Expression and Role of Vitellogenin Genes in Ovarian Development of Zeugodacus cucurbitae
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Insect Rearing
2.2. RNA Isolation and cDNA Synthesis
2.3. Molecular Cloning and Sequence Analysis
2.4. Spatio-Temporal Expression Revealed by RT-qPCR
2.5. Hormone-Induced Expression
2.6. Nutritional Stress
2.7. RNA Interference and Functional Analysis
2.8. Statistical Analysis
3. Results
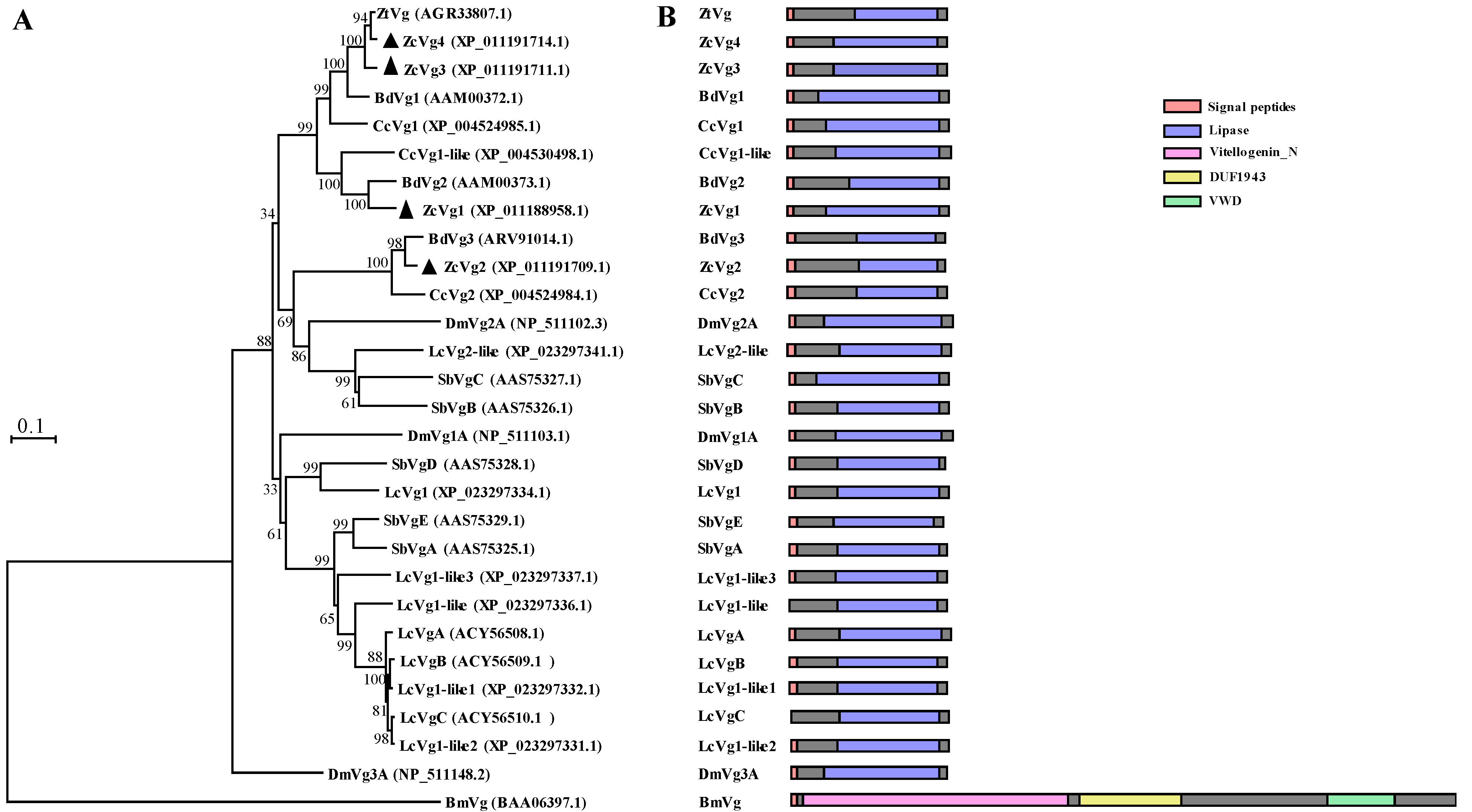
3.1. Sequence Analysis and Phylogenetic Analysis
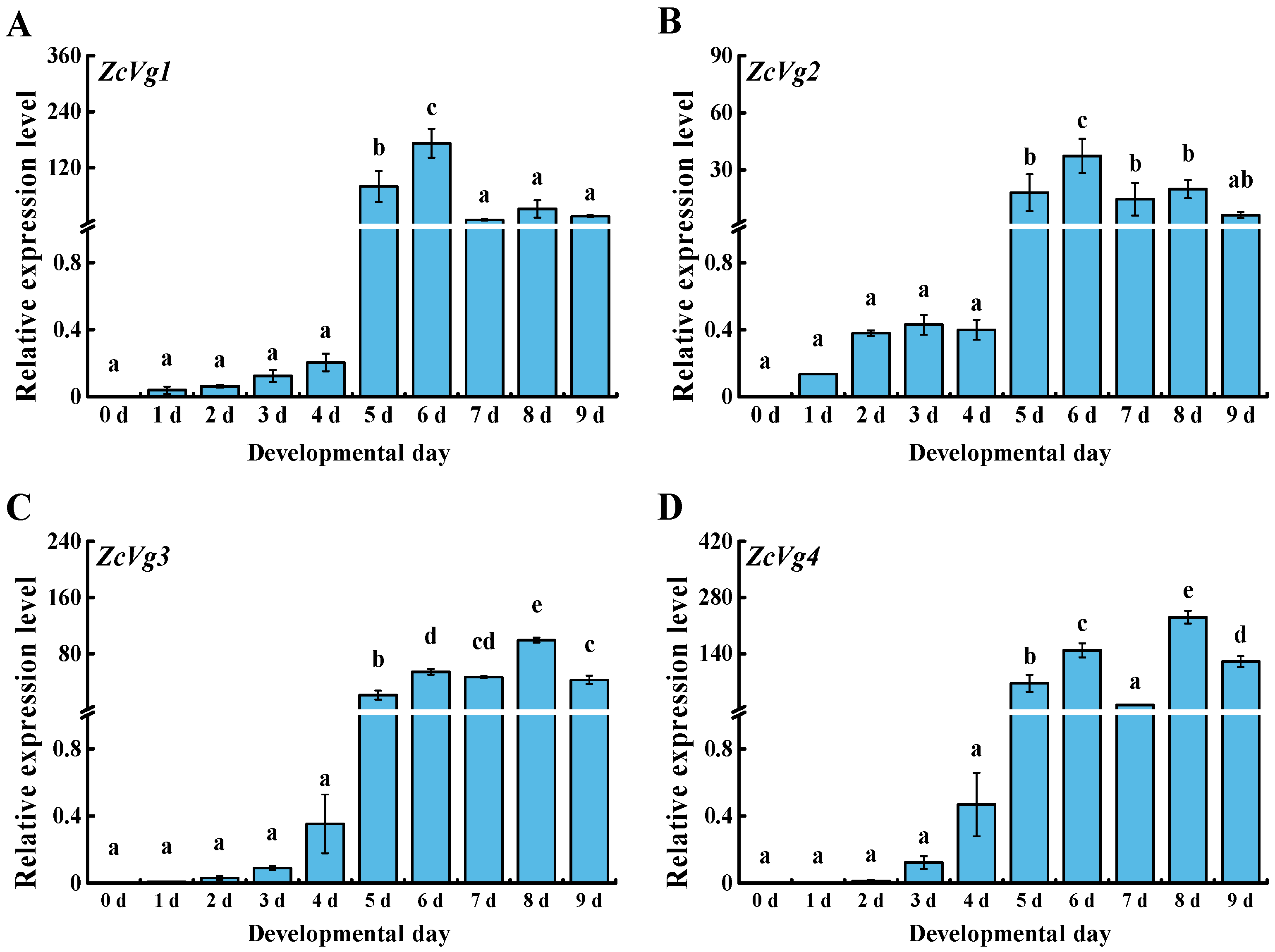
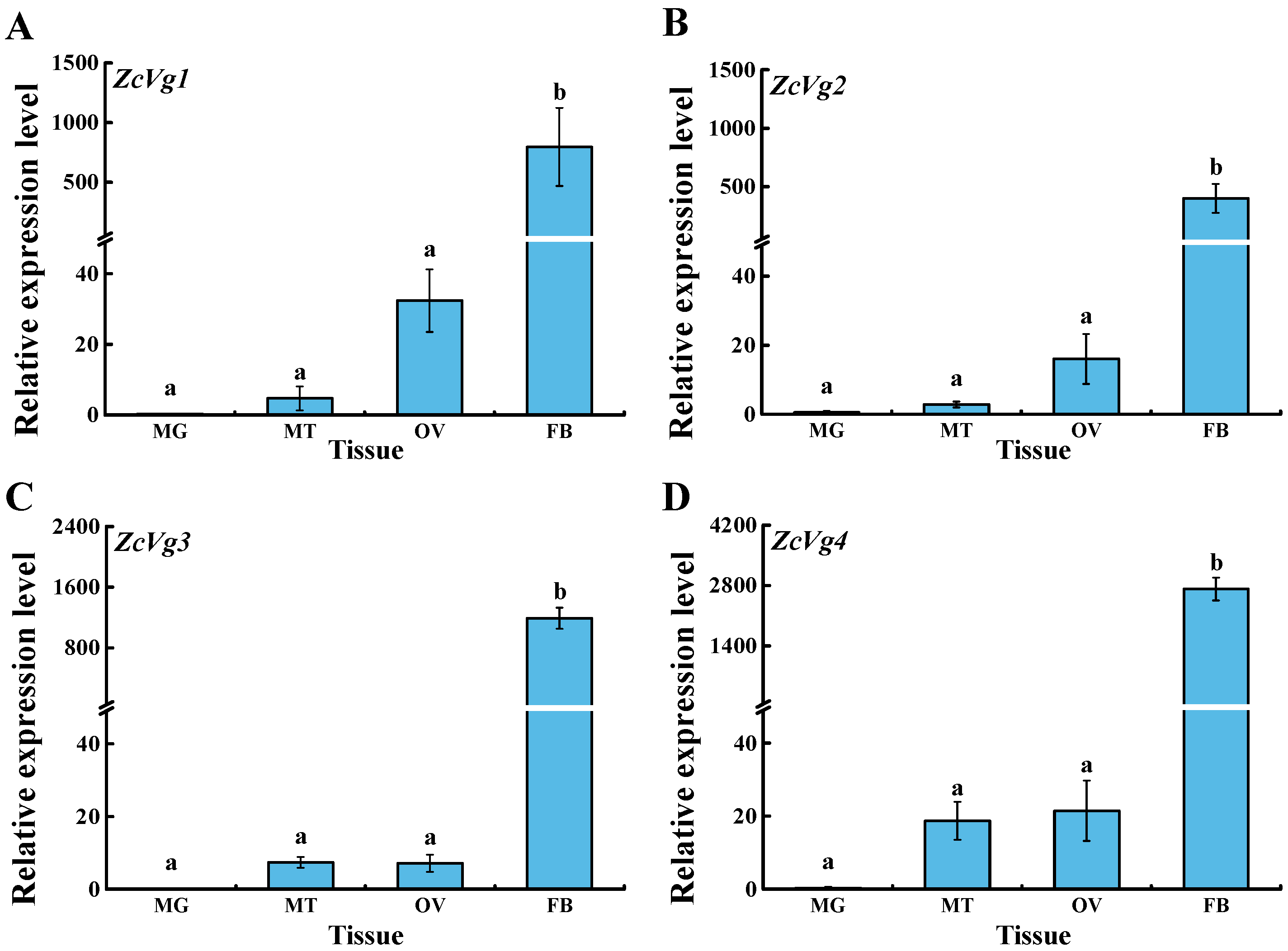
3.2. Spatio-Temporal Expression of ZcVgs
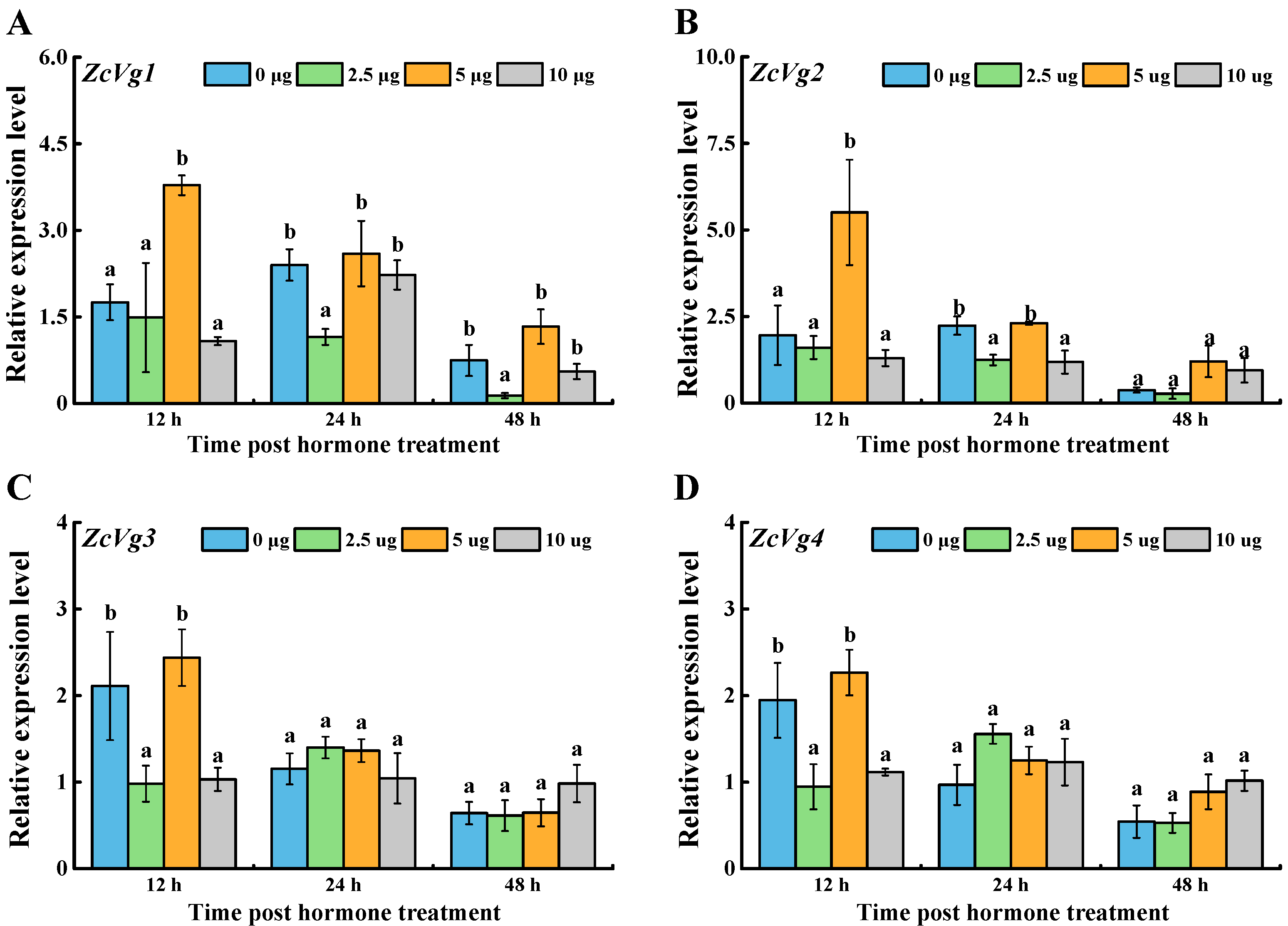
3.3. Effects of Hormone Induction on ZcVgs’ Expression
3.4. Effects of Starvation on ZcVgs’ Expression
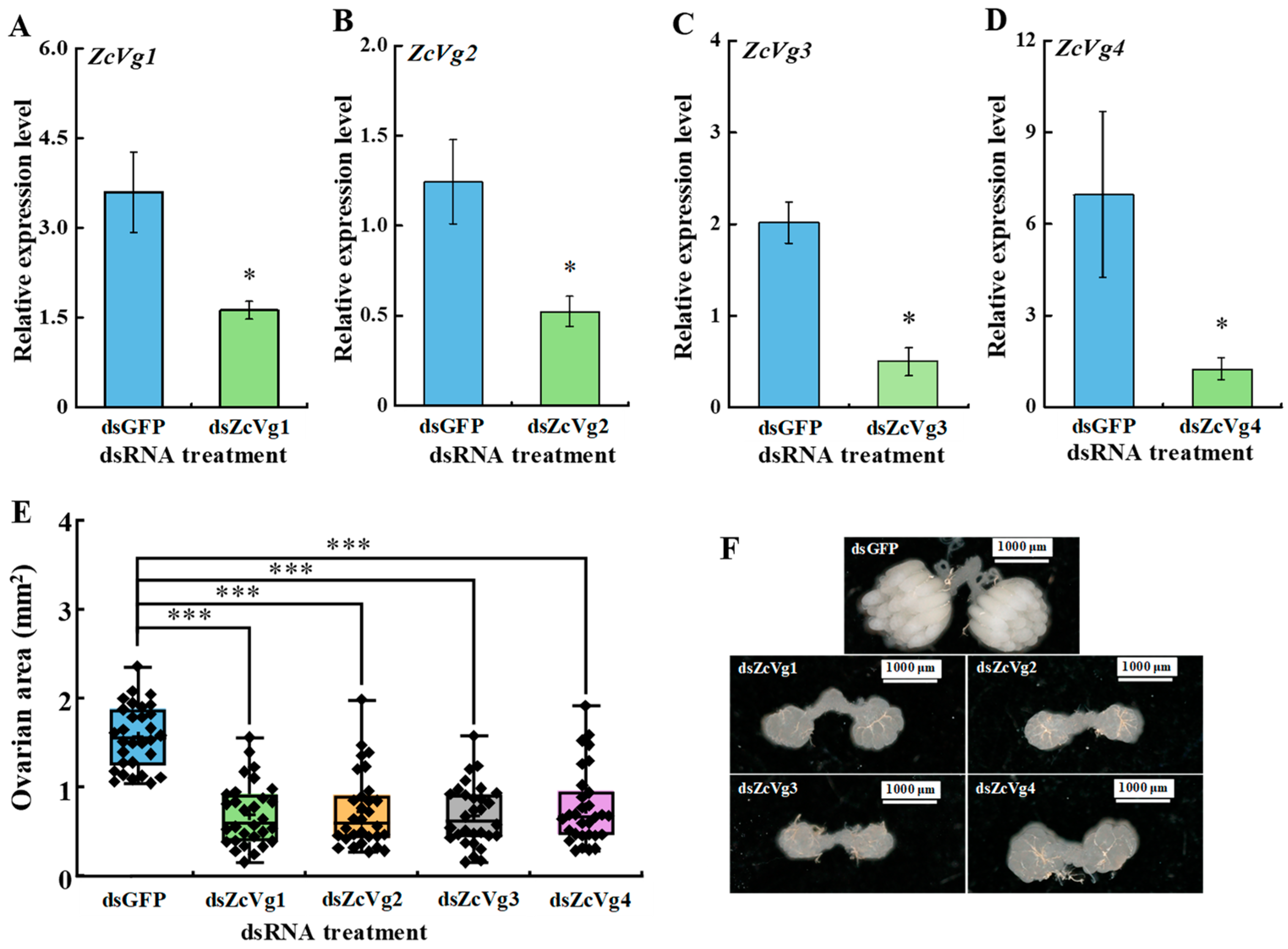
3.5. Effects of RNAi-Mediated Knockdown on Ovarian Development
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Tufail, M.; Nagaba, Y.; Elgendy, A.M.; Takeda, M. Regulation of vitellogenin genes in insects. Entomol. Sci. 2014, 17, 269–282. [Google Scholar] [CrossRef]
- Wu, Z.; Yang, L.; He, Q.; Zhou, S. Regulatory mechanisms of vitellogenesis in insects. Front. Cell Dev. Biol. 2021, 8, 1863. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.Y.; Yoon, H.J.; Jin, B.R. Osmia cornifrons vitellogenin: cDNA cloning, structural analysis and developmental expression. Entomol. Res. 2015, 45, 94–101. [Google Scholar] [CrossRef]
- Tufail, M.; Takeda, M. Molecular characteristics of insect vitellogenins. J. Insect Physiol. 2008, 54, 1447–1458. [Google Scholar] [CrossRef]
- Khalaila, I.; Peter-Katalinic, J.; Tsang, C.; Radcliffe, C.M.; Aflalo, E.D.; Harvey, D.J.; Dwek, R.A.; Rudd, P.M.; Sagi, A. Structural characterization of the N-glycan moiety and site of glycosylation in vitellogenin from the decapod crustacean Cherax quadricarinatus. Glycobiology 2004, 14, 767–774. [Google Scholar] [CrossRef] [Green Version]
- Tarone, A.M.; McIntyre, L.M.; Harshman, L.G.; Nuzhdin, S.V. Genetic variation in the Yolk protein expression network of Drosophila melanogaster: Sex-biased negative correlations with longevity. Heredity 2012, 109, 226–234. [Google Scholar] [CrossRef] [Green Version]
- Isoe, J.; Hagedorn, H.H. Mosquito vitellogenin genes: Comparative sequence analysis, gene duplication, and the role of rare synonymous codon usage in regulating expression. J. Insect Sci. 2007, 7, 1–49. [Google Scholar] [CrossRef]
- Lee, J.M.; Hatakeyama, M.; Oishi, K. A simple and rapid method for cloning insect vitellogenin cDNAs. Insect Biochem. Mol. Biol. 2000, 30, 189–194. [Google Scholar] [CrossRef]
- Tufail, M.; Hatakeyama, M.; Takeda, M. Molecular evidence for two vitellogenin genes and processing of vitellogenins in the American cockroach, Periplaneta americana. Arch. Insect Biochem. Physiol. 2001, 48, 72–80. [Google Scholar] [CrossRef]
- Adams, T.S.; Filipi, P.A.; Yi, S.X. Effect of age, diet, diapause and juvenile hormone on oogenesis and the of amount of vitellogenin and vitellin in the twospotted stink bug, Perillus bioculatus (Heteroptera: Pentatomidae). J. Insect Physiol. 2002, 48, 477–486. [Google Scholar] [CrossRef]
- Shi, X.F.; Li, Y.N.; Yi, Y.Z.; Xiao, X.G.; Zhang, Z.F. Identification and characterization of 30 k protein genes found in Bombyx mori (Lepidoptera: Bombycidae) transcriptome. J. Insect Sci. 2015, 15, 47–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rachinsky, A.; Srinivasan, A.; Ramaswamy, S.B. Regulation of juvenile hormone biosynthesis in Heliothis virescens by Manduca sexta allatotropin. Arch. Insect Biochem. Physiol. 2003, 54, 121–133. [Google Scholar] [CrossRef] [PubMed]
- Al Baki, M.A.; Lee, D.W.; Jung, J.K.; Kim, Y. Insulin signaling mediates previtellogenic development and enhances juvenile hormone-mediated vitellogenesis in a lepidopteran insect, Maruca vitrata. BMC Dev. Biol. 2019, 19, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adams, T.S.; Filipi, P.A. Interaction between juvenile-hormone, 20-hydroxyecdysone, the corpus cardiacum allatum complex, and the ovaries in regulating vitellogenin levels in the housefly, Musca domestica. J. Insect Physiol. 1988, 34, 11–19. [Google Scholar] [CrossRef]
- Hansen, I.A.; Attardo, G.M.; Rodriguez, S.D.; Drake, L.L. Four-way regulation of mosquito yolk protein precursor genes by juvenile hormone-, ecdysone-, nutrient-, and insulin-like peptide signaling pathways. Front. Physiol. 2014, 5, 103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trumbo, S.T. Juvenile hormone and parental care in subsocial insects: Implications for the role of juvenile hormone in the evolution of sociality. Curr. Opin. Insect Sci. 2018, 28, 13–18. [Google Scholar] [CrossRef] [PubMed]
- Parthasarathy, R.; Palli, S.R. Molecular analysis of nutritional and hormonal regulation of female reproduction in the red flour beetle, Tribolium castaneum. Insect Biochem. Mol. Biol. 2011, 41, 294–305. [Google Scholar] [CrossRef] [Green Version]
- Husain, M.; Rasool, K.G.; Tufail, M.; Alwaneen, W.S.; Aldawood, A.S. RNAi-mediated silencing of vitellogenin gene curtails oogenesis in the almond moth Cadra cautella. PLoS ONE 2021, 16, e0245928. [Google Scholar] [CrossRef]
- Blariza, M.J.; Grosso, C.G.; Garcia, B.A. Silencing of Two Vitellogenin Genes Inhibits Oviposition in the Chagas Disease Vector Triatoma infestans (Hemiptera: Reduviidae). Am. J. Trop. Med. Hyg. 2017, 97, 477–480. [Google Scholar] [CrossRef] [Green Version]
- Moriyama, M.; Hosokawa, T.; Tanahashi, M.; Nikoh, N.; Fukatsu, T. Suppression of bedbug’s reproduction by RNA interference of vitellogenin. PLoS ONE 2016, 11, e0153984. [Google Scholar] [CrossRef] [Green Version]
- Veerana, M.; Kubera, A.; Ngernsiri, L. Analysis of the vitellogenin gene of rice moth, Corcyra cephalonica stainton. Arch. Insect Biochem. Physiol. 2014, 87, 126–147. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Peng, L.; Wan, F.H. Pleiotropic functions of insect vitellogenin: With honey bee Apis mellifera as an example. Acta Entomol. Sin. 2010, 53, 335–348. [Google Scholar]
- De Meyer, M.; Delatte, H.; Mwatawala, M.; Quilici, S.; Vayssieres, J.F.; Virgilio, M. A review of the current knowledge on Zeugodacus cucurbitae (Coquillett) (Diptera, Tephritidae) in Africa, with a list of species included in Zeugodacus. Zookeys 2015, 540, 539–557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.J.; Song, Y.J.; Han, H.L.; Xu, H.Q.; Wei, D.; Smagghe, G.; Wang, J.J. Genome-wide analysis of long non-coding RNAs in adult tissues of the melon fly, Zeugodacus cucurbitae (Coquillett). BMC Genom. 2020, 21, 600. [Google Scholar] [CrossRef]
- McQuate, G.T.; Teruya, T. Melon fly, Bactrocera cucurbitae (Diptera: Tephritidae), infestation in host fruits in the Southwestern islands of Japan before the initiation of island-wide population suppression, as recorded in publications of Japanese public institutions. Int. J. Trop. Insect Sci. 2015, 7, 27–37. [Google Scholar] [CrossRef] [Green Version]
- Sim, S.B.; Geib, S.M. A chromosome-scale assembly of the Bactrocera cucurbitae genome provides insight to the genetic basis of white pupae. G3-Genes Genomes Genet. 2017, 7, 1927–1940. [Google Scholar] [CrossRef] [Green Version]
- Wei, D.; Xu, H.Q.; Chen, D.; Zhang, S.Y.; Li, W.J.; Smagghe, G.; Wang, J.J. Genome-wide gene expression profiling of the melon fly, Zeugodacus cucurbitae, during thirteen life stages. Sci. Data 2020, 7, 45. [Google Scholar] [CrossRef] [Green Version]
- Letunic, I.; Doerks, T.; Bork, P. SMART: Recent updates, new developments and status in 2015. Nucleic Acids Res. 2015, 43, D257–D260. [Google Scholar] [CrossRef]
- Procter, J.B.; Carstairs, G.M.; Soares, B.; Mourao, K.; Ofoegbu, T.C.; Barton, D.; Lui, L.; Menard, A.; Sherstnev, N.; Roldan-Martinez, D.; et al. Alignment of biological sequences with jalview. Methods Mol. Biol. 2021, 2231, 203–224. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Li, W.J.; Song, Y.J.; Xu, H.Q.; Wei, D.; Wang, J.J. Vitelline membrane protein gene ZcVMP26Ab and its role in preventing water loss in Zeugodacus cucurbitae (Coquillett) embryos. Entomol. Gen. 2021, 41, 279–288. [Google Scholar] [CrossRef]
- Hellemans, J.; Mortier, G.; De Paepe, A.; Speleman, F.; Vandesompele, J. QBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007, 8, R19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pereira, R.; Sivinski, J.; Teal, P.E. Influence of methoprene and dietary protein on male Anastrepha suspensa (Diptera: Tephritidae) mating aggregations. J. Insect Physiol. 2009, 55, 328–335. [Google Scholar] [CrossRef] [PubMed]
- Smolenaars, M.M.W.; Madsen, O.; Rodenburg, K.W.; Van der Horst, D.J. Molecular diversity and evolution of the large lipid transfer protein superfamily. J. Lipid Res. 2007, 48, 489–502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morandin, C.; Havukainen, H.; Kulmuni, J.; Dhaygude, K.; Trontti, K.; Helantera, H. Not only for egg yolk-functional and evolutionary insights from expression, selection, and structural analyses of formica ant vitellogenins. Mol. Biol. Evol. 2014, 31, 2181–2193. [Google Scholar] [CrossRef] [Green Version]
- Shang, F.; Niu, J.Z.; Ding, B.Y.; Zhang, Q.; Ye, C.; Zhang, W.; Smagghe, G.; Wang, J.J. Vitellogenin and its receptor play essential roles in the development and reproduction of the brown citrus aphid, Aphis (Toxoptera) citricidus. Insect Mol. Biol. 2018, 27, 221–233. [Google Scholar] [CrossRef]
- Wei, D.; Jia, H.T.; Zhang, M.Y.; Li, R.; Smagghe, G.; Wang, J.J. Comparative analysis of differential gene expression profiling of sex-bias fat body of Bactrocera dorsalis (Diptera: Tephritidae) identifying a new vitellogenin gene. Ann. Entomol. Soc. Am. 2018, 111, 43–54. [Google Scholar] [CrossRef]
- Upadhyay, S.K.; Singh, H.; Dixit, S.; Mendu, V.; Verma, P.C. Molecular characterization of vitellogenin and vitellogenin receptor of Bemisia tabaci. PLoS ONE 2016, 11, e0155306. [Google Scholar] [CrossRef] [Green Version]
- Qian, C.; Liu, C.; Zhu, B.; Meng, Y.; Wei, G. Identification and expression analysis of vitellogenin from silk-producing insect, Actias selene Hubner. Afr. J. Biotechnol. 2011, 10, 999–1010. [Google Scholar]
- Shu, Y.; Zhou, J.; Tang, W.; Lu, K.; Zhou, Q.; Zhang, G. Molecular characterization and expression pattern of Spodoptera litura (Lepidoptera: Noctuidae) vitellogenin, and its response to lead stress. J. Insect Physiol. 2009, 55, 608–616. [Google Scholar] [CrossRef]
- Swevers, L. An update on ecdysone signaling during insect oogenesis. Curr. Opin. Insect Sci. 2019, 31, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Parthasarathy, R.; Sun, Z.; Bai, H.; Palli, S.R. Juvenile hormone regulation of vitellogenin synthesis in the red flour beetle, Tribolium castaneum. Insect Biochem. Mol. Biol. 2010, 40, 405–414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elgendy, A.M.; Tufail, M.; Mohamed, A.A.; Takeda, M. A putative direct repeat element plays a dual role in the induction and repression of insect vitellogenin-1 gene expression. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2019, 234, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Comas, D.; Piulachs, M.D.; Belles, X. Fast induction of vitellogenin gene expression by juvenile hormone III in the cockroach Blattella germanica (L.) (Dictyoptera, Blattellidae). Insect Biochem. Mol. Biol. 1999, 29, 821–827. [Google Scholar] [CrossRef]
- Li, Y.; Gao, H.; Zhang, Y.; Lin, X. Role of the transcription factor Taiman in moulting and ovarian development of Nilaparvata lugens. Entomol. Gen. 2021, 41, 169–177. [Google Scholar] [CrossRef]
- Hannon, G.J. RNA interference. Nature 2002, 418, 244–251. [Google Scholar] [CrossRef]
- Maktura, G.C.; Paranhos, B.J.; Marques-Souza, H. RNAi in fruit flies (Diptera: Tephritidae): Successes and challenges. J. Appl. Entomol. 2021, 145, 740–756. [Google Scholar] [CrossRef]
- Boldbaatar, D.; Umemiya-Shirafuji, R.; Liao, M.; Tanaka, T.; Xuan, X.; Fujisaki, K. Multiple vitellogenins from the Haemaphysalis longicornis tick are crucial for ovarian development. J. Insect Physiol. 2010, 56, 1587–1598. [Google Scholar] [CrossRef]
- Sappington, T.W.; Raikhel, A.S. Molecular characteristics of insect vitellogenins and vitellogenin receptors. Insect Biochem. Mol. Biol. 1998, 28, 277–300. [Google Scholar] [CrossRef]



| Gene | Upstream Primer (5′-3′) | Downstream Primer (5′-3′) | Purpose |
|---|---|---|---|
| ZcVg1 | ATCTAGAGCACAATATGAATATACTGA | GGACACATTAGTATTGGCTGT | ORF cloning |
| ZcVg2 | CCATGAATCCTTTAAGCATT | TAACCGTAGTTTAGTTGTTCGA | |
| ZcVg3 | GATCATGAATTCACTGAAGATTT | TGTTAGCATATTAATCCTGGTTT | |
| ZcVg4 | CATGAATCCGCTGAAGAT | TGTTAGCATATCAATCCTGGT | |
| ZcVg1 | GCCAAATGATCGGCAAGACC | GAAGATCTTTGCGGGGTCCA | RT-qPCR |
| ZcVg2 | TCTAGCCGTTCAAGCAGTCA | GTGTTGCCGGTTTGACG | |
| ZcVg3 | GCTCCACACTCACCAACATG | CGAACATCTTGGCAGGGT | |
| ZcVg4 | CACTCACCAACATGAAGCGT | TTAGCAGGGTCCAAAGC | |
| Act3 | GAAACCTTCAACACACCCGC | CGGCCAAATCCAAACGAAGG | |
| Rps3 | TAAGTTGACCGGAGGTTTGG | TGGATCACCAGAGTGGATCA | |
| Rpl13 | GTTGTGCGTTGCGAGGAATT | GCTTGTCGTATGGTGGTGGA | |
| α-Tub | CGCATTCATGGTTGATAACG | GGGCACCAAGTTAGTCTGGA | |
| β-Tub1 | GAATTGATGCGACTGGTGCC | CTGAATCCATGGTGCCAGGT | |
| dsZcVg1 | taatacgactcactatagggTCATCTTTCCAAAATCGAT | taatacgactcactatagggACCAGTCCAGCGTTTGTA | dsRNA synthesis |
| dsZcVg2 | taatacgactcactatagggCAGAAGCAACTCAATACGTG | taatacgactcactatagggCCAAATCAATGACCACTAAA | |
| dsZcVg3 | taatacgactcactatagggACATGTCAATGGAAGAAGC | taatacgactcactatagggCAACATCGATAAGAGCTAAT | |
| dsZcVg4 | taatacgactcactatagggATGTCAGTGGAAGAGGCC | taatacgactcactatagggCAACAATGTGGATGATCTC | |
| dsGFP | taatacgactcactatagggTGAGCAAGGGCGAGGAGCTG | taatacgactcactatagggTCGATGCGGTTCACCAG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, D.; Han, H.-L.; Li, W.-J.; Wang, J.-J.; Wei, D. Expression and Role of Vitellogenin Genes in Ovarian Development of Zeugodacus cucurbitae. Insects 2022, 13, 452. https://doi.org/10.3390/insects13050452
Chen D, Han H-L, Li W-J, Wang J-J, Wei D. Expression and Role of Vitellogenin Genes in Ovarian Development of Zeugodacus cucurbitae. Insects. 2022; 13(5):452. https://doi.org/10.3390/insects13050452
Chicago/Turabian StyleChen, Dong, Hong-Liang Han, Wei-Jun Li, Jin-Jun Wang, and Dong Wei. 2022. "Expression and Role of Vitellogenin Genes in Ovarian Development of Zeugodacus cucurbitae" Insects 13, no. 5: 452. https://doi.org/10.3390/insects13050452






