidae) for malaria control programmes. Malar. J. 2018, 17, 43. [Google Scholar] [CrossRef] [PubMed]
- Singh, O.; Nanda, N.; Dev, V.; Bali, P.; Sohail, M.; Mehrunnisa, A.; Adak, T.; Dash, A. Molecular evidence of misidentification of Anopheles minimus as Anopheles fluviatilis in Assam (India). Acta Trop. 2010, 113, 241–244. [Google Scholar] [CrossRef]
- Hunt, R.; Mahon, R. Collections of Anopheles-Quadriannulatus (Diptera, Culicidae) From Human Habitations in Southern-Africa. J. Entomol. Soc. S. Afr. 1986, 49, 390–391. [Google Scholar]
- Burke, A.; Dandalo, L.; Munhenga, G.; Dahan-Moss, Y.; Mbokazi, F.; Ngxongo, S.; Coetzee, M.; Koekemoer, L.; Brooke, B. A new malaria vector mosquito in South Africa. Sci. Rep. 2017, 7, 43779. [Google Scholar] [CrossRef]
- Harbach, R.E. Anopheles Classification. Mosquito Taxonomic Inventory. Towards Malaria Elimination 2018. Available online: https://www.intechopen.com/books/towards-malaria-elimination-a-leap-forward (accessed on 30 August 2021).
- Ahmad, R.; Azahary, A.R.A.; Mohamed Nor, Z.; Wan Mohd Ali, W.N.; Ismail, Z.; Mohd Omar, H.; Mohd Majid, A.; Hamid, Z.; Ismail, S.; Lee, H.L. Comparative Human Landing Catch and CDC Light Trap in Mosquito Sampling in Knowlesi Malaria Endemic Areas in Peninsula Malaysia. Adv. Entomol. 2016, 4, 1–10. [Google Scholar] [CrossRef]
- Sum, J.-S.; Lee, W.-C.; Amir, A.; Braima, K.A.; Jeffery, J.; Abdul-Aziz, N.M.; Fong, M.-Y.; Lau, Y.-L. Phylogenetic study of six species of Anopheles mosquitoes in Peninsular Malaysia based on inter-transcribed spacer region 2 (ITS2) of ribosomal DNA. Parasites Vectors 2014, 7, 309. [Google Scholar] [CrossRef] [PubMed]


| Primer’s Name | Sequences | Product Sizes (bp) | |
|---|---|---|---|
| Universal forward primer | LeucogrpFwd | 5′-GCG YCG CTG GCC TGC ACG-3′ | - |
| An. balabacensis | balabaRev | 5′-CGG CGC AGC GAC TCY ACC G-3′ | 224–228 |
| An. cracens | craRev4 | 5′-GC ACC GCT CTT GGC GGG ATA T-3′ | 421–426 |
| An. introlatus | introRev3 | 5′-CG ACG AGC GCG YGA GCG A-3′ | 298–299 |
| An. latens Clade I | laten1Rev | 5′-CCC GGG CGT CCG GTG TTT-3′ | 198 |
| An. latens Clade II | laten2Rev | 5′-CCG GGC GTC YGC GGT GTA C-3′ | 197 |
| Species | No. of Specimen | No. of Sequenced Samples | Positive in Multiplex PCR Assay | % Positive Amplification |
|---|---|---|---|---|
| An. balabacensis | 21 | 21 | 18 | 85.71 |
| An. cracens | 25 | 25 | 23 | 92 |
| An. introlatus | 30 | 30 | 30 | 100 |
| An. latens | 23 | 23 | 21 | 91.30 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pramasivan, S.; Liew, J.W.K.; Jeyaprakasam, N.K.; Low, V.L.; Ngui, R.; Vythilingam, I. Multiplex PCR Assay for the Identification of Four Species of the Anopheles Leucosphyrus Sub-Group in Malaysia. Insects 2022, 13, 195. https://doi.org/10.3390/insects13020195
Pramasivan S, Liew JWK, Jeyaprakasam NK, Low VL, Ngui R, Vythilingam I. Multiplex PCR Assay for the Identification of Four Species of the Anopheles Leucosphyrus Sub-Group in Malaysia. Insects. 2022; 13(2):195. https://doi.org/10.3390/insects13020195
Chicago/Turabian StylePramasivan, Sandthya, Jonathan Wee Kent Liew, Nantha Kumar Jeyaprakasam, Van Lun Low, Romano Ngui, and Indra Vythilingam. 2022. "Multiplex PCR Assay for the Identification of Four Species of the Anopheles Leucosphyrus Sub-Group in Malaysia" Insects 13, no. 2: 195. https://doi.org/10.3390/insects13020195
APA StylePramasivan, S., Liew, J. W. K., Jeyaprakasam, N. K., Low, V. L., Ngui, R., & Vythilingam, I. (2022). Multiplex PCR Assay for the Identification of Four Species of the Anopheles Leucosphyrus Sub-Group in Malaysia. Insects, 13(2), 195. https://doi.org/10.3390/insects13020195






