3.2. Taxonomy
Megaphragma Timberlake, 1924. Proc. Haw. Entomol. Soc. 5: 412–414. Type species: Megaphragma mymaripenne Timberlake, by original designation.
Sethosiella Kryger, 1932. Bulletin de la Société Royale d’Egypte 16: 38–39. Type species: Sethosiella priesneri Kryger, by original designation. Synonymy by Ghesquière 1939, p. 36.
Paramegaphragma Lin, 1992. Entomotaxonomia 14(2): 133–135, 138. Type species: Paramegaphragma stenopterum Lin, by original designation. Synonymy by Delvare 1993, p. 151.
Diagnosis. Female (
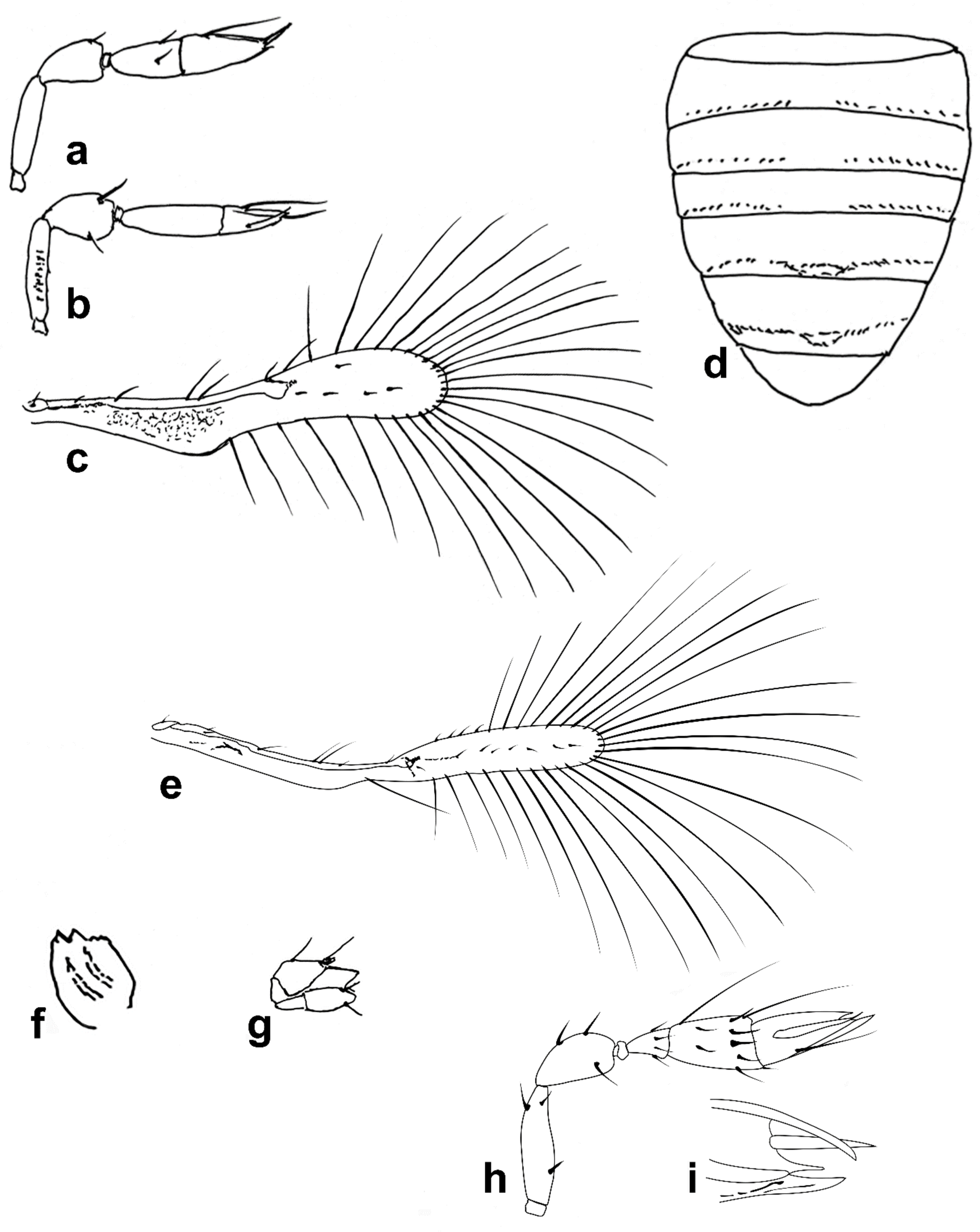
Figure 1a). Body rather compact, extremely small, length 0.16–0.3 mm. Antenna (
Figure 1b) inserted at mid level of the internal orbital line, with short radicle, scape usually elongate, pedicel, anellus, single funicle segment present or absent, clava one -, two -, or three-segmented. Antennal formula: 1 (scape), 1 (pedicel), (1) (anellus), 1 (funicle), 2 (clava); or 1,1,(1),1,1 or 1,1,(1),0,3. The antenna is counted as four- or five-segmented, since the anellus is not counted among the antennomeres. Claval segment 1 without multiporous placoid sensilla. Mandible with two small teeth. Maxillary palp very small and labial palp vestigial. Eye black unless otherwise stated. Mesosoma rather high, usually shorter than metasoma. Pronotum very short; mid lobe of mesoscutum not much longer than wide, either smooth or with polygonal or striate sculpture; one pair of adnotaular setae. Scutellum shorter than mid lobe of mesoscutum, with a pair of setae. Metanotum short; propodeum slightly longer than metanotum, or, in the middle, even longer, with a well-developed central area (disc) that may bear crenulae. Propodeal spiracle placed in an oval groove, and near the internal margin with two very small setae. Fore wing (
Figure 1c) extremely narrow compared with other Trichogrammatidae genera, 5.3–10× as long as maximum discal width, with short submarginal vein; costal cell and parastigma not distinct; marginal vein very long, with one short seta at the base and with one or two setae centrally, which when paired may be of similar or very different lengths; stigmal vein very short with one or two short setae on the stigma; disc with one or a few setae in one or two rows or glabrous (when there is one seta it is located on the dorsal surface of the wing, when discal setae are more numerous they are located on both dorsal and ventral surfaces of the wing, cf Figure 22d,e). Hind wing without discal fringe on front margin. Legs robust, often with striate sculpture on coxae, also on femora and tibiae. T7 and T8, respectively, without spiracle and cercus.
Male: As female, but often with postanellar antennomeres shaped differently. Genitalia tubular, very simple and usually small.
Relationship. The closest relatives of
Megaphragma appear to be
Prestwichia Lubbock and
Sinepalpigramma Viggiani and Pinto [
56]. Unfortunately, sequences for neither of these genera were available for comparison. We have used an
Epoligosita Girault, two
Oligosita Walker, and a
Probrachista Viggiani species as outgroups. These Oligositinae genera are close phylogenetically to
Megaphragma according to a previous molecular study [
33]. Species-group relationships are discussed below.
Distribution: Cosmopolitan.
Hosts and biology. The known species of
Megaphragma are all egg parasitoids of Thysanoptera (
Supplementary Figure S4) [
57,
58,
59]. Biological data are available only for a few species, e.g.,
M. mymaripenne, M. longiciliatum (as
M. amalphitanum) [
12,
13,
14], and are given below where available. It is interesting that at the same locality there may be more than one species of
Megaphragma, even in Europe.
Megaphragma viggianii and
M. polilovi were found in Italy at the same locality and on the same host, while in a single sample from near Barkás Lake in Hungary, there are three species (
M. longiciliatum,
M. noyesi, and the undescribed species represented by the specimen HUM4, close to
M. longiciliatum but distinct genetically).
On the basis of present knowledge, the following species group are proposed in Megaphragma:
M. mymaripenne-group: antenna with a single funicle segment that is longer than wide (this feature also shared by polychaetum- and longiciliatum- groups); T1 with longitudinal and/or transverse cells, with some denticles laterally within the cells (Figures 17f, 18f and 20b); T2–T4 each with a pair of short setae.
Included species: M. funiculatum Polaszek and Fusu sp. nov., M. mymaripenne Timberlake, M. nowickii Polaszek, Fusu, and Viggiani sp. nov., M. noyesi Polaszek and Fusu sp. nov., M. polilovi Polaszek, Fusu, and Viggiani sp. nov.
M. longiciliatum-group: same as mymaripenne-group, but without cells on T1. According to the phylogenetic analysis, the group appears to be derived from within the mymaripenne-group having lost the denticulate cells on T1.
Included species: M. longiciliatum Subba Rao, M. fanenitrakely Polaszek and Fusu sp. nov., M. priesneri Kryger, M. viggianii Polaszek, Fusu, and Polilov sp. nov. The species of the macrostigmum-group (M. caribea and M. macrostigmum) characterized by a four-segmented antenna, might be derived species within this group.
M. polychaetum-group: antenna with a long, cylindrical, funicle segment; spatulate sensilla at the end of each clava segment, and a robust terminal sensillum on C2; fore wing disc with more than seven setae, often arranged in two rows. Male antenna is particularly distinctive, with an elongate C1, short C2 usually with very long sensilla.
Included species: M. cockerilli Polaszek and Fusu sp. nov., M. giraulti sp. nov., M. polychaetum Lin, M. kinuthiae Polaszek, Fusu, and Viggiani sp. nov. Our molecular analysis also includes two males of this group, representing two undescribed species (vouchers SAM1 and SAM2, NHMUK). They have the antennal structure characteristic for males of the group, but our analyses recover them basal to the mymaripenne- and longiciliatum- groups instead of clustering them with the other two species of the polychaetum-group.
M. ghesquierei-group: antenna without funicle segment and with clava three-segmented, because the funicle is fused with the clava along an oblique suture. Fore wing disc with one seta on the dorsal surface or no setae. Propodeum characteristically produced centrally, almost always with a row of crenulae. Metafemur with a robust spine close to the connection with the tibia. Because of the intergradation in the structure of the antenna between the
ghesquierei and other groups, we do not currently consider
Paramegaphragma as a valid genus for the species in the
ghesquierei- plus
stenopterum- groups. It is possible that future analyses, especially including multigene or reduced genome representation data, may lead to the reinstatement of
Paramegaphragma Lin as a valid genus. The two species formerly assigned to
Paramegaphragma by Lin [
9],
M. stenopterum and
M. macrostigmum, are not closely related and clearly belong to different species-groups (
stenopterum-group and
macrostigmum-group, respectively), though on morphological grounds
stenopterum-group is clearly related to
ghesquierei-group or even integral part of it. This is another reason for not recognizing
Paramegaphragma.
Included species: M. breviclavum sp. nov., M. chienleei sp. nov., M. deflectum Lin, M. digitatum sp. nov., M. ghesquierei Ghesquière, M. hansoni sp. nov., M. liui sp. nov., M. pintoi Viggiani sp. nov., M. rivelloi sp. nov., M. striatum Viggiani, M. tamoi Polaszek, Fusu, and Viggiani sp. nov., M. tridens Fusu and Polaszek sp. nov., M. vanlentereni Polaszek and Fusu sp. nov.
M. stenopterum-group: same as
M. ghesquierei but with clava two-segmented. The antennal structure is very suggestive of the
ghesquierei-group, given the similarity between the apparent
C1 of the
stenopterum-group and that of the
ghesquierei-group; i.e., it is actually a funicle completely fused to the clava. In the
antecessor-group, the funicle is distinct albeit transverse and anneliform. Pending further evidence, we consider the
stenopterum-group as possibly nested within the
ghesquierei-group.
Megaphragma macrostigmum and
M. caribea (
macrostigmum-group) were considered by previous authors to belong in a group with
M. stenopterum [
60], and
M. macrostigmum with
M. stenopterum were both originally included by Lin [
9] in his genus
Paramegaphragma. However, the former two species lack any of the obvious apomorphies of the
ghesquierei-group except for the apparently lost funicle. Members of the
macrostigmum-group are otherwise similar in the structure of the fore wing and sculpture of the mesoscutum to the species in the
longiciliatum-,
mymaripenne-, and
polychaetum- groups and are probably not related to the
ghesquierei- and
stenopterum- groups. Our molecular analysis did not include
M. stenopterum, the only member of this species group.
Included species: M. stenopterum (Lin).
M. antecessor-group: antenna with a transverse funicle segment not much larger than the anellus, and clava one- or two-segmented. Metatibia with a characteristic row of setae (Figures 11d and 21j). The structure of the antenna seems intermediate between that characteristic of the longiciliatum- and mymaripenne- groups and that of the ghesquierei-group. In the latter species group, the antenna is apparently without a funicle, as the funicle is completely fused with the clava by an oblique suture and, hence, the clava appears three-segmented. Our phylogenetic analysis shows that M. antecessor and M. momookherjeeae, while resembling the ghesquierei species group in many features (including fore wing structure and the robust spine on metatibia), appear outside it, and basal to all remaining Megaphragma except members of ghesquierei-group (partitioned analyses) or the most basal species group of all Megaphragma (unpartitioned analysis).
Included species: M. antecessor Polaszek and Fusu sp. nov., M. momookherjeeae Polaszek and Fusu sp. nov., M. uniclavum Polaszek and Fusu sp. nov.
M. macrostigmum-group: as explained above, M. macrostigmum and M. caribea, while undoubtedly very closely related to each other, appear to have no connection with the ghesquierei-group (our molecular analysis did not include either of these two species). The antenna has the clava two-segmented and no funicle as in the stenopterum-group; the fore wing structure, however, is not similar to the ghesquierei-group but suggestive of the longiciliatum-group, especially M. priesneri; metafemur without the robust spine characteristic for the antecessor-, ghesquierei-, and stenopterum- groups.
Included species: M. caribea Delvare, M. macrostigmum (Lin).
Megaphragma caribea Delvare (
Figure 3a–c, Figure 12d–f and Figure 19a–c)
Diagnosis.
Female: Antenna four-segmented (excluding anellus), without funicle, clava two-segmented (
Figure 3a and Figure 12d);
C1 with 16
MT, 2 long
UST, 1
SS;
C2 with 4
MPS, 2
MT, 1 SB, 1 prominent apical
SS (
Figure 3a and Figure 12d).
Mid lobe of mesoscutum with large, but shallow polygonal cells (Figures 12e and 19c). Propodeum with central area short. Fore wing 5.5× as long as maximum discal width, marginal vein about twice length of submarginal vein, with two rather long setae (of equal length) present at midpoint of marginal vein (
Figure 3c). Metasoma with lines of microspines evident on
T2–
T6 (cf Figure 19a). Ovipositor 1.7× as long as mesotibia.
Body yellow, with the following slightly darkened: occiput, meso- and meta-coxae, apices of meso- and metafemora. Metasoma with pale brown transverse bands.
Male: Antenna with C1 longer than in female (Figure 19b).
Material examined. Paratypes: GUADELOUPE: Vieux Habitants, 17.XI.1988, coll. J. Etienne, ex eggs Selenothrips rubrocinctus (Giard) on Psidium guajava. Slides n. 8002.4, 8002.6, 8002.9 (2♀, 1♂, NHMUK).
Non-types: COLOMBIA: Cartagena, i.2015, with Heliothrips haemorrhoidalis and Selenothrips rubrocinctus on Terminalia catappa, coll. A.A. Polilov (1♂, AICF).
Species-group placement: M. macrostigmum-group—possibly a subgroup of the M. longiciliatum-group.
Distribution: Colombia, Guadeloupe.
Host: Selenothrips rubrocinctus (Giard).
DNA data: no DNA sequences.
Comments: The species was described in detail by the author. Megaphragma caribea is clearly close to M. macrostigmum (Lin). At present, their discrimination is based on the absence of long UST on the basal clava (C1) of the antenna of the latter species (Figure 6a). Since the original description did not indicate whether the species-group name caribea is a noun or an adjective, following Art. 31.2.2. of ICZN, we treat it as a noun and do not make a gender agreement.
Diagnosis.
Female: Antenna (
Figure 3e) without funicle, clava three-segmented, with
C1 and
C2 almost fused;
C1 with 1
UST;
C2 with 1
UST, and ≥4
MT;
C3 with ≥2
MPS, 1
MT, and 1 SB.
Figure 3.
Line drawings of Megaphragma species: (a) M. caribea, female antenna (Paratype); (b) M. caribea, male antenna (Paratype); (c) M. caribea, female fore wing (Paratype); (d) M. deflectum, female body dorsal (Holotype); (e) M. deflectum, female antenna (Holotype); (f) M. deflectum, female fore wing (Holotype); (g) M. ghesquierei, male body lateral (Paratype).
Figure 3.
Line drawings of Megaphragma species: (a) M. caribea, female antenna (Paratype); (b) M. caribea, male antenna (Paratype); (c) M. caribea, female fore wing (Paratype); (d) M. deflectum, female body dorsal (Holotype); (e) M. deflectum, female antenna (Holotype); (f) M. deflectum, female fore wing (Holotype); (g) M. ghesquierei, male body lateral (Paratype).
Mid lobe of mesoscutum (
Figure 3d) with longitudinal striate sculpture extending to scutellum; propodeum (
Figure 3d) with central area extended posteriorly, crenulae absent. Fore wing (
Figure 3f) 9× as long as maximum width; longest fringe seta 5× as long as maximum discal width. Fore wing disc without setae. Marginal vein with two long subequal setae centrally. Stigmal vein not enlarged, with two sensilla apically. Middle tibia with one large spine basally; metafemur with spine.
T1 with elongate cells laterally, 2–3× as long as wide;
T2–
T4 without setae laterally. Ovipositor 1.7× as long as mesotibia.
Body largely brown, the following paler: legs except coxae and metafemur. Antenna with pedicel pale; scape, C1–C3 darker. Fore wing strongly infuscate basally; stigmal and marginal veins brown; marginal vein very dark centrally.
Male: As female but C3 with fewer MPS and with ASC apically.
Material examined. Holotype ♀ (FAFU). CHINA: Wuyishan, Fujian, 19.x.1987, Wang Jiashe col.
Paratype: CHINA: Fuzhou, Fujian, 8.v.1987, N.Q. Lin col. (1♀, FAFU).
Non-type: CHINA: Wuyishan, Fujian, 10.x.1987, Wang Jiashe col. (1♂, FAFU).
Species-group placement: M. ghesquierei-group.
Distribution: China.
DNA data: no DNA sequences.
Comments: This species was correctly considered allied to
M. ghesquierei Ghesquière mostly due to features of the antenna (
Figure 3e), fore wing (
Figure 3f), and other characteristics of the body, but
M. deflectum can be distinguished easily from that species by the sculpture of the mid lobe of the mesoscutum and the central area of propodeum (
Figure 3d). The male “allotype” (paratype) of
M. deflectum is actually a male of
M. rivelloi sp. nov. (see below).
Diagnosis.
Female: Antenna without funicle, clava three-segmented,
C1 distal margin transverse (cf
Figure 3g).
C1 with ≥1
MT;
C2 with ≥2
MT, ≥2
UST;
C3 with ≥2
MPS, 1
MT, and 1
SB.
Mid lobe of mesoscutum anteriorly with reticulate sculpture, remainder with longitudinal striation continuing onto scutellum (cf
Figure 3g). Propodeum with a large subtriangular central area. Fore wing 7× as long as maximum width (
Figure 4a); the disc pointed distally, without setae. Metasoma with tergites with some short transverse striation centrally, and each with a pair of lateral setae (
Figure 4b).
Body dark brown, with the following paler: frons and occiput, scutellum and propodeum, tarsi. Metasoma with tergites and sternites appearing as dark bands (in the slide-mounted types). Fore wing basally strongly infuscate with a dark marginal vein.
Male: Similar to female in all aspects of morphology except genitalia characters.
Material examined. Holotype ♀ (MRAC). D. R. CONGO: Rutshuru, i.1938, ex eggs of Panchaetothrips noxius Priesner on Coffea arabica.
Paratypes: D. R. CONGO: 1♂, on slide with holotype; 3♀ on one slide, with data as holotype except “Neotopotype” in Ghesquière’s writing (MRAC).
Non-type: 1♂, labeled type in the Nowicki collection, no other data (DACE).
Species-group placement: M. ghesquierei-group.
Distribution: D. R. Congo.
Host: Panchaetothrips noxius Priesner
DNA data: no DNA sequences.
Comments: The species is rather easily recognizable by the combination of features of the antenna, mid lobe of mesoscutum, propodeum, fore wing, and metasomal tergites.
The species was intended to be described by Nowicki, but was published by Ghesquière [
61] (p. 36) because Nowicki’s manuscript on several African Trichogrammatidae never reached the journal Revue de zoologie et de botanique Africaines in Tervuren where Ghesquière was working. Ghesquière [
61] (p. 37) gives the date of collection as “XII.1937”, but, as given above, the holotype is labeled: “I.1938”.
Figure 4.
Line drawings of Megaphragma species: (a) M. ghesquierei, female fore wing (Holotype); (b) M. ghesquierei, male propodeum and tergites (Paratype); (c) M. giraulti, male antenna (Paratype); (d) M. giraulti, female antenna (Holotype); (e) M. giraulti, male metasoma (Paratype); (f) M. giraulti, female lateral meso- and metasoma (Holotype); (g) M. giraulti, female antenna, detail (Holotype).
Figure 4.
Line drawings of Megaphragma species: (a) M. ghesquierei, female fore wing (Holotype); (b) M. ghesquierei, male propodeum and tergites (Paratype); (c) M. giraulti, male antenna (Paratype); (d) M. giraulti, female antenna (Holotype); (e) M. giraulti, male metasoma (Paratype); (f) M. giraulti, female lateral meso- and metasoma (Holotype); (g) M. giraulti, female antenna, detail (Holotype).
Megaphragma longiciliatum Subba Rao (
Figure 5e–h and Figure 16e)
Megaphragma longiciliatum Subba Rao, 1969. Proc. R. Ent. Soc. Lond. (B) 38(7–8): 114.
Megaphragma aligarhensis Yousuf and Shafee, 1988. Indian J. syst. Ent. 4(2) [1987]: 114. Syn. nov.
Megaphragma amalphitanum Viggiani in Viggiani and Bernardo, 1997. Boll. Zool. Agr. Bach. Ser. II 29(1): 51–55. Syn. nov.
Megaphragma magniclava Yousuf and Shafee, 1988. Indian J. syst. Ent. 4(2) [1987]: 115–116. Syn. nov.
Megaphragma decochaetum Lin, 1992. Entomotaxonomia 14(2): 131–132. Syn. nov.
Megaphragma shimalianum Hayat, 2009. Oriental Insects 43: 212–213. Syn. nov.
Diagnosis. Female: Antenna (
Figure 5e and Figure 16e) with clava two-segmented. Funicle with ≥2
MT;
C2 with ≥6
MT, 1
UST;
C3 with ≥3
MPS, and 1
UST.
Mid lobe of mesoscutum anteriorly with reticulate sculpture. Propodeum with a very short central area. Fore wing 8× as long as wide (
Figure 5g). Metasoma (
Figure 5h) without subpolygonal sculpture on tergites, but with some ridges,
T2–
T4 each with a pair of long setae. Ovipositor 1.1× as long as mesotibia.
Body brown to dark brown, with the following paler: antenna, legs. Metasoma with tergites and sternites appearing as dark bands (in the slide-mounted types). Fore wing completely hyaline.
Male: Similar to female in most characters except genitalia; antennal funicle slightly more elongate than in female, and clava darker than remainder of antenna.
C2 without long
UST;
C3 shorter than in female (
Figure 5f).
Material examined. Holotype ♀ M. longiciliatum (NHMUK). INDIA: Bangalore, Avati, ex. Frankliniella lilivora Takahashi on Polyanthes tuberosa, x.1968, V. P. Rao. Paratypes: 13♀ 1♂, same data as holotype (NHMUK).
Holotype ♀ M. aligarhensis (AMU). INDIA: Aligarh, IX.1985, M. Yousuf.
Holotype ♀ M. amalphitanum (DACE). ITALY: Vietri sul mare (SA), x.1994, coll. G. Viggiani, ex egg of Heliothrips haemorrhoidalis on Viburnum tinus. Paratypes: 36♀, 32♂, mostly obtained from the same host collected in the same holotype locality (DACE).
Holotype ♀ M. decochaetum (FAFU). CHINA: Fuzhou, Fujian, 30.vi.1987, coll. Lin. Paratype: CHINA: Fuzhou, Fujian, 30.vi.1987, coll. Lin (1♂, FAFU).
Holotype ♀ M. magniclava (AMU). INDIA: Aligarh, 25.x.1985, M. Yousuf.
Paratypes M. shimalianum. INDIA: Uttar Pradesh, Mainpuri Malau, slide XIV 1, 2, 4, 6.ix.2007, F. R. Khan col. (3♀, 1♂, AMU); Firozabad, Nagla Prabhu, slide IX, 4.ix.2007, F. R. Khan col. (12♀, AMU).
Non-types: ARGENTINA: INTA Oliveros Santa Fe, v.2004, ex Caliothrips phaseoli, A. M. Molinari col. (2♀, 7♂, DACE, AICF); San Miguel de Tucuman, x–xi.2006, ex Thysanoptera eggs on corn, E. Luft col. (2♂, DACE, AICF); Salta Prov., Aguas Blancas, Routa 19, 22.72° S, 64.40° W, 447 m, 23.iii.2003, swp rainforest along Bolivia border, J. Munro 003-03-23-01 (1♀, UCRC); Salta Prov. Rosario de la Frontera (grounds of Hotel Termas), 25.84° S, 64.93° W, 447 m, 20.iii.2003, sweeping, J. Munro 003-03-20-10 (1♀, UCRC); Salta Prov., RN81, 66 km E. jct RP 24, 23.24° S, 63.40° W, 260 m, 24.iii. 2003, swp Dry Chaco, J. Munro 003-03-24-01 (6♀, AICF, UCRC). AUSTRALIA: WA, Margaret R, Warner Glen Rd, Stone Cottages, 34°04.44′ S, 115°08.14′ E, eucalyptus forest, YPT, 15–16.xi.2002, George, Owen, Hawks, Munro PEET02-010P (1♀, UCRC). CHINA: 24.v.1987 and 26.v.1987, coll. Lin, identified as M. decochaetum (1♀, 1♂, FAFU). D. R. CONGO: Province Orientale, Yangambi Biosphere Reserve 0°45.822′ N 24°30.285′ E, 15.v.2012, screen sweep primary forest, A. Polaszek col. BMNH 2012-88, DNA: COM 2.1 and COM 2.3 (2♀, AICF, NHMUK). FRANCE: Dept Gironde, St Colombe (nr Castillon-la-Bataille), Pitray, 1.viii.2000, S. Bessart, M. van Helden (3♀, UCRC); Dordogne, 3.5 km E Issigeac, 44°43′ N 0°38′ E, 100 m, 31.vii.2013, J.S. Noyes col. NHM(Ent.) 2013-144, DNA: FRM2 to FRM6 (4♀, 1♂, AICF, NHMUK). HUNGARY: Őrség Nemzeti Park, Barkás Lake, 46°52′ N 16°26′ E, 268 m, 28.vi.2010, screen-sweep, J.S. Noyes col., BMNH(Ent) 2010-63, DNA: HUM1 (1♂, NHMUK). INDIA: Uttar Pradesh, New Delhi, IARI, 220 m, 28°37′51″ N 77°09′50″ E, 5–7.xi.2003, pan trap, J. Heraty col. (1♀ 4♂, UCRC); Karnataka, W of Mudigere, 850–912 m, 13°07′05″ N 75°30′20″ E, 24.xi.2003, sweep evergreen forest, J. Heraty col. (3♀, UCRC). INDONESIA: W Java, Gunung Halimun NP, Tea-Forest Junction, 1066 m, 6°41′07″ S 106°31′16″ E, 17.ix.2015, screen-sweep, A. Polaszek col., DNA1147 (1♀, NHMUK). ITALY: Vietri sul mare, Benincasa, 40°40′ N 44°20′ E, 17.vii.2013, ex Heliothrips haemorrhoidalis on Viburnum tinus, G. Viggiani, DNA: ITM10 (1♀, NHMUK). MALAYSIA: Sarawak, Mentawai 4°14′ N 114°52′ E, ix.2011, screen sweep, A. Polaszek col., DNA: SRM1 (1♀, NHMUK). OMAN: Hajar Mts, screen-sweep, 20.i.2017 A. Polaszek col., DNA: MO3, MO13, MO20, MO22 (MO13 was destroyed during the DNA extraction) (2♀ 1♂, NHMO). PAPUA NEW GUINEA: Central Province, 15km SE Port Moresby, 1.i.1986, screen-sweep eucalyptus grassland, G. Gordh col. 86-01-01-1 (1♀, ANIC). UNITED ARAB EMIRATES: Abu Dhabi Emirate, Al Ain, Al Khabisi garden, 24°13.521N 55°41.95E, 25–30.iii.2019, yellow pan trap, A. Polaszek, B. Howarth col. (1♀, NHMUK). USA: Florida, Lake Seminole Park, Seminole, 27°50–51′ N 82°46′ W 9.vii.2015, sweep, Z. Lahey col., DNA1111, 1112, 1113 (3 specimens, NHMUK, currently misplaced).
Species-group placement: M. longiciliatum-group.
Distribution: Argentina, Australia, China, D. R. Congo, France, India, Indonesia (Java), Italy, Malaysia (Borneo, Sarawak), Oman, Papua New Guinea, Portugal [
62] (as
M. amalphitanum), UAE, and USA.
Hosts: Caliothrips phaseoli (Hood) (Argentina); Frankliniella lilivora Takahashi (India); Heliothrips haemorrhoidalis (Bouché) (Italy). The record from Argentina “ex Thysanoptera eggs on corn” could be from Frankliniella williamsi Hood.
DNA data: CO1: 8 sequences from 4 countries: D. R. Congo, France, Hungary, Malaysia (Sarawak); 28S: 18 sequences from 8 countries: D. R. Congo, France, Hungary, Indonesia (Java), Italy, Malaysia (Sarawak), Oman, USA.
Comments:
Megaphragma longiciliatum is the most widely distributed
Megaphragma species; hence the large number of synonyms. We have examined 150 specimens from 14 countries and have DNA sequences for 18 specimens from 8 very widely distributed countries. We have carefully assessed morphological variation within the specimens examined, and consider that it encompasses the morphological characteristics of the type material of the species synonymized above [
7,
9,
10,
63,
64].
The holotype of M. longiciliatum is in extremely poor condition. The mountant, presumably gum chloral, has turned black. It is to be hoped that in a few years’ time, the holotype will be destroyed completely, and one of the paratypes, all of which are still in excellent condition, can be designated a neotype. Unfortunately, there is no current provision under the Code to legitimately replace a holotype specimen that has deteriorated irremediably.
Megaphragma macrostigmum (Lin) (
Figure 6a–d)
Paramegaphragma macrostigmum Lin, 1992. Entomotaxonomia 14(2): 135–136.
Megaphragma macrostigmum: Delvare, 1993. Revue fr. Ent. (n.s.) 15(4): 151.
Diagnosis.
Female: Antenna without funicle and clava two-segmented (
Figure 6a);
C1 with two short
MT;
C2 with one
MPS, one
SB, and one
UST.
Mid lobe of mesoscutum with some large, but shallow polygonal cells. Propodeum with a very short central area. Fore wing 5.3× as long as maximum discal width, with two rather long setae in the middle of marginal vein (
Figure 6c); disc with 4–5 setae not in a row. Metasoma with a line of microspines evident on
T2–
T6 (cf
Figure 6d). Ovipositor 2.1× as long as mesotibia. The main features of the antenna and fore wing are illustrated in
Figure 6a,c.
Body uniformly pale brown, fore wing slightly to moderately infuscate below marginal vein.
Male: Similar to female in most characters except genitalia; antennal funicle slightly shorter than in female,
C1 longer,
C2 with long
UST (
Figure 6b).
Material examined. Holotype ♂ (FAFU). CHINA: Fuzhou, Fujian, 31.viii.1987, N.Q. Lin col.
Paratype: CHINA: Guangzhou, 3.xi.1985 N.Q. Lin col. (1♀, FAFU)
Non-type: CHINA: Guangzhou, 30.x.1985, N.Q. Lin col. (1♂, FAFU).
Species-group placement:
M. macrostigmum group—possibly a subgroup of the
M. longiciliatum-group.
Figure 5.
Line drawings of Megaphragma species: (a) M. giraulti, female fore wing (Holotype); (b) M. kinuthiae, female antenna; (c) M. kinuthiae, female antenna (detail); (d) M. kinuthiae, female fore wing; (e) M. longiciliatum, female antenna; (f) M. longiciliatum, male antenna; (g) M. longiciliatum, female fore wing; (h) M. longiciliatum, female dorsal meso-and metasoma.
Figure 5.
Line drawings of Megaphragma species: (a) M. giraulti, female fore wing (Holotype); (b) M. kinuthiae, female antenna; (c) M. kinuthiae, female antenna (detail); (d) M. kinuthiae, female fore wing; (e) M. longiciliatum, female antenna; (f) M. longiciliatum, male antenna; (g) M. longiciliatum, female fore wing; (h) M. longiciliatum, female dorsal meso-and metasoma.
Distribution: China.
DNA data: no DNA sequences.
Comments: This species is very similar to M. caribea; at present, the only difference from the latter species appears to be the absence of long sensilla on C2.
Megaphragma mymaripenne Timberlake, 1924. Proc. Haw. Entomol. Soc. 5: 414–415.
Megaphragma mymaripenne: Viggiani, 1997. Boll. Lab. Ent. agr. Filippo Silvestri 53: 117–122.
Diagnosis.
Female: Antenna (
Figure 6h and Figure 17d) with clava two-segmented, funicle with ≥4
MT;
C1 trapezoid in lateral view with length 1.5× maximum width or less (longer in dorsal or ventral view), with 2
UST, but without linear sensilla (
MPS), ≥9
MT;
C2 with ≥3
MPS, 1
MT, 1
SB, and 1
SS (
Figure 6i).
Mid lobe of mesoscutum anteriorly with subpolygonal sculpture, but often appearing smooth in slide-mounts. Propodeum with a very short central area. Fore wing (
Figure 6e) 9–10× as long as wide, marginal vein with two long setae in the middle, setae on disc more or less regularly in a row of 10–15 setae, and longest fringe seta 5–6× as long as maximum disc width.
T1 with sculpture represented by a combination of transverse and longitudinal cells, lateral ones twice as long as wide; sides of some cells with denticles present. The subsequent tergites show rather variable sculpture, differing from the pattern on the first tergum.
T2–
T4 each with a pair of very short setae.
Body uniformly pale brown, scutellum paler than mesoscutum. Legs pale, wings hyaline. Clava slightly darker than the remainder of the antenna.
Male (hitherto undescribed): same as female but antenna slender, with funicle twice as long as wide and
C1 about 1.7× as long as
C2.
T1 with sculpture not as complete as in the female. Genitalia simple, tubular, 4.5× as long as wide (cf
Figure 7f).
Material examined. Holotype ♀ (USNM). USA: Hawaii, Mountain View, i.1920, C.E. Pemberton col.
Paratype: same data and on the same slide with holotype (1♀, USNM).
Non-types: ARGENTINA: San Miguel de Tucuman, x–xi.2006, ex Thysanoptera eggs on corn, E. Luft col. (5♀, DACE); ix.2006, from corn, E. Vinla col. (3♂, DACE); Salta Prov., Rosario de la Frontera, 25.83° S 64.88° W, 745 m, 20.iii.2003, sweep forest, J. Munro 003-03-20-01 (1♀, UCRC); La Rioja Prov., Chuquis, 28°53′40″ S 67°00′31″ W, 1575 m, 17.iii.2003, sweep acacia scrub, J. Munro 003-03-17-05 (1♀, UCRC); Salta Prov., Orán, rd to San Andres along Rio Blanca, 23.11° S, 64.52° W, 535 m, 23.iii.2003, sweep scrub and ginger, J. Munro 003-03-23-02 (2♀, UCRC). BRAZIL: Santa Catarina, Nova Teutonia, 17.x.1949, F. Plaumann col. BM 1957-341 (1♀, NHMUK). COSTA RICA: Limón, Hitoy-Cerere Reserve, 9°40′ N 83°02′ W, 100 m, 24–26.ii.2008, J.S. Noyes col. NHM(E)2010-21AQ (1♀, NHMUK). DOMINICAN REPUBLIC: San Cristobal, S. Cristobal Manomatuey, 20 km NW valley, 500 m, 23.iii.1991, L. Masner col. (1♀, UCRC). ECUADOR: 1♀, Galapagos Is., Sta Cruz, Bellavista 2 mi N, 360 m, guava thicket, v–vii.1985, S. and J. Peck col. (CNCI). GUADELOUPE: Petit Borg, Domaine Duclos, 28.ii.1989, with Solenothrips rubrocinctus and Heliothrips haemorrhoidalis on Inga ingoides, J. Etienne col. (3♀, NHMUK). ISRAEL: Bet Dagan, ix.1996, ex Heliothrips haemorrhoidalis, M. Wysoki col. (7♀, DACE, AICF). MEXICO: Chiapas, 6.2 miles N Berriozabal, premontane rain forest, 9.viii.1990, 4000′ J.B. Woolley col. (3♀, 2♂, UCRC). USA: California, Orange Co., Irvine, 13.vi.1990, ex Heliothrips haemorrhoidalis on avocado, H.G. Johnson (1♀, NHMUK); California, Orange Co., South Coast Field Station, El Toro, ex Heliothrips haemorrhoidalis on avocado, H.G. Johnson (2♀, 1♂, UCRC); California, Orange Co., 10.ix.1989, ex Heliothrips haemorrhoidalis on avocado N. Hessein col. (1♀, DACE); Virginia, Montgomery Co., 8 km NW Blacksburg, 19–30.vi.1987, MT, rural, 1000 m, BRC HYM. TEAM (1♀, CNCI); California, San Diego Co., Valley Center, Weslilac Rd, Playa Grove, on avocado, H.G. Johnson (7♀, UCRC); Missouri, Parkville, 39°12′17″ N, 94°40′38″ W, 5.vii.2015, swept, Z. Lahey col., DNA: 1114 (1♀, NHMUK—currently misplaced).
Species-group placement: M. mymaripenne-group.
Distribution: Argentina, Brazil, Chile, Costa Rica, Dominican Republic, Ecuador, Guadeloupe, Israel, Mexico, USA, and Venezuela.
Hosts:
Megaphragma mymaripenne is a solitary egg endoparasitoid of several species of Panchaetothripinae (Thripidae). The most common host is the widespread
Heliothrips haemorrhoidalis. The populations recorded in the USA [
12] are represented mainly by females. The population reared in Argentina from maize and identified as
M.
mymaripenne [
11] differs from the known populations of the species: the reared specimens from maize appear to be normally bisexual.
DNA data: 28S: 1 sequence, Missouri (USA).
Comments: This species was described in detail by Timberlake [
65], and additional features were given by Viggiani [
6].
Megaphragma mymaripenne is extremely difficult to distinguish morphologically from the closely related species
M. polilovi, and even from the more distantly related species
M. noyesi, with which it has been previously confused. They differ, however, in the length and shape of
C1, length of the scape and colour of the radicle, and length of the ovipositor, respectively, as outlined in the key. Without the molecular data, these subtle differences would have been overlooked or treated as intraspecific variability. The correlation between the molecular clades and morphological characters indicates, however, that there are three species involved.
Records from Israel are the only Old-World records for this species; previous records of
M. mymaripenne, e.g., from Italy [
13,
14,
66], turned out to be misidentifications of the new species
M. polilovi.
Megaphragma polychaetum Lin (
Figure 8a–c)
Megaphragma polychaetum Lin, 1992. Entomotaxonomia 14(2): 132–133.
Megaphragma anomalifuniculi Yuan et Lou in Yuan et al., 1997. Journal of Northeast Normal University 4: 62–63. Syn. nov.
Diagnosis.
Female: Antenna long and narrow (
Figure 8a) with pedicel shorter than the subcylindrical funicle, which has two
MT. Clava two-segmented,
C1 twice as long as funicle, with two
MT and two long
UST;
C2 with two
MPS, two
MT, and a terminal basiconic sensillum (
SB) slightly shorter than half
C2 length.
Mid lobe of mesoscutum with subpolygonal sculpture. Propodeum with a very short central area (
Figure 8c). Fore wing (
Figure 8b) 8–9× as long as wide, with two short setae in the middle of the marginal vein, and a disc with two distinct rows of 6–8 setae (
Figure 8b). Tergites of metasoma without sculpture, but with some short and strong setae. The ovipositor is 1.1× as long as the mesotibia.
Head (including antenna), metasoma, meso-, and metacoxae are very dark. Remainder of body, including legs, pale brown. Fore wing strongly infuscate basally.
Male: Unknown.
Material examined. Paratypes: CHINA: Wuyishan, Fujian, 30.vii.1987, Wang Jiashe col. (3♀, FAFU); 10.vii.1987, 14-051, 14-052 (2♀, FAFU).
Species-group placement: M. polychaetum-group.
Distribution: China.
DNA data: no DNA sequences. DNA sequences are very likely to be close to those of M. cockerilli sp. nov. (see below).
Comments: The type material of
M. anomalifuniculi was not available to the authors. According to the illustration given by Yuan and Lou [
67],
M. anomalifuniculi appears to be similar, if not identical, to
M. polychaetum Lin. The features concerning the funicular segment appear to derive from a preparation artifact.
Megaphragma priesneri (Kryger) (
Figure 8d–g and Figure 22e)
Sethosiella priesneri Kryger, 1932. Bulletin de la Société Royale d’Egypte 16: 40.
Megaphragma priesneri: Ghesquière, 1839. Rev. Zool. Bot. Afr. 33(1): 38.
Diagnosis. Female: Antenna (
Figure 8d) with pedicel slightly shorter than scape, funicle as long as half pedicel. Clava two-segmented with two long
UST on
C1 of female;
C2 with one
MPS and two
MT.
Figure 6.
Line drawings of Megaphragma species: (a) M. macrostigmum, female antenna (Paratype); (b) M. macrostigmum, male antenna (Holotype); (c) M. macrostigmum, female fore wing (Paratype); (d) M. macrostigmum, male dorsal metasoma (Holotype); (e) M. mymaripenne, female fore wing (Holotype); (f) M. mymaripenne, mandible (Holotype); (g) M. mymaripenne, maxillary palp (Holotype); (h) M. mymaripenne, female antenna (Holotype); (i) M. mymaripenne, female antenna (detail of apex) (Holotype).
Figure 6.
Line drawings of Megaphragma species: (a) M. macrostigmum, female antenna (Paratype); (b) M. macrostigmum, male antenna (Holotype); (c) M. macrostigmum, female fore wing (Paratype); (d) M. macrostigmum, male dorsal metasoma (Holotype); (e) M. mymaripenne, female fore wing (Holotype); (f) M. mymaripenne, mandible (Holotype); (g) M. mymaripenne, maxillary palp (Holotype); (h) M. mymaripenne, female antenna (Holotype); (i) M. mymaripenne, female antenna (detail of apex) (Holotype).
Figure 7.
Line drawings of Megaphragma species: (a) M. nowickii, female propodeum and T1 (Holotype); (b) M. nowickii, male meso- and metasoma (Paratype); (c) M. nowickii, female antenna (Holotype); (d) M. nowickii, male antenna (Paratype); (e) M. nowickii, female fore wing (Holotype); (f) M. nowickii, male aedeagus (Paratype).
Figure 7.
Line drawings of Megaphragma species: (a) M. nowickii, female propodeum and T1 (Holotype); (b) M. nowickii, male meso- and metasoma (Paratype); (c) M. nowickii, female antenna (Holotype); (d) M. nowickii, male antenna (Paratype); (e) M. nowickii, female fore wing (Holotype); (f) M. nowickii, male aedeagus (Paratype).
Figure 8.
Line drawings of Megaphragma species: (a) M. polychaetum, female antenna (Paratype); (b) M. polychaetum, female fore wing (Paratype); (c) M. polychaetum, female propodeum and metasoma, lateral view (Paratype); (d) M. priesneri, female antenna (Neotype); (e) M. priesneri, male antenna (non-type); (f) M. priesneri, female dorsal meso- and metasoma (Neotype); (g) M. priesneri, female fore wing (Neotype).
Figure 8.
Line drawings of Megaphragma species: (a) M. polychaetum, female antenna (Paratype); (b) M. polychaetum, female fore wing (Paratype); (c) M. polychaetum, female propodeum and metasoma, lateral view (Paratype); (d) M. priesneri, female antenna (Neotype); (e) M. priesneri, male antenna (non-type); (f) M. priesneri, female dorsal meso- and metasoma (Neotype); (g) M. priesneri, female fore wing (Neotype).
Mid lobe of mesoscutum (
Figure 8f) anteriorly with subpolygonal sculpture; propodeum with a very short central area. Fore wing (
Figure 8g) 7× as long as wide, with maximum distal width less than 2× width measured at apex of marginal vein (
Figure 8g); maximum fringe seta length 4× maximum discal width; setae on ventral disc surface short, penultimate one not reaching to the base of the distal (Figure 22e).
T1 without sculpture, but with a row of microspines;
T2–
T4 each with a pair of setae, shorter than their corresponding tergites. Ovipositor 1.1× mesotibia.
Entire head and body are very dark. Legs and antenna paler. Wings hyaline.
Male: Similar to female in most characters except genitalia. Antenna with funicle and
C1 more elongate than in female, without
UST on
C1;
C2 much shorter than in female (
Figure 8e).
Material examined. Neotype ♀ (NHMUK), here designated. EGYPT: Tanta, 30.11.30, vine leaves with Retithrips.
Non-types: ISRAEL: Higwe Yisrael, xi.1996, M. Wysoki coll., ex eggs Retithrips syriacus on Vitis vinifera (15♀, 3♂, NHMUK, DACE, AICF).
Species-group placement: M. longiciliatum-group.
Distribution: Egypt, Israel.
Host: Retithrips syriacus (Mayet).
DNA data: no DNA sequences.
Comments: Following extensive inquiries over the decades since 1990 in Egypt and Denmark, the holotype (and indeed the remainder of the type series of four specimens) appears to be lost. A specimen with data almost identical to the holotype is in the NHMUK, but has aberrant antennae. Nevertheless, we here designate that specimen as neotype, given that the data are very similar to those of the original type [
68] (only the collection date differs by less than a month). Furthermore, all of the remaining morphology accords perfectly with the original description. Unfortunately, extensive efforts to collect fresh specimens in both Egypt and Israel failed.
The neotype designation for M. priesneri (Kryger) satisfies the provisions of Article 75.3 of the International Code of Zoological Nomenclature by: (1) clarifying the taxonomic identity of the species in its accepted modern concept (Article 75.3.1); (2) defining the combination of features of the sculpture of the mesoscutum and T1, propodeal structure and wing proportions as diagnostic for the species (Article 75.3.2); (3) providing data and description sufficient to ensure recognition of the specimen designated (Article 75.3.3); (4) giving reasons (no references available heretofore) for believing that the original type material is lost (Article 75.3.4); (5) selecting a neotype specimen consistent with the original description of the species and that was collected not long (less than 1 month) after the original description (specimen in this case) and, as such, represents the type species (Article 75.3.5); (6) choosing a neotype from the originally cited type locality, Tanta, Egypt (Article 75.3.6); and (7) recording that the neotype is the property of a recognized scientific institution, NHMUK in London (Article 75.3.7).
Megaphragma stenopterum (Lin) (
Figure 9e–h)
Paramegaphragma stenopterum Lin, 1992. Entomotaxonomia 14(2): 134–135.
Megaphragma stenopterum: Delvare, 1993. Revue fr. Ent. (n.s.) 15(4): 151.
Diagnosis.
Female: Antenna (
Figure 9e) without funicle, clava two-segmented, and
C2 twice as long as
C1.
C1 with two
MT;
C2 with one
MPS, two
MT, one
SB, and one
UST.
Mid lobe of mesoscutum, scutellum, and central area of propodeum longitudinally striate (
Figure 9g). Fore wing (
Figure 9h) 9× as long as wide, and longest fringe seta 7× as long as maximum discal width, with two long central setae on the marginal vein, one long discal seta; hind margin sinuate. Tergites of metasoma without sculpture or crenulae (
Figure 9g).
Body brown, the head darker brown. Fore wing basally strongly infuscate.
Male: Almost no discernible differences from female except genitalia characters. Even the antennae are very similar (
Figure 9e,f).
Material examined. Paratype:
CHINA: Fuzhou, Fujian, 20.xii.1987, N.Q. Lin col. (1♀, FAFU).
Figure 9.
Line drawings of Megaphragma species: (a) M. rivelloi, female antenna (Holotype); (b) M. rivelloi, female metasoma (Holotype); (c) M. rivelloi, female fore wing (Holotype); (d) M. rivelloi, female habitus (Holotype); (e) M. stenopterum, female antenna (Paratype); (f) M. stenopterum, male antenna (Paratype); (g) M. stenopterum, female dorsal meso- and metasoma (Paratype); (h) M. stenopterum, female fore wing (Paratype).
Figure 9.
Line drawings of Megaphragma species: (a) M. rivelloi, female antenna (Holotype); (b) M. rivelloi, female metasoma (Holotype); (c) M. rivelloi, female fore wing (Holotype); (d) M. rivelloi, female habitus (Holotype); (e) M. stenopterum, female antenna (Paratype); (f) M. stenopterum, male antenna (Paratype); (g) M. stenopterum, female dorsal meso- and metasoma (Paratype); (h) M. stenopterum, female fore wing (Paratype).
Non-types: CHINA: Fuzhou, Fujian, 6.x.1987, N.Q. Lin col. (1♀, FAFU); Fuzhou, Fujian, 20.vi.1987, N.Q. Lin col. (1♂, FAU).
Species-group placement: M. stenopterum-group.
Distribution: China.
DNA data: no DNA sequences.
Comments: This species, described in detail by the author, has the unique combination of a single seta on the fore wing and four-segmented antenna without any apparent funicle. A transverse, anelliform funicle is present in M. uniclavum, the only other species with a four-segmented antenna and a single seta on the fore wing.
There are differences between the Chinese text and the English text of the original description concerning the collecting dates of the type series. The examined paratype is mentioned in the English part but not in the Chinese part.
Megaphragma striatum Viggiani (
Figure 10a–d and Figure 21b,c)
Diagnosis.
Female: Antenna (
Figure 10a and Figure 21b) without funicle, clava three-segmented, with
C1 having a transverse distal margin.
C1 with ≥1
MT;
C2 ≥8
MT, and 2
UST;
C3 with 2
MPS, ≥2
MT, and 1
SB.
Mid lobe of mesoscutum and scutellum longitudinally striate (
Figure 10d). Propodeum with a pronounced subtriangular central area (Figure 21c). Fore wing 8× as long as wide, with one long central seta on the marginal vein, one discal seta, longest fringe seta 4–5× as long as maximum discal width. Metasoma with a row of crenulae on
T2 (Figure 21c).
Head and metasoma very dark, mesoscutum brown, the remainder, including legs and antenna, paler. Fore wing infuscate basally.
Male: Similar to female in most characters except genitalia.
Material examined. Holotype ♀ (DACE). MEXICO: Chiapas, Ocozocoautla, El Aquacero, 1800–2200′, 8.8.1990, coll. JB Woolley.
Paratypes: 2♀, 2♂, same data as holotype (UCRC).
Non-types: MEXICO: Tamaulipas, Alta Cima (nr Goméz Farias), 23°01′ N 99°09′ W, 2.xi.2009, screen-sweep A. Polaszek col., DNA: MXM1 (1♀, NHMUK); 1♀, 1♂, same data as holotype, but not mentioned in the original description (CNCI, UCRC).
Species-group placement: M. ghesquierei-group.
Distribution: Argentina, Belize, Costa Rica, Mexico.
DNA data: CO1: one sequence; 28S: one sequence (both Mexico).
Figure 10.
Line drawings of Megaphragma species: (a) M. striatum, female antenna (Holotype); (b) M. striatum, male antenna (Paratype); (c) M. striatum, female fore wing (Holotype); (d) M. striatum, female dorsal meso- and metasoma (Holotype).
Figure 10.
Line drawings of Megaphragma species: (a) M. striatum, female antenna (Holotype); (b) M. striatum, male antenna (Paratype); (c) M. striatum, female fore wing (Holotype); (d) M. striatum, female dorsal meso- and metasoma (Holotype).
Descriptions of new species
Megaphragma antecessor Polaszek and Fusu sp. nov. (
Figure 11a–d)
Description. Female: Head (
Figure 11c) with toruli vertical, in contact with each other medially; area below toruli with fine longitudinal sculpture, 1 min seta present laterally on each side. Antenna (
Figure 11a) five-segmented (excluding anellus), transverse funicle present; clava two-segmented but these almost completely fused.
C1 with ≥6
MT, 3
UST;
C2 with 3
MPS and 2
UST;
SB not detected but presumably present. Base of
C2 with one (apparent)
SS.
Mid lobe of mesoscutum (
Figure 11d) with fine longitudinal striation; vertical/ventral anterior mid lobe of mesoscutum with coarse, reticulate sculpture (
Figure 11d); propodeum with subtriangular area centrally (
Figure 11d) with 3–4 large crenulae; propodeum with hind margin arcuate. Fore wing (
Figure 11b) 8.5× as long as maximum width, maximum distal width is 92× the maximum basal width; disc with a single short seta, and longest fringe seta 6.5× maximum discal width. Marginal vein with four setae, the second (from the wing base) robust and blunt; central setae equal in length. Campaniform sensillum present below second seta, a line joining the sensillum to the fourth seta. Stigmal vein with a row of three campaniform sensilla apically. Mesotibia with two large spines basally; metafemur with spine; metatibia with a row of fine, blunt setae extending almost the entire inner length, increasing abruptly in length at the distal tibia (exact length not visible in
Figure 11d since setae positioned almost vertically; a similar row of setae is found in
M. momookherjeeae and
M. uniclavum, Figure 21j).
T1 with smooth area centrally, flanked by two or three longitudinal grooves and a longitudinal cell laterally, extending for the length of the tergum;
T1–
T4 with very long setae laterally, each longer than its tergum;
T2 with a curved row of 6–8 spicules on each side. Ovipositor 1.6× as long as mesotibia.
Body brown. Occiput and face entirely brown, vertex paler. Antenna pale brown, pedicel paler. Mesosoma with the following brown: mid lobe of mesoscutum centrally, side lobes, axillae, propodeum laterally; remainder of mesosoma pale. Entire metasoma brown, except T1 centrally pale. Fore wing distinctly infuscate basally, below, and including marginal vein.
Male: Unknown.
Material examined. Holotype ♀ (deposited in NHMUK). COSTA RICA: Puntarenas, Est. Biol. Monteverde, 10°19′ N 83°49′ W, 1540–1890 m, 26.ii.2007, J.S. Noyes BMNH(E) 2010-21, DNA: CRM2.2.
Species group placement: M. antecessor-group. Megaphragma antecessor clusters in different DNA sequence analyses with M. momookherjeeae sp. nov. (CO1 and combined analysis, strong support) or as basal to all other Megaphragma (28S, weak support). The shapes of the fore wing, propodeum, and to some extent the antenna, are strongly suggestive of the ghesquierei-group; presumably unique aspects of both DNA sequences prevent it from clustering with the species in that group.
Distribution: Costa Rica
DNA data: CO1: one sequence; 28S: one sequence.
Etymology: From the Latin antecessor (predecessor, precursor), in reference to the basal position of this species in the phylogenetic analyses. Noun in apposition.
Figure 11.
Photographs of Megaphragma species: (a) M. antecessor, female antenna (Holotype); (b) M. antecessor, female fore wing (Holotype); (c) M. antecessor, female head (Holotype); (d) M. antecessor, female dorsal meso- and metasoma (Holotype). Scale bars 20 µm.
Figure 11.
Photographs of Megaphragma species: (a) M. antecessor, female antenna (Holotype); (b) M. antecessor, female fore wing (Holotype); (c) M. antecessor, female head (Holotype); (d) M. antecessor, female dorsal meso- and metasoma (Holotype). Scale bars 20 µm.
Megaphragma breviclavum Polaszek and Fusu sp. nov. (
Figure 12a–c)
Description. Female: Antenna (
Figure 12a) five-segmented (excluding anellus); funicle absent; hence, clava three-segmented, with
C1 and
C2 almost fused.
C1 without
UST;
C2 with 2
UST, and abundant
MT;
C3 with 2–3
MPS,
SB, and
SS.
Mid lobe of mesoscutum (
Figure 12b) with longitudinal striate sculpture extending to scutellum; propodeum (
Figure 12b) with central area extended posteriorly, crenulae absent. Fore wing (
Figure 12c) 7.5× as long as maximum width; the disc with a single long seta; longest fringe seta 4.5× as long as maximum discal width. Marginal vein with one long seta centrally, extending to apex of marginal vein. Stigmal vein moderately enlarged, with four sensilla apically. Mesotibiae with one large spine basally; metafemora with spine.
T1 with elongate cells laterally, 2–3× as long as wide;
T2–
T4 without setae laterally;
T5 with long setae laterally. Ovipositor 1.7× as long as mesotibia.
Body largely brown, the following paler: legs except coxae and metafemur. Antenna with pedicel pale; scape and C1–C3 darker. Fore wing strongly infuscate basally; stigmal and marginal vein brown; marginal vein very dark centrally.
Male: C1 and C2 with scattered SS; C2 with 2–3 MT apically; C3 with long apical and ventral UST. Colour and morphology largely as in female.
Material examined. Holotype ♀ (deposited in NHMUK). MEXICO: Tamaulipas, Alta Cima (nr Gómez Farias), 23°01′ N 99°09′ W, 2.ii.2009, A. Polaszek col. NHM(E) 2010-21, DNA: MXM2.
Paratypes: 1♀, 1♂ with same data as holotype, DNA: MXM3 and MXM4 (NHMUK).
Species-group placement: M. ghesquierei-group.
Distribution: Mexico.
DNA data: CO1: three sequences; 28S: three sequences.
Etymology: A noun in apposition referring to the comparatively short clava.
Description. Female: Antenna (
Figure 12g) five-segmented (excluding anellus); funicle absent (though anellus extremely large); hence, clava three-segmented, with
C1 and
C2 strongly overlapping, almost fused;
C1 with 1 elongate, apical
UST;
C2 with ≥10
MT; and
C3 with 3 very long
UST.
Mid lobe of mesoscutum (cf
Figure 13b) smooth with some irregular longitudinal striate sculpture; propodeum (cf
Figure 13a) elongate and curved centrally and posteriorly, crenulae present.
T1 without elongate cells laterally;
T2–
T4 with short setae laterally. Ovipositor 2× as long as mesotibia. Mesotibia with one large spine basally; metafemur with spine. Fore wing (cf
Figure 13c) 8× as long as maximum width, maximum distal width equal to maximum basal width; discal setae absent, longest fringe seta 4.7× as long as maximum discal width. Marginal vein with two long setae centrally, approximately equal in length. Stigmal vein moderately enlarged, with three sensilla apically.
Head and body uniformly very pale brown.
C3 darker than remainder of antenna (
Figure 12g). Fore wing basally infuscate (cf
Figure 13c).
Male: Characteristics as for female (
Figure 13a–c) (except antenna and genitalia); although, metasoma darker than in female. Antenna as in
Figure 12h, with a much shorter
C3 compared to the female.
Material examined. Holotype ♀ (deposited in AICF). MALAYSIA: Sabah (Borneo), Danum Valley, 05°01′ N 117°49′ E, 16.ix.2012, fogged tree, T. Cockerill, DNA: SAM12.
Paratypes: MALAYSIA: Sabah (Borneo), Maliau Basin Studies Centre, Belian Trail, 04°44′ N 116°58′ E, 20.ix.2012, screen-sweep, A. Polaszek NHM(E) 2010-21, DNA: SAM4 to SAM8 (4♂, 1♀, AICF, NHMUK).
Species-group placement: M. ghesquierei-group.
Distribution: Malaysia (Borneo, Sabah).
DNA data: CO1: four sequences; 28S: six sequences.
Figure 12.
Photographs of Megaphragma species: (a) M. breviclavum, female antenna (Holotype); (b) M. breviclavum, female dorsal meso- and metasoma, composite image (Holotype); (c) M. breviclavum, female fore wing (Holotype); (d) M. caribea, female antenna (flagellum only) (Paratype); (e) M. caribea, female dorsal mesosoma (Paratype); (f) M. caribea, female fore wing (Paratype); (g) M. chienleei, female antenna (Holotype); (h) M. chienleei, male antenna (Paratype). Scale bars 20 µm except 50 µm for b, c, and f.
Figure 12.
Photographs of Megaphragma species: (a) M. breviclavum, female antenna (Holotype); (b) M. breviclavum, female dorsal meso- and metasoma, composite image (Holotype); (c) M. breviclavum, female fore wing (Holotype); (d) M. caribea, female antenna (flagellum only) (Paratype); (e) M. caribea, female dorsal mesosoma (Paratype); (f) M. caribea, female fore wing (Paratype); (g) M. chienleei, female antenna (Holotype); (h) M. chienleei, male antenna (Paratype). Scale bars 20 µm except 50 µm for b, c, and f.
Etymology: Named for pitcher-plant (Nepenthes) botanist and wildlife photographer Chien C. Lee (Sarawak, Malaysian Borneo).
Megaphragma cockerilli Polaszek and Fusu sp. nov. (
Figure 13d–f)
Description.
Female: Antenna (
Figure 13d) five-segmented (excluding anellus); pedicel as long as funicle; funicle 4× as long as wide;
C2 longer than
C1.
C1 with two prominent dorsal
UST, proximal
UST almost as long as entire clava;
C2 with two
MT and an
SB, which is only slightly shorter than
C2.
MPS apparently absent.
Mid lobe of mesoscutum (
Figure 13e) entirely with large, coarse reticulation; propodeum with a rhomboid, laterally arcuate central area, its hind margin truncate, with fine crenulae.
T1–
T4 largely smooth, with scattered denticles and no setae laterally.
T5 and
T6 with a pair of long setae centrally. Ovipositor 1.1× as long as mesotibia. Mesotibia without spines basally; metafemur without spine; metatibia with a row of five spines within the distal inner half; a robust spine towards the apex of the outer surface. Fore wing (
Figure 13f) 8.5× as long as maximum width, maximum distal width 1.4× maximum basal width; discal setae arranged in 3–4 rows, of 4–6 setae per row, longest fringe seta 5× as long as the maximum discal width. Marginal vein with two long setae centrally, of equal length. Stigmal vein with two sensilla apically.
Body largely dark brown, mesosoma paler laterally; antenna very dark brown. Fore wing infuscate basally.
Male: Unknown.
Material examined: Holotype ♀ (deposited in AICF): MALAYSIA: Sabah (Borneo), Danum Valley, 05°01′ N 117°49′ E, 16.ix.2012, fogged tree, T. Cockerill, DNA: SAM11.
Species-group placement: polychaetum-group. Very close to M. polychaetum, differing by the extremely elongate terminal sensillum basiconicum.
Distribution: Malaysia (Borneo, Sabah)
DNA data: CO1: one sequence; 28S: one sequence.
Etymology: Named for our colleague and friend, Dr Tim Cockerill, collector of this species (Falmouth University, UK).
Megaphragma digitatum Polaszek and Fusu sp. nov. (
Figure 14a–c)
Description. Female: Antenna (
Figure 14a) five-segmented (excluding anellus); funicle absent; hence, clava three-segmented, with
C1 and
C2 almost fused;
C1 with ≥4
MT, without
UST;
C2 with 2
UST and abundant
MT;
C3 with 2–3
MPS,
SB, and
SS.
Mid lobe of mesoscutum (
Figure 14b) with longitudinal striate sculpture extending to scutellum; propodeum with central area extending posteriorly, crenulae present;
T1 with one elongate cell or groove laterally, 2–3× as long as wide;
T2–
T4 without setae laterally;
T5 with long setae laterally. Ovipositor 1.5× as long as mesotibia. Mesotibia with one large spine basally; metafemur with spine. Fore wing (
Figure 14c) 8.5× as long as maximum width, maximum distal width equal to maximum basal width; the disc with a single long seta; longest fringe seta 5× as long as maximum discal width. Marginal vein with two setae centrally; proximal seta 5–7× as long as distal seta, extending to the end of the marginal vein (in
Figure 14c, the distal seta is barely visible in the space between the proximal one and the marginal vein). Stigmal vein moderately enlarged, with two sensilla apically.
Body largely brown, the following paler: legs except coxae and metafemora. Antenna with pedicel pale; scape and C1–C3 darker. Fore wing strongly infuscate basally; stigmal and marginal vein brown; marginal vein very dark centrally.
Male: Largely as in female. C1 and C2 with scattered SS; C2 with 2–3 MT apically; C3 with long apical and ventral UST.
Material examined. Holotype ♀ (deposited in NHMUK).
COSTA RICA: Puntarenas, Est. Biol. Monteverde, 10°19′ N 83°49′ W, 1540–1890 m, 26.ii.2007, J.S. Noyes BMNH(E) 2010-21, DNA: CRM2.1.
Figure 13.
Photographs of Megaphragma species: (a) M. chienleei, male dorsal meso- and metasoma (Paratype); (b) M. chienleei, male dorsal mesosoma (Paratype); (c) M. chienleei, male fore wing (Paratype); (d) M. cockerilli, female antenna (Holotype); (e) M. cockerilli, female dorsal mesosoma (Holotype); (f) M. cockerilli, female fore wing (Holotype). Scale bars 20 µm.
Figure 13.
Photographs of Megaphragma species: (a) M. chienleei, male dorsal meso- and metasoma (Paratype); (b) M. chienleei, male dorsal mesosoma (Paratype); (c) M. chienleei, male fore wing (Paratype); (d) M. cockerilli, female antenna (Holotype); (e) M. cockerilli, female dorsal mesosoma (Holotype); (f) M. cockerilli, female fore wing (Holotype). Scale bars 20 µm.
Paratypes: COSTA RICA: same data as holotype except DNA: CRM2.3, 2.6, 2.9, 2.12 (2♂, 2♀, AICF, MZUCR); Cartago, 12.5 km S Turrialba Rancho Naturalista, 1000 m, 9°50″ N 83°34″ W, 12-14.ii.2017, J.S. Noyes BMNH(E) 2017-39, DNA1681 (1♀, NHMUK). ECUADOR: Km 26.5 road Dura–Tambo, Estación Experimental Litoral Sur, INIAP, 21.xi.2017, ex Chaetanaphothrips signipennis on Musa paradisiaca, M. Arias col., DNA: ECU3 (3♀, 1♂ AICF, NHMUK, UCRC); same data except DNA: ECU4 (3♀ 2♂, AICF, NHMUK, UCRC); same data but ex Frankliniella parvula, DNA: ECU1 (2♀, NHMUK).
Non-type: same data as holotype (1♀, without wings, NHMUK).
Species-group placement: ghesquierei-group.
Distribution: Costa Rica, Ecuador.
Hosts: Chaetanaphothrips signipennis (Bagnall); Frankliniella parvula Hood.
DNA data: CO1: two sequences (Costa Rica); 28S: eight sequences (six Costa Rica, two Ecuador).
Etymology: The species name refers to the digitate C3.
Megaphragma fanenitrakely Polaszek and Fusu sp. nov. (
Figure 14d–f)
Description. Female: Antenna (
Figure 14d) five-segmented (excluding anellus), with pedicel slightly longer than funicle (12:8); clava two-segmented,
C1 with two
UST; one
SB at the apex of
C1 and
C2; apex of
C2 (
Figure 14d) also with two elongate
MPS and a long
SB.
Mesoscutum with mid lobe (
Figure 14e) entirely with coarse, reticulate sculpture; metanotum and propodeum medially short. Metasoma with a row of microspines on each segment.
T1 without cells. Ovipositor 1.1× as long as mesotibia. Mesotibia without spines basally. Metafemur without prominent spine. Fore wing (
Figure 14f) 9× as long as wide, maximum distal width 1.5× maximum basal width; the disc with 10 setae is irregularly arranged in 1–2 rows, and the longest fringe seta 6× maximum discal width. Marginal vein with two long setae centrally, of equal length. Stigmal vein moderately enlarged, with three sensilla apically.
Body largely pale brown, mesosoma paler laterally; antenna brown. Wings hyaline.
Male: Unknown.
Material examined. Holotype ♀ (deposited in NHMUK). MADAGASCAR: Nosy Komba, 13°27′45″ S 48°20′18″ E, 460 m, 22.vi.2015, screen-sweep, A. Polaszek col. BMNH(E)2015-122.
Paratypes: MADAGASCAR: Nosy Komba, closed canopy forest, 13°27′11″ S 48°20′4″ E, 170 m, 19.vi.2015, yellow pan trap, A. Polaszek col. BMNH(E)2015-122 (2♀, NHMUK).
Species-group placement: longiciliatum-group.
Distribution: Madagascar.
DNA data: no DNA sequences.
Etymology: A noun in apposition; “fanenitra kely” = “tiny wasp” (Malagasy).
Megaphragma funiculatum Fusu, Polaszek, and Viggiani sp. nov. (
Figure 14g,h and
Figure 15a,b)
Description. Female: Antenna (
Figure 14g) five-segmented (excluding anellus), pedicel twice as long as funicle, the latter trapezoid, and slightly longer than wide;
C1 slightly shorter than
C2;
C1 with 2 dorsal
UST;
C2 with ≥3
MPS 1
SB and a short
SS.
Figure 14.
Photographs of Megaphragma species: (a) M. digitatum, female antenna (Holotype); (b) M. digitatum, female dorsal mesosoma (Holotype); (c) M. digitatum, female fore wing (Holotype); (d) M. fanenitrakely, female antenna (Holotype); (e) M. fanenitrakely, female mesosoma (Holotype); (f) M. fanenitrakely, female fore wing (Holotype); (g) M. funiculatum, female antenna (Holotype); (h) M. funiculatum, female mesosoma (Holotype). Scale bars 20 µm except 50 µm for c and f.
Figure 14.
Photographs of Megaphragma species: (a) M. digitatum, female antenna (Holotype); (b) M. digitatum, female dorsal mesosoma (Holotype); (c) M. digitatum, female fore wing (Holotype); (d) M. fanenitrakely, female antenna (Holotype); (e) M. fanenitrakely, female mesosoma (Holotype); (f) M. fanenitrakely, female fore wing (Holotype); (g) M. funiculatum, female antenna (Holotype); (h) M. funiculatum, female mesosoma (Holotype). Scale bars 20 µm except 50 µm for c and f.
Mid lobe of mesoscutum smooth, with weakly impressed large subpolygonal sculpture (not visible in paratype which has been strongly macerated). Metanotum and propodeum relatively long centrally, each about half the length of scutellum. Propodeum with short central area, without crenulae.
T1 (
Figure 15a) sculpture with cells converging centrally, lateral cells 2–3× as long as wide, without denticles (some denticles present on innermost cells).
T2–
T4 without long setae laterally, each with similar sculpture comprising a central irregular oval cell and elongate lateral cells; those on
T3 and
T4 divided medially. Ovipositor 1.3× as long as mesotibia. Mesotibia with one large spine basally; metafemur without spine. Fore wing (
Figure 15b) 10× as long as wide, maximum distal width 1.2× maximum basal width; the disc with a single irregular row of five setae; longest fringe seta 5.5× maximum discal width. Marginal vein with two long setae centrally.
Head, including antenna, pale; mesosoma largely pale, anterior half of mesoscutal mid lobe brown; metasoma entirely brown, T1 darker than the remainder. Wings hyaline.
Male: Unknown.
Material examined. Holotype ♀ (deposited in NHMUK). COSTA RICA: Limón, Hitoy-Cerere Reserve, 100 m, 9°40′ N 83°02′ W, 24–26.ii.2008, J.S. Noyes col. NHM(E) 2010-21AQ, DNA: CRM 3.103.
Paratypes: same data as holotype, DNA: CRM 3.46 (1♀, AICF), CRM 3.100 (1♀, MZUCR).
Species-group placement: mymaripenne-group.
Distribution: Costa Rica.
DNA data: 28S: two sequences.
Etymology: Named for the distinctive funicle.
Description. Female: Antenna (
Figure 4d and
Figure 15c) five-segmented (excluding anellus), with pedicel slightly longer than funicle (12:8); clava two-segmented,
C1 1.5× as
C2, with ≥10
MT and 2
UST; one
SB at the apex of
C1 and
C2; apex of
C2 (
Figure 4g) also with 2 elongate
MPS, a long
SB and
UST.
Mid lobe of mesoscutum and scutellum without apparent sculpture; metanotum and propodeum medially short. Metasoma with a row of microspines on each segment.
T1 without cells (
Figure 4f). Ovipositor 1.1× as long as mesotibia. Mesotibia without spines basally. Metafemur without prominent spine. Fore wing (
Figure 5a) 9× as long as wide, maximum distal width 1.5× maximum basal width; disc with 10 setae irregularly arranged in 1–2 rows; fringe with longest seta 6× maximum discal width. Marginal vein with two long setae centrally, of equal length. Stigmal vein moderately enlarged, with three sensilla apically.
Body brown/yellow. Mesosoma largely pale, but mid lobe of mesoscutum brown anteriorly. Scape and pedicel pale, C1–C3 brown. Fore wing slightly infuscate basally.
Male: As female, but antenna (
Figure 4c) with
C1 approximately 2×
C2. Metasoma (
Figure 4e) with a row of microspines on each segment.
Material examined. Holotype ♀ (deposited in NHMUK). COSTA RICA: Puntarenas, Est. Biol. Monteverde, 10°19′ N 83°49′ W, 1540–1890 m, 26.ii.2007, J.S. Noyes BMNH(E) 2010-21, DNA: CRM2.4.
Paratypes: COSTA RICA: same data as holotype, DNA: CRM2.5 (1♀, AICF), CRM2.7, 2.8, 2.10, 2.11 (4♀, DACE, MZUCR, NHMUK); Limón, Hitoy-Cerere Reserve, 9°40′ N 83°02′ W, 100 m, 24–26.ii.2008, J.S. Noyes NHM(E) 2010-21AQ, DNA: CRM 3.23/B11, CRM 3.105, B9, E1 (2♀, 2♂, NHMUK); Cartago, 12.5 km S Turrialba, Rancho Naturalista, 1000 m, 9°50″ N 83°34″ W, 12–14.ii.2017, J.S. Noyes BMNH(E) 2017-39, DNA1683 (1♀, NHMUK); Heredia, La Selva Biol. Sta., 10°26′ N 84°01′ W, 75 m, 27–28.ii.2003, J. S. Noyes (1♂, DACE).
Non-types: ARGENTINA: Salta Prov. Orán, road to San Andres along Rio Blanca, 399 m, 23.09° S 63.37° W, 23.iii.2003, J. Munro 003-03-23-02 (1♀, UCRC). USA: Northampton, 7 km S Jackson, 23.ix–14.xi.1987, MT, Bald Cypress Swamp, BRC Hym Team (1♀, UCRC).
Species-group placement: polychaetum-group. The male antenna is very distinctive in the group, and M. giraulti male antenna agrees very well with several other species that definitely belong to the polychaetum-group (but without sequence data to back up this assertion).
Distribution: Argentina, Costa Rica, USA.
DNA data: CO1: four sequences; 28S: eight sequences (all Costa Rica).
Etymology: The species is named for A.A. Girault for his pioneering studies on the Trichogrammatidae.
Megaphragma hansoni Polaszek, Fusu, and Viggiani sp. nov. (
Figure 15d–f)
Description.
Female: Antenna (
Figure 15d) five-segmented (excluding anellus); funicle absent; hence clava three-segmented, with
C1 and
C2 almost fused;
C1 with 1–2
MT;
C2 with some
MT and 2
UST;
C3 with 2–3
MT, 2
MPS, and prominent
SB and
SS.
Mid lobe of mesoscutum (
Figure 15e) with longitudinal striate sculpture extending to scutellum; propodeum (
Figure 15e) with central area extended posteriorly, crenulae present;
T1 with elongate cells laterally, 2–3× as long as wide;
T2–
T4 without setae laterally;
T5 with long setae laterally. Ovipositor 2× as long as mesotibia. Mesotibia with one large spine basally; metafemur with spine. Fore wing (
Figure 15f) 7× as long as maximum width, maximum distal width equal to maximum basal width; disc with a single short seta; longest fringe seta 5× as long as maximum discal width. Marginal vein with one long seta centrally, extending almost to the end of the marginal vein; a minute additional seta next to it. Stigmal vein moderately enlarged, with four sensilla apically.
Body largely brown, the following paler: most of mesosoma except anterior half of mid lobe of mesoscutum, anterior half of T1 and antenna. Fore wing slightly infuscate basally; stigmal and marginal vein pale brown.
Male: Largely as in female. C1 and C2 with scattered SS; C2 with 2–3 MT, 1 apically; C3 with long apical and ventral UST.
Material examined. Holotype ♀ (deposited in NHMUK). COSTA RICA: Limón, Hitoy-Cerere Reserve, 9°40′ N 83°02′ W, 100 m, 24–26.ii.2008, J.S. Noyes NHM(E) 2010-21AQ, DNA: CRM 3.101 (but no associated DNA sequence).
Paratypes: COSTA RICA: same data as holotype except DNA: CRM3.4, 3.17/B5, 3.40/D4, 3.97, 3.99, 3.104 (CRM3.40 lost after DNA extraction) (1♀, 4♂, AICF, NHMUK); Puntarenas, Est. Biol. La Gamba, 8°42′ N 83°12′ W, 150 m, 13–14.ii.2008, J.S. Noyes BMNH(E) 2010-21AQ, DNA: CRM1.1, 1.2, 1.4, 1.7, 1.12-1.19 (12♂, DACE, MZUCR, NHMUK, UCRC).
Species-group placement: ghesquierei-group.
Distribution: Costa Rica.
DNA data: 28S: seven sequences.
Etymology: Named for our colleague and co-author on this paper, Professor Paul Hanson, University of Costa Rica, San José.
Megaphragma kinuthiae Polaszek, Fusu, and Viggiani sp. nov. (
Figure 5b–d)
Description. Female: Antenna (
Figure 5b) five-segmented (excluding anellus); pedicel as long as funicle; funicle 3× as long as wide;
C1 slightly longer or as long as
C2 with two dorsal
UST; three elongate
MPS extending beyond clava tip.
Figure 15.
Photographs of Megaphragma species: (a) M. funiculatum, female metasoma (Holotype); (b) M. funiculatum, female fore wing (Holotype); (c) M. giraulti, female antenna (Holotype); (d) M. hansoni, female antenna (Holotype); (e) M. hansoni, female dorsal meso- and metasoma (Holotype); (f) M. hansoni, female fore wing (Holotype). Scale bars 20 µm.
Figure 15.
Photographs of Megaphragma species: (a) M. funiculatum, female metasoma (Holotype); (b) M. funiculatum, female fore wing (Holotype); (c) M. giraulti, female antenna (Holotype); (d) M. hansoni, female antenna (Holotype); (e) M. hansoni, female dorsal meso- and metasoma (Holotype); (f) M. hansoni, female fore wing (Holotype). Scale bars 20 µm.
Head with toruli separated by about their own width. Mid lobe of mesoscutum smooth; propodeum with straight hind margin, without crenulae;
T1 non-reticulate;
T2–
T4 with short setae laterally. Mesotibia with two large spines basally; metafemur with spine; metatibia with a row of fine, blunt setae extending almost their entire inner length, increasing abruptly in length distally. Fore wing (
Figure 5d) about 9× as long as maximum width, maximum distal width 1.3× maximum basal width; discal setae arranged in two rows, each with 4–5 setae, longest fringe seta 10× as long as maximum discal width. Marginal vein with two long setae centrally. Stigmal vein with two sensilla apically. Ovipositor 1.1× as long as mesotibia.
Body entirely yellow, anterior mesosoma brown, posterior metasoma slightly darker. Wings hyaline.
Male: Unknown.
Material examined. Holotype ♀ (deposited in NHMUK). KENYA: Meru, vi. 1965, ID. No. 2851, CIE 233, BM 196. T. F. Crowe, ex tea leaves.
Paratypes: same data as holotype (6♀, NHMUK). All specimens are on the same slide; the holotype is circled in red.
Species-group placement: polychaetum-group. The species appears closest to M. giraulti based on morphology.
Distribution: Kenya.
Host: Not identified, but possibly Scirtothrips dorsalis (Hood), a species common on tea in Kenya.
DNA data: no DNA sequences.
Etymology: Named for our colleague and friend Dr Wanja Kinuthia, National Museums of Kenya, Nairobi.
Megaphragma liui Polaszek and Fusu sp. nov. (
Figure 16a–d)
Description. Female: Antenna (
Figure 16a) five-segmented (excluding anellus),
C1 and
C2 strongly overlapping;
C3 elongate, more than half the length of
C1 and
C2; 1 min
UST on
C1, two long
UST on
C2; three elongate
MPS extending beyond clava tip.
Head with toruli very close together, separated by about one-third their own width. Mid lobe of mesoscutum (
Figure 16b) with fine longitudinal striations, but also with distinct large reticulate cells; propodeum medially with strongly produced hind margin, with two crenulae (
Figure 16b).
T1 smooth centrally, but with 8–10 elongate cells laterally (
Figure 16b);
T2–
T4 with short setae laterally, lateral cells present. Mesotibia without large spines basally, but a robust spine present at the apex of mesofemur; metafemur with spine; metatibia with a group of fine, sharp setae on inner surface apically. Metacoxa and metafemur (
Figure 16c) with distinct longitudinal sculpture ventrally, contrasting with transverse sculpture dorsally. Fore wing (
Figure 16d) 7× as long as maximum width, maximum distal width 1× maximum basal width; disc distally pointed, without setae (but one wing with a possible indication of a minute seta); longest fringe seta 4× as long as maximum discal width. Marginal vein with two setae centrally, the proximal one very robust, about 1.5× as long as distal. Stigmal vein with one elongate sensillum apically. Ovipositor 1.9× as long as mesotibia.
Body entirely brown, mesosoma pale posteriorly, T1 with pale areas laterally. C1 very dark, pedicel paler than the remainder of the antenna. Fore wing strongly infuscate basally. Legs dark, tarsi pale.
Male: Unknown.
Material examined. Holotype ♀ (deposited in UCRC).
BRUNEI: Temburong Dist., Bukit Patoi trail, 41–290 m, 4°45′21″ N 115°10′30″ E, 4 July 2010, swp dipterocarp forest, J. Mottern M10-065, DNA1656.
Figure 16.
Photographs of Megaphragma species: (a) M. liui, female antenna; (b) M. liui, female dorsal meso- and metasoma (Holotype); (c) M. liui, female metacoxa and metafemur (Holotype); (d) M. liui, female fore wing (Holotype); (e) M. longiciliatum, female antenna (Holotype). Scale bars 20 µm except 50 µm for d.
Figure 16.
Photographs of Megaphragma species: (a) M. liui, female antenna; (b) M. liui, female dorsal meso- and metasoma (Holotype); (c) M. liui, female metacoxa and metafemur (Holotype); (d) M. liui, female fore wing (Holotype); (e) M. longiciliatum, female antenna (Holotype). Scale bars 20 µm except 50 µm for d.
Species-group placement:
ghesquierei-group. In the concatenated and partitioned analysis, this species is not included in the group (
Figure 2b), though in an unpartitioned analysis with a simple substitution model it is one of the most basal species of the
ghesquierei-group (
Figure 2a). It is also retrieved as part of the group in the tree based on the 28S sequences alone, where it is sister to
M. rivelloi but on a very long branch (
Supplementary Figure S3). Hence, morphology, and partly molecular analyses, indicate that our inclusion of the species is correct.
Distribution: Brunei.
DNA data: CO1: one sequence; 28S: one sequence.
Etymology: Named for our colleague and friend Prof. Shu-sheng Liu, Zhejiang University, Hangzhou, China.
Megaphragma momookherjeeae Polaszek and Fusu sp. nov. (
Figure 17a–c)
Description. Female: Antenna (
Figure 17a) five-segmented (excluding anellus); transverse funicle present, clava two-segmented;
C1 longer than
C2;
C1 with ≥5
MT, 2
UST, and with fine, longitudinal striation;
C2 with basal
SS, 2
UST, and ≥4
MPS.
Mid lobe of mesoscutum smooth; propodeum without distinct central area.
T1–
T4 largely smooth,
T2–
T4 with long setae laterally. Ovipositor exserted, exceptionally long for the genus, more than 3× as long as mesotibia (
Figure 17b). Mesotibia with a very robust spine basally, 0.4× tibial length; metafemur without spine; metatibia with a row of about 17 spines along almost the entire inner length, and 4 robust spines toward the apex of the outer surface. Fore wing (
Figure 17c) 9× as long as maximum width; maximum distal width 1× maximum basal width; disc with a single, minute seta; longest fringe seta 6.5× as long as maximum discal width. Marginal vein apparently with three long setae centrally, of equal length. Stigmal vein with three sensilla apically.
Body largely dark brown; scutellum, propodeum, and lateral mesosoma paler (
Figure 17b); antenna pale brown,
C2 darker. Fore wing basally infuscate.
Male: Unknown.
Material examined. Holotype ♀ (deposited in NHMUK). COSTA RICA: Cartago, 12.5 km S. Turrialba, Rancho Naturalista, 1000 m, 9°50″ N 83°34″ W, 12–14.ii.2017, J.S. Noyes BMNH(E) 2017-39, DNA1680.
Species-group placement: M. antecessor-group. Resembling also the ghesquierei-group in some features (e.g., fore wing with one seta), but clearly not clustering with that group in any molecular analyses. It is recovered as sister to M. antecessor, and the two clustered together as basal to all species-groups except the ghesquierei-group (partitioned analysis), or as the most basal species group of Megaphragma (unpartitioned analysis).
Distribution: Costa Rica.
DNA data: CO1: one sequence.
Etymology: Named for Mo Mookherjee, a friend of the first author (AP).
Megaphragma nowickii Polaszek, Fusu, and Viggiani sp. nov. (
Figure 7a–f and
Figure 17e–g)
Description. Female: Antenna (
Figure 7c and
Figure 17g) five-segmented (excluding anellus); pedicel twice as long as funicle; funicle twice as long as wide;
C1 slightly shorter or as long as
C2;
C1 with ≥7
MT; 2
UST; 3 elongate
MPS extending beyond clava apex.
Mid lobe of mesoscutum smooth, anteriorly with subpolygonal sculpture (cf
Figure 7b). Metanotum and propodeum narrow centrally, the latter without an extension or crenulae.
T1 sculpture (
Figure 7a and
Figure 17f) with cells converging centrally, lateral cells 3× as long as wide, each with 3–5 inward-pointing denticles;
T2–
T4 without long setae laterally, all with coarse reticulate sculpture becoming lateral distally. Ovipositor 1.7× as long as mesotibia. Mesotibia with two large spines basally; metafemur with spine; metatibiae with a row of fine, blunt setae extending almost their entire inner length, increasing abruptly in length distally. Fore wing (
Figure 7e) 9.5× as long as wide, maximum distal width 1.1× maximum basal width; disc with a single row of six setae; longest fringe seta 4.5× maximum discal width. Marginal vein with two long setae centrally.
Body entirely brown, metasoma slightly darker posteriorly. Pleural parts of mesosoma and hind legs except for tarsi lighter. Wings hyaline.
Male: Largely as in female. Antenna with
C1 slightly longer than
C2;
T1 (
Figure 7b) with an incomplete pattern of cells. Aedeagus as in
Figure 7f.
Material examined. Holotype ♀ (deposited in NHMUK). D. R. CONGO: Province Orientale, Yangambi Biosphere Reserve, 15.v.2012, N 0°48.837′ E 24°30.287′, screen sweep, primary forest, A. Polaszek col, BMNH(E) 2012-88, DNA: COM1.16.
Paratypes: D. R. CONGO: same data as holotype except DNA: COM1.1–1.15, 1.17–1.23, 1.26, 1.27 (2♀, 22♂, AICF, DACE, IITA, NHMUK). UGANDA: Mabira Forest, N0°23′22″ E33°00′22″, 1250 m, 1.iii.2015, A. Polaszek, screen sweep, DNA1132/F11 (1♀, NHMUK); Mabira Forest, N0°23′22″ E33°00′22″, 1250 m, 1.iii.2015 A. Polaszek, screen sweep, DNA1116–1118, 1120–1124 (1♀, 7♂, NHMUK).
Non-types: BENIN: Dept. Zou, Zogbodomey, Massi, 18.xii.1989, ex egg Megalurothrips sjostedti on Pueraria, M. Tamo col. 275 (1♂, 1♀, NHMUK, IITA).
Species-group placement: mymaripenne-group.
Distribution: Benin, D. R. Congo, Uganda.
Host: Megalurothrips sjostedti (Trybom).
DNA data: CO1: 25 sequences from D. R. Congo; 28S: 30 sequences from 2 countries: D. R. Congo (21), Uganda (9).
Etymology: Named for S. Nowicki for his outstanding contribution to the knowledge of the Trichogrammatidae.
Megaphragma noyesi Polaszek and Fusu sp. nov. (
Figure 18a)
Description.
Female: Antenna (
Figure 18a) five-segmented (excluding anellus); pedicel almost twice as long as funicle; funicle slightly longer than wide;
C1 shorter than
C2;
C1 with two dorsal
UST;
C2 with three elongate
MPS extending beyond clava apex.
Mid lobe of mesoscutum and scutellum smooth. Metanotum and propodeum narrow centrally, the latter without an extension or crenulae.
Mesotibia without large spines basally; metafemur without spine; metatibia with a row of robust spines extending along the inner surface of distal half.
Fore wing 9× as long as wide, maximum distal width 1.3× maximum basal width; disc with a single row of six setae, and longest fringe seta 5× maximum discal width. Marginal vein with two long setae centrally, subequal in length (cf
Figure 18g).
T1 sculpture with cells converging centrally, six lateral cells present, 2–3× as long as wide, each with 1–3 inward-pointing denticles;
T2–
T4 with lateral cells indicated, with lateral setae not detected (cf
Figure 18f). Ovipositor as long as mesotibia. Body largely brown, mesosoma largely pale.
T1 very dark brown in contrast to rest of body. Legs pale. Wings hyaline.
Male: Largely as in female. Antenna with C1 longer than C2; T1 with an incomplete pattern of cells. Genitalia tubular as in other species, 3× as long as wide.
Material examined. Holotype ♀ (deposited in NHMUK). UK: England, Surrey, Coulsdon Common, Happy Valley, 51°17′ N 0°07′ W, 168 m, viii.2013, J.S. Noyes BMNH(E) 2013-, DNA: UKM14.
Paratypes:
UK: same data as holotype except DNA: UKM8–13 (UKM9 has DNA sequences but was lost during extraction) (4♀, 1♂, AICF, DACE, NHMUK); East Sussex, Brede High Wood, TQ79432018/19, 30.viii.2019–20.ix.2019, D. Binns col., DNA1612–1619 (1612, 1614 and 1618 have DNA sequences but were lost during extraction) (10♀, 3♂, NHMUK, UCRC).
HUNGARY: Őrség Nemzeti Park, Barkás Lake, 46°52′ N 16°26′ E, 268 m, 28.vi.2010, J.S. Noyes screen sweeping, BMNH(Ent) 2010-63, DNA: HUM2 (1♀, NHMUK) (HUM3 has DNA sequences but was lost during extraction), HUM5 (1♀, AICF); 4–5 km SW Kőszeg, Meszes Völgy, 47°22′ N 16°31′ E, 431 m, 26.vi.2010, screen sweeping, J.S. Noyes BMNH(Ent) 2010-63, DNA: HUM6 (1♂, AICF); Őrség National Park, Lugosi Valley, 46°54′ N 16°27′ E, 231 m, 28.vi.2010, J.S. Noyes, DNA: HUM 8–14, BMNH(Ent) 2010-63 (3♀ 4♂, NHMUK).
Figure 17.
Photographs of Megaphragma species: (a) M. momookherjeeae, female antenna (Holotype); (b) M. momookherjeeae, female lateral meso-and metasoma (Holotype); (c) M. momookherjeeae, female fore wing (Holotype); (d) M. mymaripenne, female antenna (Holotype); (e) M. nowickii, female dorsal meso- and metasoma (Holotype); (f) M. nowickii, female dorsal base of metasoma (Holotype); (g) M. nowickii, female antenna (Holotype). Scale bars 20 µm except 100 µm for c.
Figure 17.
Photographs of Megaphragma species: (a) M. momookherjeeae, female antenna (Holotype); (b) M. momookherjeeae, female lateral meso-and metasoma (Holotype); (c) M. momookherjeeae, female fore wing (Holotype); (d) M. mymaripenne, female antenna (Holotype); (e) M. nowickii, female dorsal meso- and metasoma (Holotype); (f) M. nowickii, female dorsal base of metasoma (Holotype); (g) M. nowickii, female antenna (Holotype). Scale bars 20 µm except 100 µm for c.
Non-type: CZECH REPUBLIC: Moravia, Vranov, River Dyje, ss riparian forest, 13.viii.1991, L. Masner (1♀, UCRC).
Species-group placement: mymaripenne-group.
Distribution: UK (England); Czech Republic, Hungary.
DNA data: CO1: 21 sequences from 2 countries; 28S: 23 sequences from 2 countries: Hungary, UK.
Etymology: Named for Dr John Noyes of the Natural History Museum, London, for his outstanding contribution to our knowledge of Chalcidoidea. As a chalcid collector, John is unmatched so far, and a major proportion of the material in this study was collected by him.
Megaphragma pintoi Viggiani sp. nov. (
Figure 18b–d)
Description.
Female: Antenna (
Figure 18b) five-segmented (excluding anellus); pedicel slightly longer than funicle; funicle 3× as long as wide;
C1 slightly longer or as long as
C2;
C1 with two
UST; three
MPS extending beyond
C2 apex.
Mid lobe of mesoscutum (
Figure 18c) smooth, with 5–6 deep striae anteriorly; propodeum with straight hind margin, without crenulae.
T1 non-reticulate;
T2–
T4 with short setae laterally. Ovipositor 1.1× as long as the mesotibia. Metafemur without spine but a prominent seta present. Fore wing (
Figure 18d) 10× as long as maximum width, and maximum distal width 1.4× maximum basal width; discal setae arranged in 2–3 rows, each with 3–5 setae, longest fringe seta 5.5× as long as maximum discal width. Marginal vein with two long setae centrally. Stigmal vein with two sensilla apically.
Body entirely pale brown (eyes deep purple); legs and antenna pale. Wings hyaline.
Male: Unknown.
Material examined. Holotype ♀ (deposited in PFRNZ). NEW ZEALAND: Auckland, Mt Albert, Science Center, 17.v.1997, ex Heliothrips haemorrhoidalis ex acmena leaves, P. Stevens.
Non-type: COLOMBIA: Supata, 5°04′07″ N 74°16′48″ W, 1800 m, 31.12.2018, sweep, A.A. Polilov col. (1♀, AICF).
Species-group placement: polychaetum-group.
Distribution: Colombia, New Zealand.
DNA data: no DNA sequences.
Etymology: Named for Emeritus Professor John Pinto, formerly of University of California, Riverside, in recognition of his monumental contribution to our understanding of Trichogrammatidae.
Comments: Known so far from only two specimens to date; this species has an apparently extraordinary distribution, being known from Colombia and New Zealand. It seems very likely that it will turn up elsewhere and is probably another cosmopolitan Megaphragma species.
Megaphragma mymaripenne: Viggiani and Bernardo, 1998. Boll. Zool. agr. Bach. Ser. II 29: 51–55; Bernardo and Viggiani, 2003. Boll. Lab. Entomol. agr. Filippo Silvestri 58 [2002]: 77–85; Polilov, 2012. Arthropod Struct. Dev. 41(1): 29–34; Makarova et al., 2015. Arthropod Struct. Dev. 44(1): 21–32; Polilov, 2016. At the Size Limit—Effects of Miniaturization in Insects; Polilov, 2017. PLoS ONE 12(5): e0175566; Diakova et al., 2018. PeerJ 6: e6005 (misidentifications).
Description.
Female: Antenna (
Figure 18e and
Figure 20a) five-segmented (excluding anellus); pedicel almost twice as long as funicle; funicle slightly longer than wide, with 3
MT and 1 ASC;
C1 shorter than
C2;
C1 with 17
MT,
SS, and 2
UST;
C2 with
SB, 2
MT,
SS, and 4 elongate
MPS extending beyond clava apex.
Mid lobe of mesoscutum and scutellum smooth (
Figure 18f and
Figure 19d). Propodeum with a distinct central area with lateral boundaries in line with those of mesoscutum and scutellum (
Figure 19d); two lateral lobes present behind propodeum central area (
Figure 19d); propodeum without crenulae.
T1 sculpture (
Figure 18f and
Figure 20b) with cells converging centrally, about six lateral cells, 2× as long as wide, mesal cells each with 2–3 inward-pointing denticles.
T2–
T4 with 2–3 lateral cells, with short setae laterally. Ovipositor 1.3× as long as mesotibia. Mesotibia with two large spines basally; metafemur without spine; metatibia with a row of fine, blunt setae extending almost its entire inner length, increasing abruptly in length distally. Fore wing (
Figure 18g) 9.5× as long as wide, maximum distal width 1.4× maximum basal width; disc with a single row of six setae, longest seta of fringe 5.8× maximum disc width. Marginal vein with two long setae centrally.
Antenna (
Figure 18e) with radicle brown, very dark compared to the remainder of the antenna; remainder of body largely brown, mesosoma largely pale, mid lobe of mesoscutum brown anteriorly, an indistinct brown spot on the scutellum. Legs pale. Wings hyaline.
Male: Largely as in female. Antenna with C1 slightly longer than C2; T1 with an incomplete pattern of cells. Genitalia tubular as in other species, 3× as long as wide.
Material examined. Holotype ♀ (deposited in DACE). ITALY: Vietri sul Mare, Benincasa, 40°40′ N 14°44′ E, 17.vii.2013, G. Viggiani ex Heliothrips haemorrhoidalis on Viburnum tinus, DNA: ITM9.
Paratypes: ITALY: same data as holotype, DNA: ITM8, 11, 12, 14 (4♀, AICF, DACE, NHMUK).
Species group placement: mymaripenne-group.
Distribution: Italy.
Host:
Heliothrips haemorrhoidalis. Males are very rare, and reproduction is normally thelytokous [
14] (as
M. mymaripenne).
DNA data: CO1: five sequences; 28S: five sequences; all from Italy.
Etymology: Named for our colleague Alexey Polilov, co-author of this paper, for his outstanding contribution to our knowledge of the Trichogrammatidae and miniaturization in insects.
Comments: Found in Italy at the same locality and on the same host as M. viggianii.
Description.
Female: Antenna (
Figure 9a and
Figure 21a) five-segmented (excluding anellus), pedicel approximately two-thirds the length of
C1 and
C2;
C3 one-quarter shorter than
C2;
C2 with a single
UST;
C3 with
SB, two
MPS,
MT, and apical
SB.
Mid lobe of mesoscutum and scutellum with longitudinally striate sculpture; propodeum (
Figure 9b) with a large subtriangular and crenulated central area. Metasomal tergites without reticulation but with transverse striations, laterally with a row of 2–7 microspines. Ovipositor approximately 1.3× as long as mesotibia. Fore wing (
Figure 9c) 6–7× as long as wide; disc without setae; longest fringe seta 4–4.5× as long as the maximum discal width.
Body yellow with brown mostly on mesosoma; fore wing infuscate behind the venation.
Male: Largely as in female except for genitalia.
Material examined. Holotype ♀ (deposited in DACE):
ITALY: Basilicata, Rivello (PZ), vii.2002, yellow sticky traps in a vineyard.
Figure 18.
Photographs of Megaphragma species: (a) M. noyesi, female antenna (Holotype); (b) M. pintoi, female antenna (Holotype); (c) M. pintoi, female mesoscutum (Holotype); (d) M. pintoi, female fore wing (Holotype); (e) M. polilovi, female antenna (Holotype); (f) M. polilovi, female dorsal mesosoma (part) and metasoma (Holotype); (g) M. polilovi, female fore wing (Holotype). Scale bars 20 µm except 50 µm for a, d, and g.
Figure 18.
Photographs of Megaphragma species: (a) M. noyesi, female antenna (Holotype); (b) M. pintoi, female antenna (Holotype); (c) M. pintoi, female mesoscutum (Holotype); (d) M. pintoi, female fore wing (Holotype); (e) M. polilovi, female antenna (Holotype); (f) M. polilovi, female dorsal mesosoma (part) and metasoma (Holotype); (g) M. polilovi, female fore wing (Holotype). Scale bars 20 µm except 50 µm for a, d, and g.
Figure 19.
SEM micrographs of Megaphragma species (non-types): (a) M. caribea, male habitus; (b) M. caribea, male head and antennae; (c) M. caribea, male mesoscutum; (d) M. polilovi, female dorsal habitus. Scale bars 50 µm for a, 20 µm for b and c, and 100 µm for d.
Figure 19.
SEM micrographs of Megaphragma species (non-types): (a) M. caribea, male habitus; (b) M. caribea, male head and antennae; (c) M. caribea, male mesoscutum; (d) M. polilovi, female dorsal habitus. Scale bars 50 µm for a, 20 µm for b and c, and 100 µm for d.
Figure 20.
SEM micrographs of Megaphragma species (non-types): (a) M. polilovi, female antenna; (b) M. polilovi, female metasoma; (c) M. viggianii, female habitus; (d) M. viggianii, female antenna. Scale bars 20 µm for a, b and d, and 100 µm for c.
Figure 20.
SEM micrographs of Megaphragma species (non-types): (a) M. polilovi, female antenna; (b) M. polilovi, female metasoma; (c) M. viggianii, female habitus; (d) M. viggianii, female antenna. Scale bars 20 µm for a, b and d, and 100 µm for c.
Paratypes: ITALY: same data as holotype (19♀, DACE, NHMUK). INDIA: UP, New Delhi, IARI; 220 m 28°37′51″ N, 77°09′50″ E, xi.5.2003, pan trap, J. Heraty col. H03-106 (1♀, UCRC). INDONESIA: W Java, Gunung Halimun NP, Tea-Forest Junction, 1066 m, 6°41′07″ S 106°31′16″ E, screen-sweep, 17.ix.2015, A. Polaszek, DNA1148 (1♀, NHMUK). JAPAN: Tokyo area, ?1984, Takagi col., ex Scirtothrips dorsalis on tea, A. Loomans leg. (5♀, AICF, NHMUK). UK: Surrey, Coulsdon Common, 25.viii.2002, J.S. Noyes, screen sweep (1♂, NHMUK). VIETNAM: Cat Tien NP, sweeping, N11°24′45″ E107°25′23″, 21.xi.2018, leg. A.A. Polilov, DNA1687 (1♀, AICF); Cat Tien NP, sweeping, N11°24′45″ E107°25′23″, 25.xi.2016, leg. A.A. Polilov (3♀, 2♂, AICF, UCRC).
Non-type: CHINA: 1♂ misidentified as M. deflectum, Wuyishan, Fujian, 19.x.1987 Wang Jiashe col. (FAU).
Species group placement: ghesquierei-group.
Distribution: China, India, Indonesia, Italy, Japan, UK, Vietnam.
Host: Scirtothrips dorsalis Hood.
DNA data: 28S: 1 sequence (Vietnam).
Etymology: Named for the ancient village of Rivello in Italy, where this extremely widespread species was first discovered. As noted above under M. deflectum, one paratype (male “allotype”) of that species is in fact M. rivelloi.
Megaphragma tamoi Polaszek, Fusu, and Viggiani sp. nov. (
Figure 21d,e)
Description.
Female: Antenna (
Figure 21d) five-segmented (excluding anellus); funicle absent; hence, clava three-segmented, with
C1 and
C2 almost fused;
C1 without
UST;
C2 with one elongate
UST, reaching more than half the length of
C3;
C3 with
MPS,
SB, and
SS.
Mid lobe of mesoscutum with longitudinal striate sculpture extending to scutellum (cf
Figure 14b); propodeum with central area extended posteriorly, crenulae present (cf
Figure 15e).
T2–
T4 without setae laterally. Ovipositor 1.7× as long as mesotibia. Mesotibia with one large spine basally; metafemur with spine (cf
Figure 16c, upper). Fore wing 6× as long as the maximum width, maximum distal width 1.2× maximum basal width; disc with a single long seta (
Figure 21e), longest fringe seta 3× maximum discal width. Marginal vein with two setae centrally, equal in length, extending to the end of the marginal vein. Stigmal vein moderately enlarged, with two sensilla apically.
Largely brown, the following paler: legs except coxae and metafemur. Pedicel pale; scape, C1–C3 darker. Fore wing strongly infuscate basally; stigmal and marginal vein brown; marginal vein very dark centrally.
Male: Largely as in female. C1 and C2 with scattered SS; C2 with 2–3 MT apically; C3 with long apical and ventral UST. C3 is darker than preceding segments.
Material examined. Holotype ♀ (deposited NHMUK). BENIN: Agbotagon, 6°49′ N 2°12′ E, 22.iv.1990, M. Tamo 351.
Paratypes: BENIN: same data as holotype (8♀, DACE, IITA); Mono Province, 7.x.1988, M. Tamo, D-Vac on cowpea Vigna unguiculata (1♀, NHMUK); Mono Province, 25.ii.1988, M. Tamo, emergence cage Megalurothrips sjostedti on Vigna unguiculata (1♀, IITA); Dept Zou, Zogbodomey, 22.i.1990, ex egg Megalurothrips sjostedti, M. Tamo col. 275, 342 (2♀, IITA); Cotonou, IITA Station, 16.i.1990, ex egg Megalurothrips sjostedti on Pueraria, M. Tamo col. 336 (1♀, NHMUK).
Species group placement: ghesquierei-group.
Distribution: Benin.
Host: Megalurothrips sjostedti.
DNA data: no DNA sequences.
Etymology: Named for our colleague and friend Manu Tamo (IITA, Benin), collector of many
Megaphragma specimens.
Figure 21.
Photographs of Megaphragma species: (a) M. rivelloi, female antenna (Holotype); (b) M. striatum, female antenna (MXM1 above and Paratype below); (c) M. striatum, female metasoma (MXM1); (d) M. tamoi, female antenna (Holotype); (e) M. tamoi, female fore wing (Holotype); (f) M. tridens, female antenna (Holotype); (g) M. tridens, male antenna (Paratype); (h) M. tridens, female metasoma (Holotype); (i) M. uniclavum, female antenna (Holotype); (j) M. uniclavum, female metatibia (Holotype); (k) M. vanlentereni, female antenna (Holotype); (l) M. vanlentereni, female mesosoma (Holotype); (m) M. vanlentereni, female fore wing (Holotype). Scale bars 20 µm.
Figure 21.
Photographs of Megaphragma species: (a) M. rivelloi, female antenna (Holotype); (b) M. striatum, female antenna (MXM1 above and Paratype below); (c) M. striatum, female metasoma (MXM1); (d) M. tamoi, female antenna (Holotype); (e) M. tamoi, female fore wing (Holotype); (f) M. tridens, female antenna (Holotype); (g) M. tridens, male antenna (Paratype); (h) M. tridens, female metasoma (Holotype); (i) M. uniclavum, female antenna (Holotype); (j) M. uniclavum, female metatibia (Holotype); (k) M. vanlentereni, female antenna (Holotype); (l) M. vanlentereni, female mesosoma (Holotype); (m) M. vanlentereni, female fore wing (Holotype). Scale bars 20 µm.
Megaphragma tridens Fusu and Polaszek sp. nov. (
Figure 21f–h)
Description.
Female: Antenna (
Figure 21f) five-segmented (excluding anellus); funicle absent; hence, clava three-segmented, with
C1 and
C2 almost fused;
C1 apparently without
UST;
C2 with 1 prominent
UST, abundant
MT, and 1 apical
MPS;
C3 with
MT, 2-3
UST,
SB, and
SS.
Mid lobe of mesoscutum with longitudinal striate sculpture (cf
Figure 14b); propodeum with central area extended posteriorly, crenulae absent (cf
Figure 21l).
T1 with elongate cells laterally, 2–3× as long as wide (cf
Figure 12b);
T2–
T4 with a short, robust seta near lateral margin;
T5 centrally with subparallel striations and with long setae laterally (
Figure 21h). Ovipositor 1.7× as long as the mesotibia. Mesotibia with one large spine basally; metafemur with spine. Fore wing 8.5× as long as the maximum width, maximum distal width equal to maximum basal width; disc with a single long seta (cf
Figure 12c), longest fringe seta 3.5× as long as maximum discal width. Marginal vein with one long seta centrally, extending to the end of the marginal vein. Stigmal vein moderately enlarged, with four sensilla apically.
Body largely brown, the following paler: legs except coxae and metafemur. Pedicel pale; scape, C1–C3 darker. Fore wing slightly infuscate basally; stigmal and marginal vein brown.
Male: Largely as in female. Antennal clava (
Figure 21g) with
C1 and
C2 with scattered
SS;
C2 with 2-3
MT apically;
C3 with long apical and ventral
UST.
Material examined. Holotype ♀ (deposited in NHMUK). COSTA RICA: Puntarenas, La Gamba Biol. Sta., 150 m, 8°42′ N 83°12′ W, 13–14.ii.2008, J.S. Noyes BMNH(E) 2010-21AQ, DNA: CRM1.10.
Paratypes: COSTA RICA: same data as holotype, DNA: CRM1.5, 1.8 (1♀, 1♂, AICF, MZUCR); Limón, Hitoy-Cerere Reserve, 9°40′ N 83°02′ W, 100 m, 24–26.ii.2008, J.S. Noyes NHM(E) 2010-21AQ, DNA: CRM 3.41, 3.102 (1♀ 1♂, AICF, NHMUK).
Species-group placement: ghesquierei-group.
Distribution: Costa Rica.
DNA data: no DNA sequences.
Etymology: From the Latin word tridens, in reference to the three apical sensilla on C3, resembling a longer central and two shorter lateral teeth of a trident (from Latin tri=three and dens=tooth). Noun in apposition.
Megaphragma uniclavum Polaszek and Fusu sp. nov. (
Figure 21i,j)
Description.
Female: Antenna (
Figure 21i) four-segmented (excluding anellus); transverse funicle present; clava one-segmented (unique so far for the genus, though
antecessor approaches this condition); 2
UST; 3–4 long
MPS extending beyond half the clava.
Head with few features discernible due to mounting position, and extensively obscured by eye pigment. Mid lobe of mesoscutum with fine longitudinal striation (cf
Figure 14b); vertical/ventral anterior mid lobe of mesoscutum with coarse, reticulate sculpture (cf
Figure 3g); propodeum elongate centrally, longitudinally striate with three large crenulae.
T2–
T4 with very long setae laterally, each longer than its tergum;
T3 and
T4 with large irregular cells laterally. Mesotibia with two large spines basally; metafemur with spine; metatibia with a row of fine, elongate setae extending almost their entire inner length (
Figure 21j). Fore wing 5× as long as maximum width; longest fringe seta 6.7× as long as maximum discal width, maximum distal width versus maximum basal width is unclear; disc (cf
Figure 11b) with single short seta. Other details of disc and venation are unclear. Stigmal vein with a row of three sensilla apically. Ovipositor 2× as long as the mesotibia.
Body uniformly pale. Distal metasoma darker. Wings hyaline.
Male: Unknown.
Material examined. Holotype ♀ (deposited in NHMUK). COSTA RICA: Heredia, La Selva BS., 10°26′ N 84°01′ W, 75 m, 28–29.ii.2008, J.S. Noyes BMNH(E) 2010-21 AQ.
Species-group placement: antecessor-group. Megaphragma uniclavum is so far unique for the genus having a single claval segment. Despite the very unusual fore-wing structure, it is clearly affiliated with M. antecessor (similar mesoscutal sculpture and row of setae on metatibia).
Distribution: Costa Rica.
DNA data: no DNA sequences.
Etymology: This species is so far the only Megaphragma known with a single claval segment; hence, the name uniclavum. Noun in apposition.
Megaphragma vanlentereni Polaszek and Fusu sp. nov. (
Figure 21k–m)
Description.
Female: Antenna (
Figure 21k) five-segmented (excluding anellus); funicle absent; hence, clava three-segmented, with
C1 and
C2 almost fused;
C1 with 2
UST;
C3 with three
MPS extending beyond apex of clava.
Mid lobe of mesoscutum (
Figure 21l) with irregular longitudinal striate sculpture; propodeum (
Figure 21l) elongate, curved centrally and posteriorly; crenulae present.
T1 without cells;
T2–
T4 without setae laterally. Ovipositor 1.7× as long as mesotibia. Mesotibia with two large spines basally. Fore wing (
Figure 21m)6.5× as long as maximum width, maximum distal width 1.1× maximum basal width; disc with one short seta, longest fringe seta 4× as long as maximum discal width. Marginal vein with two long setae centrally, of equal length. Stigmal vein moderately enlarged, with three sensilla apically.
Body largely pale with dorsal mesosoma, including propodeum, pale brown. Scape and pedicel pale, C1 and C2 brown, C3 very dark brown in contrast. Fore wing infuscate basally; stigmal and marginal vein distally brown.
Male: Unknown.
Material examined: Holotype ♀ (deposited in NHMUK). MALAYSIA, Sabah, Maliau Basin Studies Centre, Knowledge Trail, 04°44′ N 116°58′ E, 22.ix.2012, A. Polaszek, screen-sweep, NHM(E) 2010-21, DNA: SAM3.
Species group placement: ghesquierei-group.
Distribution: Malaysia (Borneo, Sabah).
DNA data: CO1: one sequence; 28S: one sequence (both Malaysia, Sabah).
Etymology: Named for our colleague and friend Joop van Lenteren, a pioneer of biocontrol, especially in greenhouses.
Megaphragma viggianii Fusu, Polaszek, and Polilov sp. nov. (
Figure 20c,d and
Figure 22a–d)
Megaphragma amalphitanum: Nedoluzhko et al. 2016. Mitochondrial DNA Part A 27(6): 4526–4527; Polilov, 2016. At the Size Limit—Effects of Miniaturization in Insects; Nedoluzhko et al., 2017. Genom. Data 11: 87–88; Polilov, 2017. PLoS ONE 12(5): e0175566; Prokhortchouk et al., 2017. Mosc. Univ. Biol. Sci. Bull. 72(1): 30–32; Diakova et al., 2018. PeerJ 6: e6005; Sharko et al., 2019. PLoS ONE 14(12): e0226485; Polilov et al., 2021. Sci. Rep. 11(1): 4717; Boudinot et al., 2020. J. Zool. 313(2): 99–113; Diakova and Polilov, 2021. J. Hymenopt. Res. 84: 69–73 (misidentifications).
Description.
Female: Antenna (
Figure 20d and
Figure 22a) five-segmented (excluding anellus); pedicel 2× as long as funicle; funicle slightly longer than wide;
C2 longer than
C1;
C1 with 1 ASC, 2
MT;
C2 with 13
MT, 2
UST;
C3 with 2
MPS, 1
SS, and 3
UST extending beyond clava tip.
Mid lobe of mesoscutum (
Figure 22b) posteriorly smooth, anteriorly with large, coarse reticulation; propodeum with broad, truncate hind margin, with two widely separated lobes distally and laterally, without crenulae.
T1 with a central “V” composed of minute denticles, a row of coarser denticles laterally (
Figure 20c), and one elongate cell laterally, about 2× as long as wide. Metasoma dorsally with rows of denticles laterally on
T2 and
T3;
T2–
T4 with moderately long setae laterally. Ovipositor as long as the mesotibia.
Mesotibia without spines basally, a single seta present; metafemur without spine but with three robust setae; metatibia with a row of five or six fine setae extending along half its inner length. Fore wing (
Figure 22c) 6× as long as maximum width; maximum distal width 1.5× maximum basal width and more than 2× width measured at the apex of the marginal vein; longest fringe seta 4× as long as the maximum discal width; disc with setae in one or two rows of 4–6 setae, and setae on ventral surface long, the penultimate one reaching to the base of the distal (
Figure 22d). Marginal vein with two setae centrally, the proximal approximately 2× the length of the distal. Stigmal vein has four sensilla apically.
Head and metasoma dark brown; central/posterior mesoscutal mid lobe and lateral scutellum paler. Remainder of body, including legs, pale. Flagellum darker than remainder; wings hyaline.
Male: Antenna with pedicel almost as long as scape; funicle with two small setae; clava two-segmented; C1 >2× C2; C1 with three setae; C2 with three flagelliform setae; two multiporous plate sensilla and a short terminal process present. Metasoma dorsally with rows of denticles laterally on T1–T3. One individual with funicle apparently fused with C1.
Material examined: Holotype ♀ (deposited in DACE). ITALY: Naples, Massa Lubrense, S. Agata sui Due Golfi, 24.x.2012, 40°36′ N 14°20′ E, 114 m, ex Heliothrips haemorrhoidalis in leaves of Viburnum tinus (G. Viggiani), DNA: ITM1.
Paratypes: ITALY: same data as holotype except DNA: ITM2 (1♂), ITM3 (1♂), ITM4 (1♂), ITM5 (1♂), ITM6 (1♀), ITM7 (1♀) (AICF, NHMUK); same data but without DNA extraction codes (2♀ NHMUK); Vietri sul mare, Benincasa, 40°40′ N 44°20′ E, 17.vii.2013, ex Heliothrips haemorrhoidalis on Viburnum tinus (G. Viggiani), DNA: ITM13 (1♀, NHMUK); Liguria, Santa Margherita, iv.2015, A. Polilov, ex Heliothrips haemorrhoidalis on Viburnum tinus, DNA1095, DNA1096 (2♀, NHMUK). GREECE: Koutsoupia, 39.81N 22.80E, 22.viii.2016, Leg. A.A. Polilov, ex eggs Heliothrips haemorrhoidalis on Arbutus sp. (2♀, 1♂, AICF).
Species-group placement: longiciliatum-group.
Distribution: Greece, Italy.
Host: Heliothrips haemorrhoidalis.
DNA data: CO1: eight sequences; 28S: eight sequences.
Etymology: Named for Professor Gennaro Viggiani, specialist of microhymenoptera and biocontrol, and instigator and co-author of this paper.
Comments: Found in Italy at the same locality and on the same host as
M. polilovi. According to the CO1 sequence of the mitochondrial genome deposited on GenBank by Nedoluzhko et al. [
23], their species is not
M.
amalphitanum (=
M. longiciliatum) but
M. viggianii. Our molecular analysis retrieves the two as sister species. Both have the mid lobe of the mesoscutum reticulate anteriorly, and long lateral setae on
T2–
T4, though these are longer in the former. In
M. longiciliatum T2–
T4 have very long setae, at least as long as the tergite (
Figure 5h), whereas in
M. viggianii T2–
T4 have shorter setae, much shorter than the tergite (cf
Figure 8f).
Figure 22.
Photographs of Megaphragma species: (a) M. viggianii, female antenna (Holotype); (b) M. viggianii, female mesosoma (Holotype); (c) M. viggianii, female fore wing (Holotype); (d) M. viggianii, female ventral (above) and dorsal (below) wing surface, (Holotype); (e) M. priesneri, female ventral (above) and dorsal (below) wing surface (Neotype). Arrows point at the penultimate seta on ventral surface. Scale bars 20 µm.
Figure 22.
Photographs of Megaphragma species: (a) M. viggianii, female antenna (Holotype); (b) M. viggianii, female mesosoma (Holotype); (c) M. viggianii, female fore wing (Holotype); (d) M. viggianii, female ventral (above) and dorsal (below) wing surface, (Holotype); (e) M. priesneri, female ventral (above) and dorsal (below) wing surface (Neotype). Arrows point at the penultimate seta on ventral surface. Scale bars 20 µm.
Key to species of Megaphragma| 1 | Fore wing with a single discal seta (Figure 9h and Figure 10c) or without any discal setae (Figure 3f and Figure 4a)……………………………2 |
| - | Fore wing with at least one line of 3 or more setae (Figure 3c and Figure 5d)…………………………………………………………………………17 |
| 2 | Fore wing without discal seta (Figure 3f)…………….………………………………….……………………………………………………………………3 |
| - | Fore wing with one discal seta (Figure 9h)……………………………………….…….……………………………………………………………………6 |
| 3 | Mid lobe of mesoscutum entirely with regular longitudinally striate sculpture (Figure 3d)…………………………………………………………4 |
| - | Mid lobe of mesoscutum with reticulate sculpture anteriorly (Figure 3g), or with irregular longitudinal sculpture (Figure 13b)………………5 |
| 4 | Female: ovipositor more than 1.5× as long as mesotibia. Male: C3 with terminal sensillum less than 2× length C3……………………deflectum |
| - | Female: ovipositor less than 1.5× as long as mesotibia. Male: C3 with terminal sensillum more than 2× length C3…………………………rivelloi |
| 5 | Mid lobe of mesoscutum with reticulate sculpture anteriorly (Figure 3g)…………………………………………………………………ghesquierei |
| - | Mid lobe of mesoscutum with irregular longitudinal sculpture (Figure 13b)…………………………………………………………………chienleei |
| 6 | Fore wing with discal seta short, ≤distance between 2 proximal wing fringe setae (Figure 11b)……………………………………………………7 |
| - | Fore wing with discal seta long, ≥distance between 3 proximal wing fringe setae (Figure 10c)……………………………………………………12 |
| 7 | Female: ovipositor strongly exserted, more than 3× mesotibia (Figure 17b)…………………………………………………………momookherjeeae |
| - | Ovipositor slightly or not exserted, less than 2.5× mesotibia……………………………………………………………………………………………8 |
| 8 | Clava 1-segmented (Figure 21i). Marginal vein with both central setae minute……………………………………………………………uniclavum |
| - | Clava 2- or 3-segmented (if 2-segmented, C1 partially fused with C2). Marginal vein with at least one of the central setae elongate…………9 |
| 9 | Marginal vein with 2 central setae, the distal one minute, less than 0.2× as long as the proximal seta (Figure 15f)………………………hansoni |
| - | Marginal vein with 2 central setae, either subequal in length (Figure 11b), or the distal one about 0.5× as long as the proximal seta (Figure 16d).……………………………………………………………………………………………………………………………………………………………10 |
| 10 | Funicle present, transverse (Figure 11a); clava 2-segmented…………………………………………………………………………………antecessor |
| - | Funicle absent; clava 3-segmented…………………………………………………………………………………………………………………………11 |
| 11 | Mid lobe of mesoscutum entirely with evident regular longitudinally striate sculpture while reticulate cells hardly visible (Figure 21l)…………………………………………………………………………………………………………………………………………………vanlentereni |
| - | Mid lobe of mesoscutum entirely with fine regular longitudinally striate sculpture, surface divided into obvious reticulate cells (Figure 16b)……………………………………………………………………………………………………………………………………………………………liui |
| 12 | Marginal vein with one central seta (Figure 12c)…………………………………………………………………………………………………………13 |
| - | Marginal vein with two central setae………………………………………………………………………………………………………………………15 |
| 13 | T2–T4 of metasoma each with a robust seta near lateral margin; T5 centrally with subparallel striations (Figure 21h). Male: C3 elongate, length more than 3× basal width, with very long sensilla (Figure 21g)…………………………………………………………………………………………tridens |
| - | T2–T4 of metasoma without a seta near lateral margin; T5 centrally with reticulate sculpture (Figure 12b and Figure 21c). Male: C3 elongate or short, with less prominent sensilla (Figure 10b)……………………………………………………………………………………………………………14 |
| 14 | T2–T5 each with rows of prominent denticles laterally; T3 and T4 centrally with reticulate sculpture (Figure 21c). Female: C3 long (Figure 21b). Male: C3 long, length more than 2× basal width (Figure 10b)…………………………………………………………………………………striatum |
| - | T2–T5 with denticles reduced to faint protrusions, especially on T3 and T4; T3 and T4 centrally with sculpture consisting of faint striae (Figure 12b). Female: C3 short (Figure 12a). Male: C3 short, length less than 2× basal width……………………………………………………………breviclavum |
| 15 | Clava 2-segmented………………………………………………………………………………………………………………………………stenopterum |
| - | Clava 3-segmented……………………………………………………………………………………………………………………………………………16 |
| 16 | Marginal vein proximal central seta 2–3× length of distal central seta (Figure 21e)………………………………………………………………tamoi |
| - | Marginal vein proximal central seta about 7× longer than distal seta (Figure 14c)……………………………………………………………digitatum |
| 17 | Antenna 4-segmented (excluding anellus)…………………………………………………………………………………………………………………18 |
| - | Antenna 5-segmented (excluding anellus)…………………………………………………………………………………………………………………19 |
| 18 | Female: C1 with 2 very short UST, much shorter than C1 (Figure 6a). Male: C1 2× as long as maximum width …………………macrostigmum |
| - | Female: C1 with 2 long UST, each as long as C1 (Figure 3a). Male: C1 3× as long as maximum width………………………………………caribea |
| 19 | T1 with characteristic sculpture consisting of distinct cells with denticulate margins (Figure 1a, Figure 17f and Figure 18f)…………………20 |
| - | T1 various, but not as above; elongate cells may be present on sides of T1 (Figure 12b and Figure 16b)…………………………………………24 |
| 20 | Female: T2 sculpture with many closed cells (Figure 17f); ovipositor 1.7× mesotibia…………………………………………………………nowickii |
| - | Female: T2 sculpture without closed cells (occasionally a single cell in noyesi); ovipositor 1.3× mesotibia or shorter…………………………21 |
| 21 | Mid lobe of mesoscutum reticulate with comparatively large cells (Figure 14h)…………………………………………………………funiculatum |
| - | Mid lobe of mesoscutum smooth, or appearing longitudinally striate…………………………………………………………………………………22 |
| 22 | Female: C1 trapezoid in lateral view, length 1.5× maximum width or less. Male: funicle trapezoid, slightly longer than wide………………………………………………………………………………………………………………………………………………mymaripenne |
| - | Female: C1 parallel sided and elongate, 2× as long as wide or more. Male: funicle elongate, approximately 2× as long as wide (male unknown in M. polilovi)……………………………………………………………………………………………………………………………………………………………23 |
| 23 | Scape 4× maximum width; radicle concolorous with scape, both pale (Figure 18a); ovipositor length 1× mesotibia……………………………noyesi |
| - | Scape 6× maximum width; radicle darker compared to scape (Figure 18e); ovipositor length 1.3× mesotibia…………………………………polilovi |
| 24 | Mid lobe of mesoscutum coarsely reticulate over most of its surface (Figure 13e and Figure 14e)………………………………………25 |
| - | Mid lobe of mesoscutum reticulate only anteriorly (Figure 3g), or sculpture different (smooth Figure 4f, striate Figure 3d)………………………27 |
| 25 | Fore wing with base hyaline (not infuscate; Figure 14f)………………………………………………………………………………………fanenitrakely |
| - | Fore wing with base infuscate (Figure 13f)……………………………………………………………………………………………………………………26 |
| 26 | C2 with apical SB only slightly shorter than C2 (Figure 13d) (male unknown)…………………………………………………………………cockerilli |
| - | C2 with apical SB less than ½ length of C2 (Figure 8a)…………………………………………………………………………………………polychaetum |
| 27 | T2–T4 with very long setae laterally (Figure 5h), at least as long as the tergite…………………………………………………………………………28 |
| - | T2–T4 with short setae laterally (Figure 8f), much shorter than the tergite………………………………………………………………………………29 |
| 28 | T1 with long lateral setae. Female: funicle with long, robust UST (Figure 15c)……………………………………………………………………giraulti |
| - | T1 without long lateral setae (Figure 5h). Female: funicle without long, robust ventral UST (Figure 5e and Figure 16e)………………longiciliatum |
| 29 | Mid lobe of mesoscutum reticulate anteriorly (Figure 8f and Figure 22b)…………………………………………………………………………………30 |
| - | Mid lobe of mesoscutum with different sculpture (Figure 18c), or smooth………………………………………………………………………………31 |
| 30 | Fore wing with maximum distal width <2× width measured at apex of marginal vein (Figure 8g); setae on ventral fore wing disc short, penultimate one not reaching to the base of the distal (Figure 22e)…………………………………………………………………………………………………priesneri |
| - | Fore wing with maximum distal width >2× width measured at apex of marginal vein (Figure 22c); setae on ventral fore wing disc long, penultimate one reaching to the base of the distal (Figure 22d)……………………………………………………………………………………………………viggianii |
| 31 | Mid lobe of mesoscutum smooth. Female: proximal UST attached close to the mid point of C1, shorter than C1 (Figure 5b)……………kinuthiae |
| - | Mid lobe of mesoscutum with anterior striae (Figure 18c). Female: proximal UST attached close to the base of C1, longer than C1 (Figure 18b)……………………………………………………………………………………………………………………………………………………………pintoi |