What Drives Caterpillar Guilds on a Tree: Enemy Pressure, Leaf or Tree Growth, Genetic Traits, or Phylogenetic Neighbourhood?
Abstract
:Simple Summary
Abstract
1. Introduction
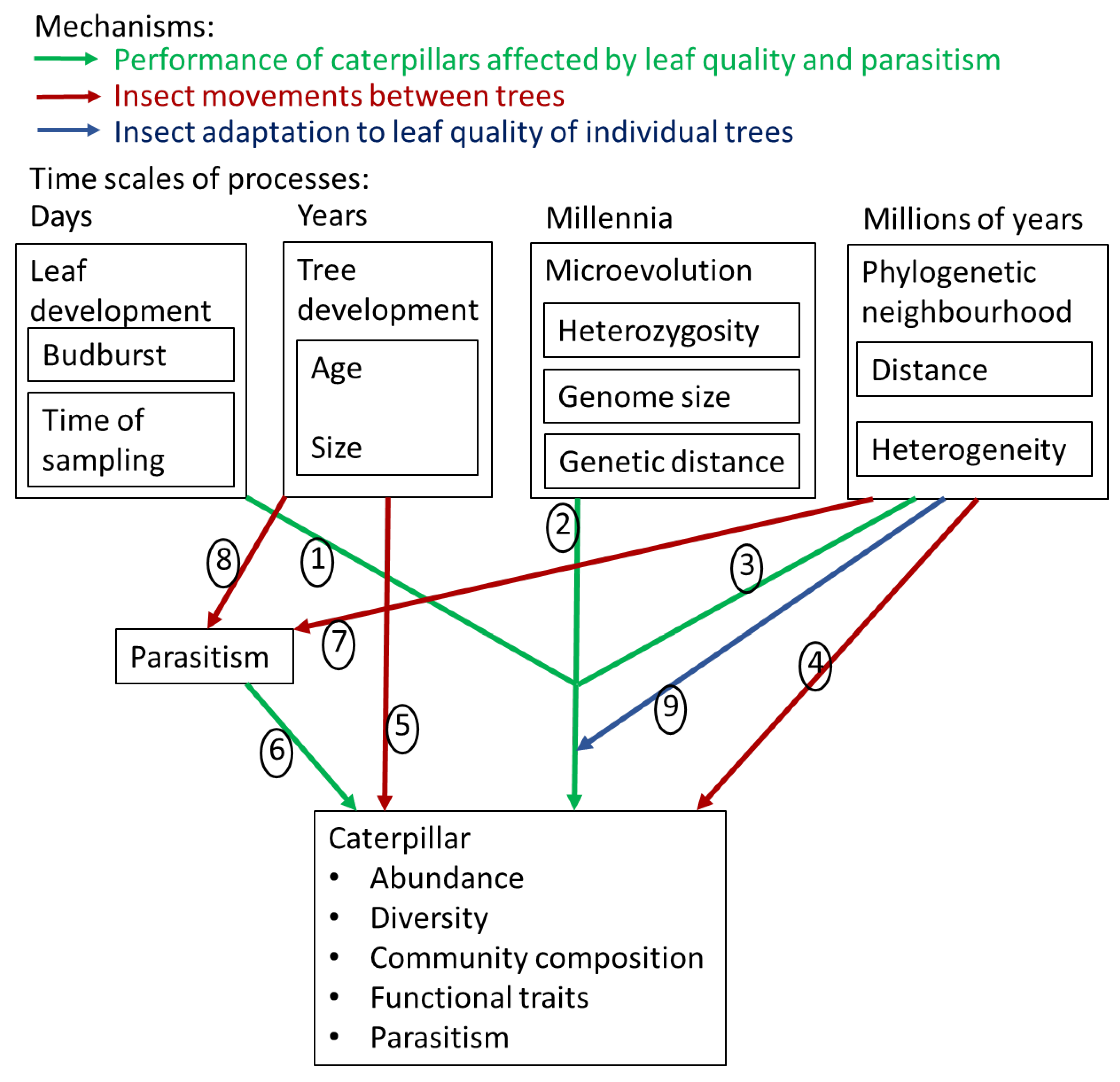
2. Material and Methods
2.1. Study Area of Focal Trees
2.2. Phylogenetic Neighbourhood of Focal Trees
2.3. Budburst Phenology and Tree Size
2.4. Genetic Characterization of Oaks
2.5. Caterpillar Collection and Rearing
2.6. Data Analysis
3. Results
3.1. Overview of the Data
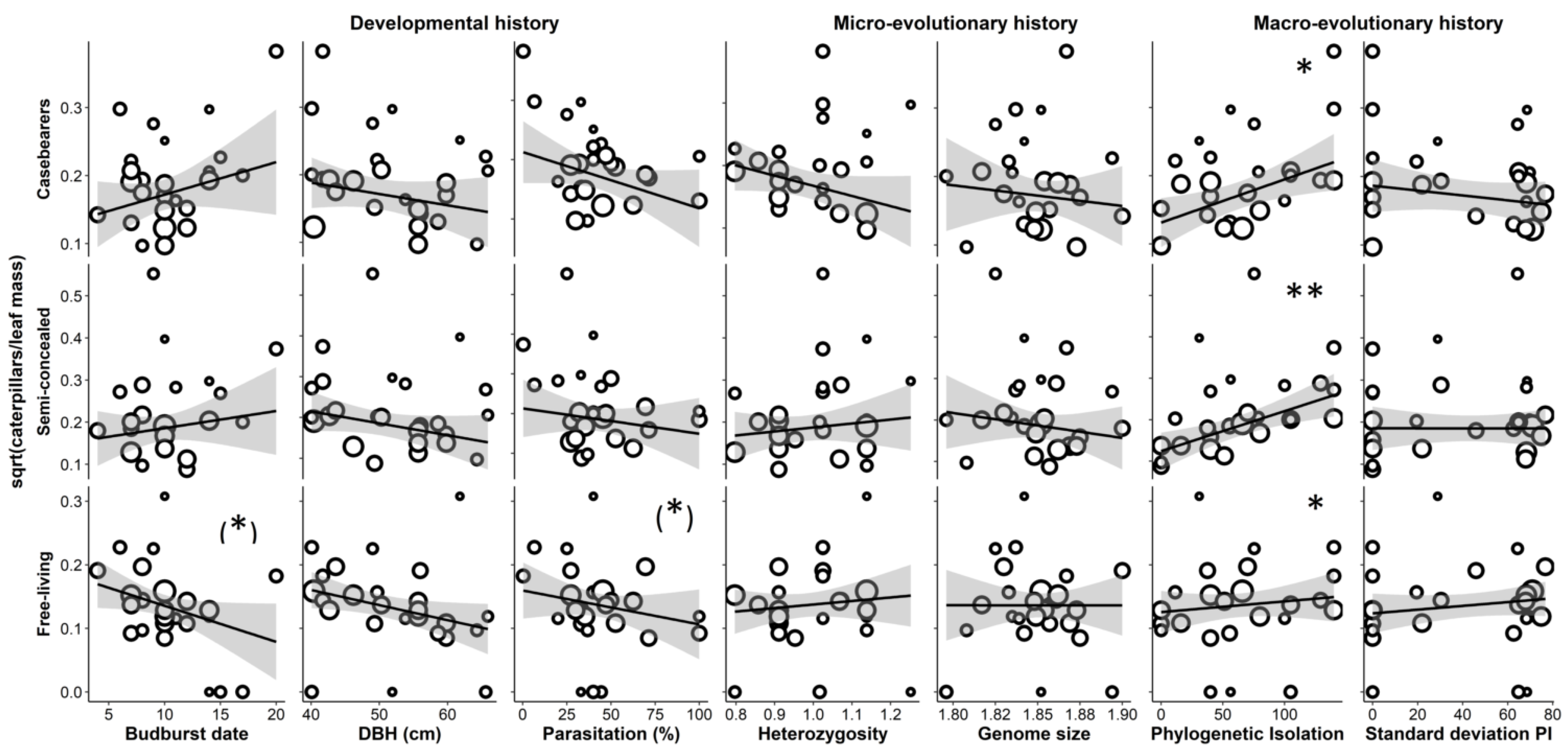
3.2. Predictors of Caterpillar Abundance, Parasitism, Simpson Diversity, and Functional Traits
3.3. Predictors of Caterpillar Community Composition
4. Discussion
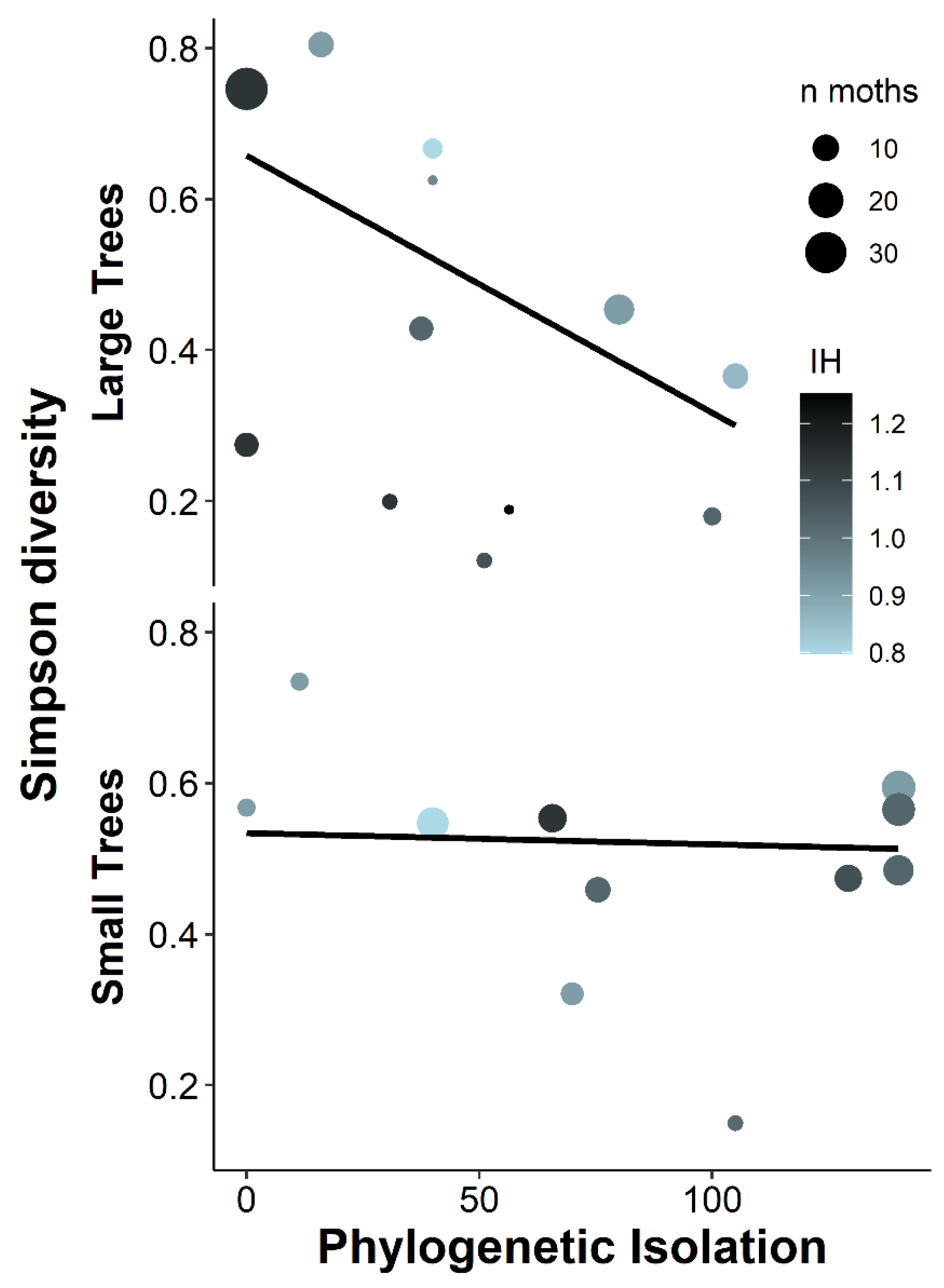
4.1. Why Was Caterpillar Abundance Higher on More Phylogenetically Isolated Trees, Opposite to Findings of Most Previous Studies?
4.2. How Could Neighbourhood Have Affected Community Composition?
4.3. Why Is the Effect of Phylogenetic Isolation on Caterpillar Abundance Strong for Entire Caterpillar Guilds, but Not for the Dominant Species or Dominant Caterpillar Family?
4.4. Why Did Only Casebearers Respond to Genetic Traits?
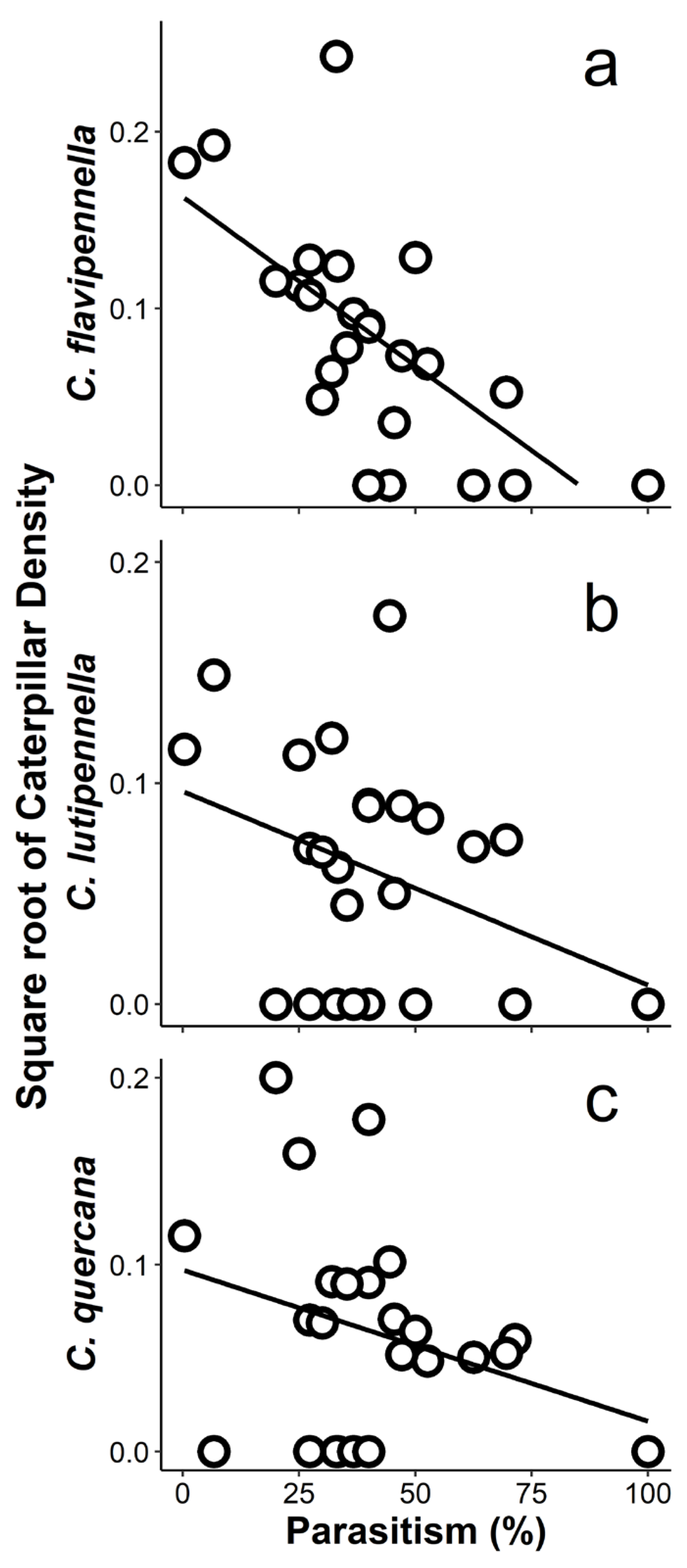
4.5. Why Is the Effect of Parasitism on Caterpillar Abundance Stronger for the Dominant Species Than for All Species Combined?
4.6. Why Was Parasitism Rate High on Large Trees?
4.7. Why Does the Abundance of Free-Living Caterpillars Rapidly Decrease over Time?
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Feeny, P.P. Effect of oak leaf tannins on larval growth of the winter moth Operophtera brumata. J. Insect Physiol. 1968, 14, 805–817. [Google Scholar] [CrossRef]
- Forkner, R.E.; Marquis, R.J.; Lill, J.T. Feeny revisited: Condensed tannins as anti-herbivore defences in leaf-chewing herbivore communities of Quercus. Ecol. Entomol. 2004, 29, 174–187. [Google Scholar] [CrossRef]
- Turčáni, M.; Patočka, J.; Kulfan, M. How do lepidopteran seasonal guilds differ on some oaks (Quercus spp.)—A case study. J. For. Sci. 2009, 55, 578–590. [Google Scholar] [CrossRef] [Green Version]
- Seifert, C.L.; Jorge, L.R.; Volf, M.; Wagner, D.L.; Lamarre, G.P.A.; Miller, S.E.; Gonzalez-Akre, E.; Anderson-Teixeira, K.J.; Novotný, V. Seasonality affects specialisation of a temperate forest herbivore community. Oikos 2021, 130, 1450–1461. [Google Scholar] [CrossRef]
- Crawley, M.; Akhteruzzaman, M. Individual variation in the phenology of oak trees and its consequences for herbivorous insects. Funct. Ecol. 1988, 2, 409–415. [Google Scholar] [CrossRef]
- Eliason, E.A.; Potter, D.A. Budburst phenology, plant vigor, and host genotype effects on the leaf-galling generation of Callirhytis cornigera (Hymenoptera: Cynipidae) on pin oak. Env. Entomol. 2000, 29, 1199–1207. [Google Scholar] [CrossRef]
- Chen, Z.; Clancy, K.M.; Kolb, T.E. Variation in budburst phenology of Douglas-fir related to western spruce budworm (Lepidoptera: Tortricidae) fitness. J. Econ. Entomol. 2003, 96, 377–387. [Google Scholar] [CrossRef]
- Barker, H.L.; Holeski, L.M.; Lindroth, R.L. Genotypic variation in plant traits shapes herbivorous insect and ant communities on a foundation tree species. PLoS ONE 2018, 13, e0200954. [Google Scholar] [CrossRef]
- Ekholm, A.; Tack, A.J.; Pulkkinen, P.; Roslin, T. Host plant phenology, insect outbreaks and herbivore communities–The importance of timing. J. Anim. Ecol. 2020, 89, 829–841. [Google Scholar] [CrossRef]
- Faticov, M.; Ekholm, A.; Roslin, T.; Tack, A.J.M. Climate and host genotype jointly shape tree phenology, disease levels and insect attacks. Oikos 2019, 129, 391–401. [Google Scholar] [CrossRef] [Green Version]
- Hunter, M.D. A variable insect-plant interaction: The relationship between tree budburst phenology and population levels of insect herbivores among trees. Ecol. Entomol. 1992, 17, 91–95. [Google Scholar] [CrossRef]
- Sthultz, C.M.; Gehring, C.A.; Whitham, T.G. Deadly combination of genes and drought: Increased mortality of herbivore-resistant trees in a foundation species. Glob. Change Biol. 2009, 15, 1949–1961. [Google Scholar] [CrossRef]
- Pihain, M.; Gerhold, P.; Ducousso, A.; Prinzing, A. Evolutionary response to coexistence with close relatives: Increased resistance against specialist herbivores without cost for climatic-stress resistance. Ecol. Lett. 2019, 22, 1285–1296. [Google Scholar] [CrossRef] [PubMed]
- Bertić, M.; Schroeder, H.; Kersten, B.; Fladung, M.; Orgel, F.; Buegger, F.; Schnitzler, J.-P.; Ghirardo, A. European oak chemical diversity–from ecotypes to herbivore resistance. New Phytol. 2021, 232, 818–834. [Google Scholar] [CrossRef] [PubMed]
- Wimp, G.M.; Martinsen, G.D.; Floate, K.D.; Bangert, R.K.; Whitham, T.G. Plant genetic determinants of arthropod community structure and diversity. Evolution 2005, 59, 61–69. [Google Scholar] [CrossRef]
- Bangert, R.; Turek, R.; Rehill, B.; Wimp, G.; Schweitzer, J.; Allan, G.J.; Bailey, J.; Martinsen, G.; Keim, P.; Lindroth, R. A genetic similarity rule determines arthropod community structure. Mol. Ecol. 2006, 15, 1379–1391. [Google Scholar] [CrossRef]
- Whitham, T.G.; Bailey, J.K.; Schweitzer, J.A.; Shuster, S.M.; Bangert, R.K.; LeRoy, C.J.; Lonsdorf, E.V.; Allan, G.J.; DiFazio, S.P.; Potts, B.M.; et al. A framework for community and ecosystem genetics: From genes to ecosystems. Nat. Rev. Genet. 2006, 7, 510–523. [Google Scholar] [CrossRef]
- Tack, A.J.; Roslin, T. The relative importance of host-plant genetic diversity in structuring the associated herbivore community. Ecology 2011, 92, 1594–1604. [Google Scholar] [CrossRef]
- Pohjanmies, T.; Tack, A.J.M.; Pulkkinen, P.; Elshibli, S.; Vakkari, P.; Roslin, T. Genetic diversity and connectivity shape herbivore load within an oak population at its range limit. Ecosphere 2015, 6, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Barker, H.L.; Riehl, J.F.; Bernhardsson, C.; Rubert-Nason, K.F.; Holeski, L.M.; Ingvarsson, P.K.; Lindroth, R.L. Linking plant genes to insect communities: Identifying the genetic bases of plant traits and community composition. Mol. Ecol. 2019, 28, 4404–4421. [Google Scholar] [CrossRef]
- Eisenring, M.; Unsicker, S.B.; Lindroth, R.L.; Barton, K. Spatial, genetic and biotic factors shape within-crown leaf trait variation and herbivore performance in a foundation tree species. Funct. Ecol. 2020, 35, 54–66. [Google Scholar] [CrossRef]
- Tack, A.J.M.; Johnson, M.T.J.; Roslin, T. Sizing up community genetics: It′s a matter of scale. Oikos 2012, 121, 481–488. [Google Scholar] [CrossRef]
- Gossner, M.M.; Brandle, M.; Brandl, R.; Bail, J.; Muller, J.; Opgenoorth, L. Where is the extended phenotype in the wild? The community composition of arthropods on mature oak trees does not depend on the oak genotype. PLoS ONE 2015, 10, e0115733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Field, E.; Schönrogge, K.; Barsoum, N.; Hector, A.; Gibbs, M. Individual tree traits shape insect and disease damage on oak in a climate-matching tree diversity experiment. Ecol. Evol. 2019, 9, 8524–8540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pearse, I.S.; Baty, J.H. The predictability of traits and ecological interactions on 17 different crosses of hybrid oaks. Oecologia 2012, 169, 489–497. [Google Scholar] [CrossRef] [PubMed]
- Mopper, S.; Mitton, J.B.; Whitham, T.G.; Cobb, N.S.; Christensen, K.M. Genetic differentiation and heterozygosity in pinyon pine associated with resistance to herbivory and environmental stress. Evolution 1991, 45, 989–999. [Google Scholar] [CrossRef]
- Walker, F.M.; Durben, R.; Shuster, S.M.; Lindroth, R.L.; Whitham, T.G. Heterozygous trees rebound the fastest after felling by beavers to positively affect arthropod community diversity. Forests 2021, 12, 694. [Google Scholar] [CrossRef]
- Šmarda, P.; Bureš, P. Understanding intraspecific variation in genome size in plants. Preslia 2010, 82, 41–61. [Google Scholar]
- Dzialuk, A.; Chybicki, I.; Welc, M.; Śliwińska, E.; Burczyk, J. Presence of triploids among oak species. Ann. Bot. 2007, 99, 959–964. [Google Scholar] [CrossRef]
- Zoldoš, V.; Papeš, D.; Brown, S.; Panaud, O.; Siljak-Yakovlev, S. Genome size and base composition of seven Quercus species: Inter-and intra-population variation. Genome 1998, 41, 162–168. [Google Scholar] [CrossRef]
- Guignard, M.S.; Crawley, M.J.; Kovalenko, D.; Nichols, R.A.; Trimmer, M.; Leitch, A.R.; Leitch, I.J. Interactions between plant genome size, nutrients and herbivory by rabbits, molluscs and insects on a temperate grassland. Proc. Biol. Sci. 2019, 286, 20182619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meyerson, L.A.; Cronin, J.T.; Bhattarai, G.P.; Brix, H.; Lambertini, C.; Lučanová, M.; Rinehart, S.; Suda, J.; Pyšek, P. Do ploidy level and nuclear genome size and latitude of origin modify the expression of Phragmites australis traits and interactions with herbivores? Biol. Invasions 2016, 18, 2531–2549. [Google Scholar] [CrossRef]
- Kunstler, G.; Lavergne, S.; Courbaud, B.; Thuiller, W.; Vieilledent, G.; Zimmermann, N.E.; Kattge, J.; Coomes, D.A. Competitive interactions between forest trees are driven by species′ trait hierarchy, not phylogenetic or functional similarity: Implications for forest community assembly. Ecol. Lett. 2012, 15, 831–840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pretzsch, H.; Schütze, G.; Uhl, E. Resistance of European tree species to drought stress in mixed versus pure forests: Evidence of stress release by inter-specific facilitation. Plant Biol. 2013, 15, 483–495. [Google Scholar] [CrossRef]
- Correia, A.; Galla, A.; Nunes, A.; Pereira, J. Ecological interactions between cork oak (Quercus suber L.) and stone pine (Pinus pinea L.): Results from a pot experiment. Forests 2018, 9, 534. [Google Scholar] [CrossRef] [Green Version]
- Fernandez, C.; Monnier, Y.; Santonja, M.; Gallet, C.; Weston, L.A.; Prévosto, B.; Saunier, A.; Baldy, V.; Bousquet-Mélou, A. The impact of competition and allelopathy on the trade-off between plant defense and growth in two contrasting tree species. Front. Plant Sci. 2016, 7, 594. [Google Scholar] [CrossRef] [Green Version]
- Sykes, M.T.; Prentice, I.C.; Cramer, W. A bioclimatic model for the potential distributions of north European tree species under present and future climates. J. Biogeogr. 1996, 23, 203–233. [Google Scholar]
- Seifert, C.L.; Volf, M.; Jorge, L.R.; Abe, T.; Carscallen, G.; Drozd, P.; Kumar, R.; Lamarre, G.P.; Libra, M.; Losada, M.E. Plant phylogeny drives arboreal caterpillar assemblages across the Holarctic. Ecol. Evol. 2020, 10, 14137–14151. [Google Scholar] [CrossRef]
- Brändle, M.; Brandl, R. Is the composition of phytophagous insects and parasitic fungi among trees predictable? Oikos 2006, 113, 296–304. [Google Scholar] [CrossRef]
- Vialatte, A.; Bailey, R.I.; Vasseur, C.; Matocq, A.; Gossner, M.M.; Everhart, D.; Vitrac, X.; Belhadj, A.; Ernoult, A.; Prinzing, A. Phylogenetic isolation of host trees affects assembly of local Heteroptera communities. Proc. R. Soc. B Biol. Sci. 2010, 277, 2227–2236. [Google Scholar] [CrossRef] [Green Version]
- Yguel, B.; Bailey, R.; Tosh, N.D.; Vialatte, A.; Vasseur, C.; Vitrac, X.; Jean, F.; Prinzing, A. Phytophagy on phylogenetically isolated trees: Why hosts should escape their relatives. Ecol. Lett. 2011, 14, 1117–1124. [Google Scholar] [CrossRef] [PubMed]
- Grandez-Rios, J.M.; Lima Bergamini, L.; Santos de Araujo, W.; Villalobos, F.; Almeida-Neto, M. The effect of host-plant phylogenetic isolation on species richness, composition and specialization of insect herbivores: A comparison between native and exotic hosts. PLoS ONE 2015, 10, e0138031. [Google Scholar] [CrossRef] [PubMed]
- Whittaker, R.J.; Triantis, K.A.; Ladle, R.J. A general dynamic theory of oceanic island biogeography. J. Biogeogr. 2008, 35, 977–994. [Google Scholar] [CrossRef]
- Feeny, P. Plant apparency and chemical defense. In Biochemical Interaction between Plants and Insects; Springer: Berlin/Heidelberg, Germany, 1976; pp. 1–40. [Google Scholar]
- Castagneyrol, B.; Giffard, B.; Péré, C.; Jactel, H.; Sipes, S. Plant apparency, an overlooked driver of associational resistance to insect herbivory. J. Ecol. 2013, 101, 418–429. [Google Scholar] [CrossRef]
- Campos, R.I.; Vasconcelos, H.L.; Ribeiro, S.P.; Neves, F.S.; Soares, J.P. Relationship between tree size and insect assemblages associated with Anadenanthera macrocarpa. Ecography 2006, 29, 442–450. [Google Scholar] [CrossRef]
- Barker, H.L.; Holeski, L.M.; Lindroth, R.L.; Koricheva, J. Independent and interactive effects of plant genotype and environment on plant traits and insect herbivore performance: A meta-analysis with Salicaceae. Funct. Ecol. 2018, 33, 422–435. [Google Scholar] [CrossRef]
- Barr, A.E.; van Dijk, L.J.A.; Hylander, K.; Tack, A.J.M. Local habitat factors and spatial connectivity jointly shape an urban insect community. Landsc. Urban Plan. 2021, 214, 104177. [Google Scholar] [CrossRef]
- Ghirardo, A.; Heller, W.; Fladung, M.; Schnitzler, J.P.; Schroeder, H. Function of defensive volatiles in pedunculate oak (Quercus robur) is tricked by the moth Tortrix viridana. Plant Cell Environ. 2012, 35, 2192–2207. [Google Scholar] [CrossRef]
- Castagneyrol, B.; Kozlov, M.V.; Poeydebat, C.; Toïgo, M.; Jactel, H. Associational resistance to a pest insect fades with time. J. Pest Sci. 2020, 93, 427–437. [Google Scholar] [CrossRef]
- Yguel, B.; Bailey, R.I.; Villemant, C.; Brault, A.; Jactel, H.; Prinzing, A. Insect herbivores should follow plants escaping their relatives. Oecologia 2014, 176, 521–532. [Google Scholar] [CrossRef] [Green Version]
- Perfecto, I.; Vet, L.E.M. Effect of a nonhost plant on the location behavior of two parasitoids: The tritrophic system of Cotesia spp. (Hymenoptera: Braconidae), Pieris rapae (Lepidoptera: Pieridae), and Brassica oleraceae. Environ. Entomol. 2003, 32, 163–174. [Google Scholar] [CrossRef]
- Cronin, J.T. Host-parasitoid extinction and colonization in a fragmented prairie landscape. Oecologia 2004, 139, 503–514. [Google Scholar] [CrossRef] [PubMed]
- Janzen, D.H. Herbivores and the number of tree species in tropical forests. Am. Nat. 1970, 104, 501–528. [Google Scholar] [CrossRef]
- Forkner, R.E.; Hunter, M.D. What goes up must come down? Nutrient addition and predation pressure on oak herbivores. Ecology 2000, 81, 1588–1600. [Google Scholar] [CrossRef]
- Hunter, M.D.; Price, P.W. Playing chutes and ladders: Heterogeneity and the relative roles of bottom-up and top-down forces in natural communities. Ecology 1992, 73, 724–732. [Google Scholar]
- Du Merle, P. Phenological resistance of oaks to the green oak leafroller, Tortrix viridana (Lepidoptera: Tortricidae). In Mechanisms of Woody Plant Defenses against Insects; Springer: Berlin/Heidelberg, Germany, 1988; pp. 215–226. [Google Scholar]
- Van Dongen, S.; Backeljau, T.; Matthysen, E.; Dhondt, A.A. Synchronization of hatching date with budburst of individual host trees (Quercus robur) in the winter moth (Operophtera brumata) and its fitness consequences. J. Anim. Ecol. 1997, 66, 113–121. [Google Scholar] [CrossRef]
- Mopper, S.; Stiling, P.; Landau, K.; Simberloff, D.; Van Zandt, P. Spatiotemporal variation in leafminer population structure and adaptation to individual oak trees. Ecology 2000, 81, 1577–1587. [Google Scholar] [CrossRef]
- Zandt, P.A.V.; Mopper, S. A meta-analysis of adaptive deme formation in phytophagous insect populations. Am. Nat. 1998, 152, 595–604. [Google Scholar] [CrossRef]
- Schroeder, H.; Degen, B. Genetic structure of the green oak leaf roller (Tortrix viridana L.) and one of its hosts, Quercus robur L. For. Ecol. Manag. 2008, 256, 1270–1279. [Google Scholar] [CrossRef]
- Serra, G.; Maestrale, G.B.; Tore, S.; Casula, S.; Baratti, M. Host plant budburst and male-biased dispersal affect the genetic structure of the green oak leaf roller moth, Tortrix viridana (Lepidoptera: Tortricidae). Biol. J. Linn. Soc. 2019, 127, 56–74. [Google Scholar] [CrossRef]
- Edmunds, G.F.; Alstad, D.N. Coevolution in insect herbivores and conifers: Scale insects adapt to intraspecific variation in pine defenses with a differentiated deme on each tree. Science 1978, 199, 941–945. [Google Scholar] [CrossRef] [PubMed]
- Gandon, S.; Michalakis, Y. Local adaptation, evolutionary potential and host–parasite coevolution: Interactions between migration, mutation, population size and generation time. J. Evol. Biol. 2002, 15, 451–462. [Google Scholar] [CrossRef]
- Jensen, J.D.; Bachtrog, D. Characterizing the influence of effective population size on the rate of adaptation: Gillespie’s Darwin domain. Genome Biol. Evol. 2011, 3, 687–701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tack, A.J.; Roslin, T. Overrun by the neighbors: Landscape context affects strength and sign of local adaptation. Ecology 2010, 91, 2253–2260. [Google Scholar] [CrossRef]
- Lester, S.E.; Ruttenberg, B.I.; Gaines, S.D.; Kinlan, B.P. The relationship between dispersal ability and geographic range size. Ecol. Lett. 2007, 10, 745–758. [Google Scholar] [CrossRef]
- Richardson, J.L.; Urban, M.C.; Bolnick, D.I.; Skelly, D.K. Microgeographic adaptation and the spatial scale of evolution. Trends Ecol. Evol. 2014, 29, 165–176. [Google Scholar] [CrossRef]
- Van Dongen, S.; Backeljau, T.; Matthysen, E.; Dhondt, A. Genetic population structure of the winter moth (Operophtera brumata L.)(Lepidoptera, Geometridae) in a fragmented landscape. Heredity 1998, 80, 92–100. [Google Scholar] [CrossRef]
- Schneider, I. Untersuchungen zur überwachung des Eichenwicklers, Tortrix viridana L.(Lepid., Tortricidae), mit seinem Pheromon. Z. Für Angew. Entomol. 1984, 98, 474–483. [Google Scholar] [CrossRef]
- Gutzwiller, F.; Dedeine, F.; Kaiser, W.; Giron, D.; Lopez-Vaamonde, C. Correlation between the green-island phenotype and Wolbachia infections during the evolutionary diversification of Gracillariidae leaf-mining moths. Ecol. Evol. 2015, 5, 4049–4062. [Google Scholar] [CrossRef] [Green Version]
- Cornell, H. Endophage-ectophage ratios and plant defense. Evol. Ecol. 1989, 3, 64–76. [Google Scholar] [CrossRef]
- Connor, E.F.; Taverner, M.P. The evolution and adaptive significance of the leaf-mining habit. Oikos 1997, 79, 6–25. [Google Scholar] [CrossRef] [Green Version]
- Hering, E. Biology of Leaf Miners; W. Junk: The Hague, The Netherlands, 1951. [Google Scholar]
- Novotny, V.; Miller, S.E.; Baje, L.; Balagawi, S.; Basset, Y.; Cizek, L.; Craft, K.J.; Dem, F.; Drew, R.A.; Hulcr, J.; et al. Guild-specific patterns of species richness and host specialization in plant-herbivore food webs from a tropical forest. J. Anim. Ecol. 2010, 79, 1193–1203. [Google Scholar] [CrossRef] [PubMed]
- Cornell, H.V.; Hawkins, B.A. Survival patterns and mortality sources of herbivorous insects: Some demographic trends. Am. Nat. 1995, 145, 563–593. [Google Scholar] [CrossRef]
- Hawkins, B.A.; Cornell, H.V.; Hochberg, M.E. Predators, parasitoids, and pathogens as mortality agents in phytophagous insect populations. Ecology 1997, 78, 2145–2152. [Google Scholar] [CrossRef]
- Connahs, H.; Aiello, A.; Van Bael, S.; Rodríguez-Castañeda, G. Caterpillar abundance and parasitism in a seasonally dry versus wet tropical forest of Panama. J. Trop. Ecol. 2011, 27, 51–58. [Google Scholar] [CrossRef] [Green Version]
- Tvardikova, K.; Novotny, V. Predation on exposed and leaf-rolling artificial caterpillars in tropical forests of Papua New Guinea. J. Trop. Ecol. 2012, 28, 331–341. [Google Scholar] [CrossRef] [Green Version]
- Hrcek, J.; Miller, S.E.; Whitfield, J.B.; Shima, H.; Novotny, V. Parasitism rate, parasitoid community composition and host specificity on exposed and semi-concealed caterpillars from a tropical rainforest. Oecologia 2013, 173, 521–532. [Google Scholar] [CrossRef]
- Yguel, B.; Courty, P.-E.; Jactel, H.; Pan, X.; Butenschoen, O.; Murray, P.J.; Prinzing, A. Mycorrhizae support oaks growing in a phylogenetically distant neighbourhood. Soil Biol. Biochem. 2014, 78, 204–212. [Google Scholar] [CrossRef] [Green Version]
- Perot, T.; Balandier, P.; Couteau, C.; Delpierre, N.; Jean, F.; Perret, S.; Korboulewsky, N. Budburst date of Quercus petraea is delayed in mixed stands with Pinus sylvestris. Agric. For. Meteorol. 2021, 300, 108326. [Google Scholar] [CrossRef]
- Castagneyrol, B.; Jactel, H.; Vacher, C.; Brockerhoff, E.G.; Koricheva, J. Effects of plant phylogenetic diversity on herbivory depend on herbivore specialization. J. Appl. Ecol. 2014, 51, 134–141. [Google Scholar] [CrossRef] [Green Version]
- Bacilieri, R.; Ducousso, A.; Petit, R.J.; Kremer, A. Mating system and asymmetric hybridization in a mixed stand of European oaks. Evolution 1996, 50, 900–908. [Google Scholar] [CrossRef] [PubMed]
- Southwood, T. The number of species of insect associated with various trees. J. Anim. Ecol. 1961, 30, 1–8. [Google Scholar] [CrossRef]
- Brändle, M.; Brandl, R. Species richness of insects and mites on trees: Expanding Southwood. J. Anim. Ecol. 2001, 70, 491–504. [Google Scholar] [CrossRef]
- Lang, T.; Abadie, P.; Léger, V.; Decourcelle, T.; Frigerio, J.-M.; Burban, C.; Bodénès, C.; Guichoux, E.; Le Provost, G.; Robin, C. High-quality SNPs from genic regions highlight introgression patterns among European white oaks (Quercus petraea and Q. robur). bioRxiv 2021, 388447. [Google Scholar] [CrossRef] [Green Version]
- Chybicki, I.; Burczyk, J. Realized gene flow within mixed stands of Quercus robur L. and Q. petraea (Matt.) L. revealed at the stage of naturally established seedling. Mol. Ecol. 2010, 19, 2137–2151. [Google Scholar] [CrossRef] [PubMed]
- Petit, R.J.; Wagner, D.B.; Kremer, A. Ribosomal DNA and chloroplast DNA polymorphisms in a mixed stand of Quercus robur and Q. petraea. Ann. Des Sci. For. 1993, 50, 41–47. [Google Scholar] [CrossRef]
- Forest Data Bank. Forests on Maps. Available online: https://www.bdl.lasy.gov.pl/portal/mapy-en (accessed on 1 October 2018).
- Magallon, S.; Crane, P.R.; Herendeen, P.S. Phylogenetic pattern, diversity, and diversification of eudicots. Ann. Mo. Bot. Gard. 1999, 86, 297–372. [Google Scholar] [CrossRef]
- Manos, P.S.; Doyle, J.J.; Nixon, K.C. Phylogeny, biogeography, and processes of molecular differentiation in Quercus subgenus Quercus (Fagaceae). Mol. Phylogenetics Evol. 1999, 12, 333–349. [Google Scholar] [CrossRef] [Green Version]
- Wikström, N.; Savolainen, V.; Chase, M.W. Evolution of the angiosperms: Calibrating the family tree. Proc. R. Soc. Lond. Ser. B Biol. Sci. 2001, 268, 2211–2220. [Google Scholar] [CrossRef]
- APG. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG II. Bot. J. Linn. Soc. 2003, 141, 399–436. [Google Scholar] [CrossRef] [Green Version]
- Poinar, G., Jr.; Chambers, K.L.; Buckley, R. Eoëpigynia burmensis gen. and sp. nov., an Early Cretaceous eudicot flower (Angiospermae) in Burmese amber. J. Bot. Res. Inst. Tex. 2007, 1, 91–96. [Google Scholar]
- Gossner, M.M.; Chao, A.; Bailey, R.I.; Prinzing, A. Native fauna on exotic trees: Phylogenetic conservatism and geographic contingency in two lineages of phytophages on two lineages of trees. Am. Nat. 2009, 173, 599–614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corff, J.; Marquis, R. Difference between understory and canopy in herbivore community composition and leaf quantity for two oak species in Missouri. Ecol. Entomol. 2001, 24, 46–58. [Google Scholar] [CrossRef]
- Youngentob, K.N.; Zdenek, C.; Gorsel, E.; Anderson, B. A simple and effective method to collect leaves and seeds from tall trees. Methods Ecol. Evol. 2016, 7, 1119–1123. [Google Scholar] [CrossRef] [Green Version]
- Razowski, J. Zwójkówki–Tortricidae. Wstęp oraz podrodziny Tortricinae i Sparganothinae. In Klucze do Oznaczania Owadów Polski; Motyle—Lepidoptera Zeszyt 41b; PWN: Warszawa, Poland, 1969; p. 27. [Google Scholar]
- Slamka, F. Die Zünslerartigen (Pyraloidea) Mitteleuropas; F. Slamka: Bratislava, Slovakia, 1997. [Google Scholar]
- Nowacki, J. The Noctuids (Lepidoptera, Noctuidae) of Central Europe; F. Slamka: Bratislava, Slovakia, 1998. [Google Scholar]
- Lepiforum, e.V. Bestimmungshilfe für die in Europa Nachgewiesenen Schmetterlingsarten. Available online: https://www.lepiforum.org (accessed on 20 November 2020).
- Razowski, J. Motyle (Lepidoptera) Polski. Część XVI—Coleophoridae. In Monografie Fauny Polski; PWN: Warszawa, Poland; Kraków, Poland, 1990; Volume 18. [Google Scholar]
- Ellis, W.N. Plant Parasites of Europe. Available online: https://bladmineerders.nl/ (accessed on 20 November 2020).
- Jenkins, D.G.; Brescacin, C.R.; Duxbury, C.V.; Elliott, J.A.; Evans, J.A.; Grablow, K.R.; Hillegass, M.; Lyon, B.N.; Metzger, G.A.; Olandese, M.L. Does size matter for dispersal distance? Glob. Ecol. Biogeogr. 2007, 16, 415–425. [Google Scholar] [CrossRef]
- Stevens, V.M.; Whitmee, S.; Le Galliard, J.F.; Clobert, J.; Bohning-Gaese, K.; Bonte, D.; Brandle, M.; Matthias Dehling, D.; Hof, C.; Trochet, A.; et al. A comparative analysis of dispersal syndromes in terrestrial and semi-terrestrial animals. Ecol. Lett. 2014, 17, 1039–1052. [Google Scholar] [CrossRef] [PubMed]
- Ellers, J.; Van Alphen, J.J.; Sevenster, J.G. A field study of size–fitness relationships in the parasitoid Asobara tabida. J. Anim. Ecol. 1998, 67, 318–324. [Google Scholar] [CrossRef]
- De Prins, J.; Steeman, C. Catalogue of the Lepidoptera of Belgium. Available online: https://projects.biodiversity.be/lepidoptera/ (accessed on 20 November 2020).
- Jonko, K. Lepidoptera Mundi. Available online: https://lepidoptera.eu/start/pl (accessed on 20 November 2020).
- Bates, D.; Mächler, M.; Bolker, B.; Walker, S. lme4: Linear Mixed-Effects Models Using Eigen and S4. R Package Version 1.1–7. 2014. 2015. Available online: https://cran.r-project.org/web/packages/lme4/index.html (accessed on 20 June 2021).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Oksanen, J.; Kindt, R.; Legendre, P.; O’Hara, B.; Stevens, M.H.H.; Oksanen, M.J.; Suggests, M. The vegan package. Community Ecol. Package 2007, 10, 719. [Google Scholar]
- Mantel, N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967, 27, 209–220. [Google Scholar]
- Barton, K. MuMIn, Multi-Model Inference. Available online: https://CRAN.R-project.org/package=MuMIn (accessed on 20 June 2021).
- Dray, S.; Dufour, A.-B. The ade4 package: Implementing the duality diagram for ecologists. J. Stat. Softw. 2007, 22, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Oksanen, J. Vegan: An Introduction to Ordination. Available online: http://cran.r-project.org/web/packages/vegan/vignettes/introvegan.pdf (accessed on 20 June 2021).
- Anderson, M.J. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001, 26, 32–46. [Google Scholar] [CrossRef]
- Jactel, H.; Moreira, X.; Castagneyrol, B. Tree diversity and forest resistance to insect pests: Patterns, mechanisms, and prospects. Annu. Rev. Entomol. 2021, 66, 277–296. [Google Scholar] [CrossRef] [PubMed]
- Alalouni, U.; Brandl, R.; Auge, H.; Schädler, M. Does insect herbivory on oak depend on the diversity of tree stands? Basic Appl. Ecol. 2014, 15, 685–692. [Google Scholar] [CrossRef]
- Hidasi-Neto, J.; Bailey, R.I.; Vasseur, C.; Woas, S.; Ulrich, W.; Jambon, O.; Santos, A.M.C.; Cianciaruso, M.V.; Prinzing, A. A forest canopy as a living archipelago: Why phylogenetic isolation may increase and age decrease diversity. J. Biogeogr. 2019, 46, 158–169. [Google Scholar] [CrossRef] [Green Version]
- Deniau, M.; Pihain, M.; Béchade, B.; Jung, V.; Brunellière, M.; Gouesbet, V.; Prinzing, A. Seeds and seedlings of oaks suffer from mammals and molluscs close to phylogenetically isolated, old adults. Ann. Bot. 2021, 127, 787–798. [Google Scholar] [CrossRef]
- Moore, R.; Francis, B. Factors influencing herbivory by insects on oak trees in pure stands and paired mixtures. J. Appl. Ecol. 1991, 8, 305–317. [Google Scholar] [CrossRef]
- Castagneyrol, B.; Bonal, D.; Damien, M.; Jactel, H.; Meredieu, C.; Muiruri, E.W.; Barbaro, L. Bottom-up and top-down effects of tree species diversity on leaf insect herbivory. Ecol. Evol. 2017, 7, 3520–3531. [Google Scholar] [CrossRef]
- Haynes, K.J.; Cronin, J.T. Matrix composition affects the spatial ecology of a prairie planthopper. Ecology 2003, 84, 2856–2866. [Google Scholar] [CrossRef]
- Haynes, K.J.; Diekötter, T.; Crist, T.O. Resource complementation and the response of an insect herbivore to habitat area and fragmentation. Oecologia 2007, 153, 511–520. [Google Scholar] [CrossRef]
- Charnov, E.L. Optimal foraging, the marginal value theorem. Theor. Popul. Biol. 1976, 9, 129–136. [Google Scholar] [CrossRef] [Green Version]
- Asplen, M.K. Dispersal strategies in terrestrial insects. Curr. Opin. Insect Sci. 2018, 27, 16–20. [Google Scholar] [CrossRef] [PubMed]
- Connor, E.F.; Faeth, S.H.; Simberloff, D. Leafminers on oak: The role of immigration and in situ reproductive recruitment. Ecology 1983, 64, 191–204. [Google Scholar] [CrossRef]
- Bruce, T.J. Interplay between insects and plants: Dynamic and complex interactions that have coevolved over millions of years but act in milliseconds. J. Exp. Bot. 2015, 66, 455–465. [Google Scholar] [CrossRef] [PubMed]
- Morrison, G.; Strong, D.R., Jr. Spatial variations in host density and the intensity of parasitism: Some empirical examples. Environ. Entomol. 1980, 9, 149–152. [Google Scholar] [CrossRef]
- Veldtman, R.; McGeoch, M.A. Spatially explicit analyses unveil density dependence. Proc. R. Soc. Lond. Ser. B Biol. Sci. 2004, 271, 2439–2444. [Google Scholar] [CrossRef]
- Fernández-arhex, V.; Corley, J.C. The functional response of parasitoids and its implications for biological control. Biocontrol Sci. Technol. 2003, 13, 403–413. [Google Scholar] [CrossRef]
- Klapwijk, M.J.; Björkman, C. Mixed forests to mitigate risk of insect outbreaks. Scand. J. For. Res. 2018, 33, 772–780. [Google Scholar] [CrossRef]
- Vinson, S.B. Host selection by insect parasitoids. Annu. Rev. Entomol. 1976, 21, 109–133. [Google Scholar] [CrossRef]
- Nieminen, M. Migration of moth species in a network of small islands. Oecologia 1996, 108, 643–651. [Google Scholar] [CrossRef]

| Number of Linear Models out of Top 10 with: | Mantel Tests | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Main Effect: | Interactions with PI | Range | p-Values | |||||||||||||||||
| Dependent Variable | n | Cluster | Day | BB | DBH | Par | IH | Fcyt | PI | sdPI | Day | BB | Diam | Par | IH | Fcyt | sdPI | Delta | Raw | Residuals |
| All caterpillars | 612 | 1 | 3− | 0 | 1− | 4 | 0 | 1+ | 10+ ** | 1− | 0 | 0 | 0 | 2− | 0 | 0 | 0 | 2.8 | 0.400 | 0.541 |
| Casebearers | 214 | 3 | 2− | 0 | 0 | 9− | 8− | 0 | 10 * | 1− | 0 | 0 | 0 | 8− | 5+ | 0 | 0 | 3.3 | 0.153 | 0.094 |
| C. flavipennella | 48 | 0 | 1− | 3− | 1− | 10− *** | 3− | 1− | 4 | 0 | 0 | 0 | 0 | 0 | 2+ | 0 | 0 | 2.7 | 0.212 | 0.230 |
| C. lutipennella | 37 | 10 | 1− | 5+ | 1− | 7− *** | 2− | 2− | 1+ | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2.3 | 0.044 * | 0.152 |
| Semi-concealed | 261 | 1 | 2− | 1+ | 2+ | 1− | 1+ | 1+ | 10+ ** | 1− | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2.9 | 0.436 | 0.614 |
| C. quercana | 35 | 0 | 1+ | 3+ | 1− | 5− | 0 | 0 | 3+ | 1+ | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2.7 | 0.130 | 0.191 |
| Free living | 136 | 1 | 8− | 1− | 1+ | 1− | 2+ | 1+ | 9+ * | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.1 | 0.469 | 0.462 |
| Geometrids | 57 | 3 | 3− | 1− | 4− | 4− * | 0 | 0 | 0 | 8+ * | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1.7 | 0.278 | 0.379 |
| Parasitism rates | 126 | 2 | 1+ | 2− | 6+ | 1+ | 1− | 2− | 9+ * | 0 | 0 | 0 | 0 | 0 | 1+ | 2.6 | 0.342 | 0.182 | ||
| Simpson diversity | 214 | 9 | 2+ | 1+ | 8 ** | 2+ | 9− * | 2− | 9 *** | 4− | 2− | 0 | 8− *** | 0 | 1+ | 2+ | 0 | 3.3 | 0.38 | 0.767 |
| Wingspan | 214 | 5 | 3− | 1+ | 1− | 1− | 1+ | 0 | 1+ | 1+ | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2.4 | 0.377 | 0.411 |
| P. Specialists | 214 | 1 | 1− | 1− | 1− | 4− | 2+ | 1− | 0 | 1+ | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2.1 | 0.196 | 0.271 |
| P. Particular Spec. | 214 | 4 | 2− | 0 | 2− | 1− | 1− | 2− | 1− | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2.4 | 0.619 | 0.529 |
| Day | BB | Diam | Par | IH | Fcyt | PI | sdPI | Residuals | Total | |
|---|---|---|---|---|---|---|---|---|---|---|
| Df | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 16 | 24 |
| SS | 0.51 | 0.027 | 0.13 | 0.217 | 0.352 | 0.098 | 0.276 | 0.26 | 1.461 | 3.33 |
| MS | 0.51 | 0.027 | 0.129 | 0.217 | 0.352 | 0.098 | 0.276 | 0.26 | 0.091 | 1 |
| Pseudo-F | 5.584 | 0.292 | 1.418 | 2.374 | 3.856 | 1.073 | 3.026 | 2.844 | 0.439 | |
| R2 | 0.15 | 0.01 | 0.04 | 0.07 | 0.11 | 0.03 | 0.08 | 0.08 | ||
| p | 0.006 ** | 0.886 | 0.210 | 0.077 | 0.017 * | 0.35 | 0.039 * | 0.047 * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Molleman, F.; Walczak, U.; Melosik, I.; Baraniak, E.; Piosik, Ł.; Prinzing, A. What Drives Caterpillar Guilds on a Tree: Enemy Pressure, Leaf or Tree Growth, Genetic Traits, or Phylogenetic Neighbourhood? Insects 2022, 13, 367. https://doi.org/10.3390/insects13040367
Molleman F, Walczak U, Melosik I, Baraniak E, Piosik Ł, Prinzing A. What Drives Caterpillar Guilds on a Tree: Enemy Pressure, Leaf or Tree Growth, Genetic Traits, or Phylogenetic Neighbourhood? Insects. 2022; 13(4):367. https://doi.org/10.3390/insects13040367
Chicago/Turabian StyleMolleman, Freerk, Urszula Walczak, Iwona Melosik, Edward Baraniak, Łukasz Piosik, and Andreas Prinzing. 2022. "What Drives Caterpillar Guilds on a Tree: Enemy Pressure, Leaf or Tree Growth, Genetic Traits, or Phylogenetic Neighbourhood?" Insects 13, no. 4: 367. https://doi.org/10.3390/insects13040367
APA StyleMolleman, F., Walczak, U., Melosik, I., Baraniak, E., Piosik, Ł., & Prinzing, A. (2022). What Drives Caterpillar Guilds on a Tree: Enemy Pressure, Leaf or Tree Growth, Genetic Traits, or Phylogenetic Neighbourhood? Insects, 13(4), 367. https://doi.org/10.3390/insects13040367







