The Impact of Fast Radiation on the Phylogeny of Bactrocera Fruit Flies as Revealed by Multiple Evolutionary Models and Mutation Rate-Calibrated Clock
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Datasets
2.2. Orthologous Gene Set Identification and Alignment
2.3. Phylogenetic Analyses
2.4. Dating Analysis
3. Results
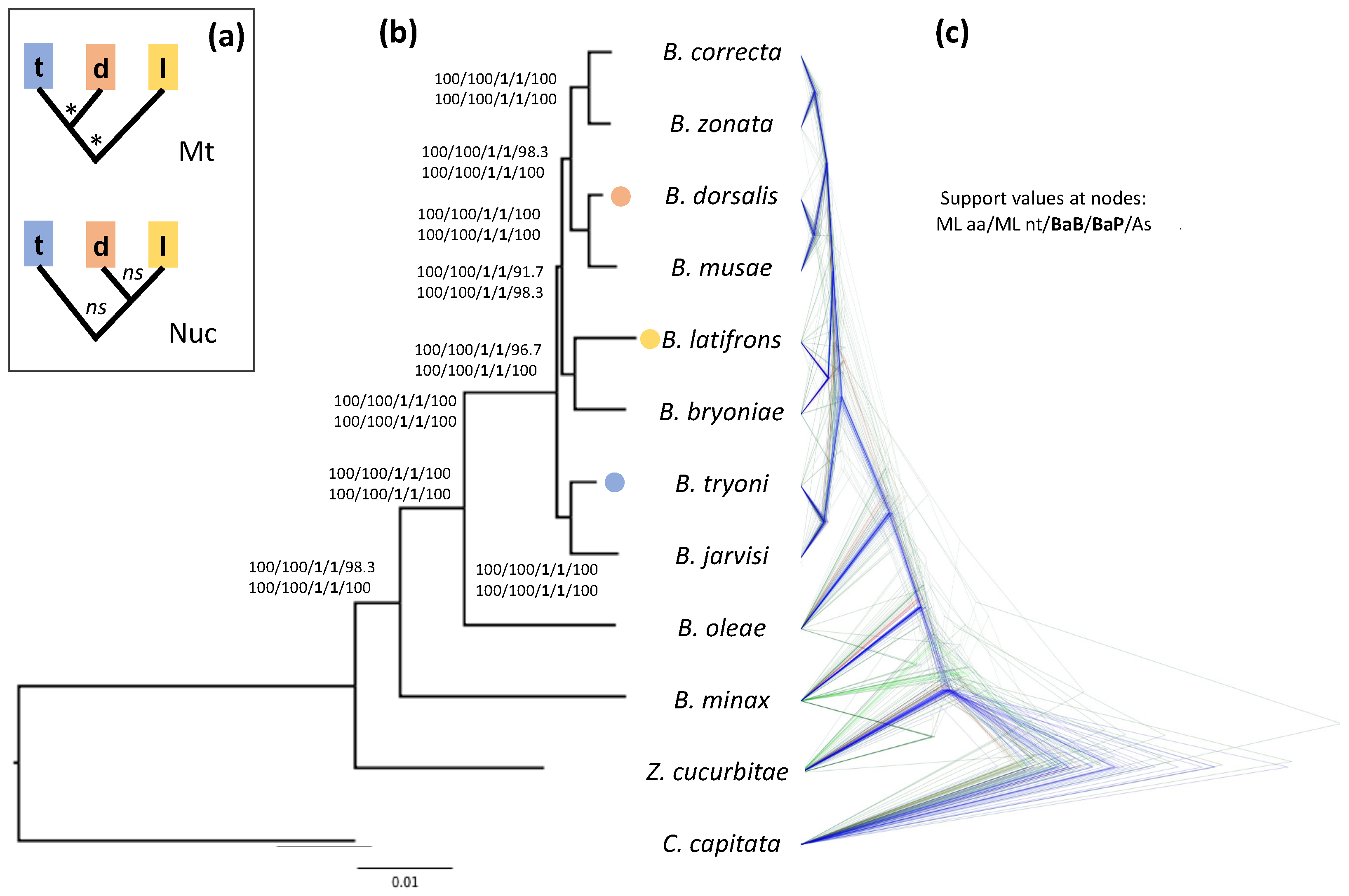
3.1. Phylogenetic Analysis
3.2. Dating Analysis
4. Discussion
4.1. Phylogenetic Analyses Reveal a Closer Affinity of B. dorsalis to B. latifrons Than to B. tryoni
4.2. Dating Analysis Suggests Fast and Recent Radiation in Bactrocera
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bickel, D.; Pape, T.; Meier, R. Diptera Diversity: Status, Challenges and Tools; Brill: Leiden, The Netherlands, 2009; ISBN 978-90-04-18100-7. [Google Scholar]
- Daane, K.M.; Johnson, M.W. Olive Fruit Fly: Managing an Ancient Pest in Modern Times. Annu. Rev. Entomol. 2010, 55, 151–169. [Google Scholar] [CrossRef] [PubMed]
- Barr, N.B.; Ledezma, L.A.; Leblanc, L.; San Jose, M.; Rubinoff, D.; Geib, S.M.; Fujita, B.; Bartels, D.W.; Garza, D.; Kerr, P.; et al. Genetic Diversity of Bactrocera dorsalis (Diptera: Tephritidae) on the Hawaiian Islands: Implications for an Introduction Pathway Into California. J. Econ. Entomol. 2014, 107, 1946–1958. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vargas, R.I.; Nishida, T. Survey for Dacus latifrons (Diptera: Tephritidae). J. Econ. Entomol. 1985, 78, 1311–1314. [Google Scholar] [CrossRef]
- Lux, S.A.; Copeland, R.S.; White, I.M.; Manrakhan, A.; Billah, M.K. A New Invasive Fruit Fly Species from the Bactrocera dorsalis (Hendel) Group Detected in East Africa. Int. J. Trop. Insect Sci. 2003, 23, 355–361. [Google Scholar] [CrossRef]
- Drew, R.A.I.; Tsuruta, K.; White, I. A New Species of Pest Fruit Fly (Diptera: Tephritidae: Dacinae) from Sri Lanka and Africa. Afr. Entomol. 2004, 13, 149–154. [Google Scholar]
- Vayssières, J.-F.; Goergen, G.; Lokossou, O.; Dossa, P.; Akponon, C. A New Bactrocera Species in Benin among Mango Fruit Fly (Diptera: Tephritidae) Species. Fruits 2005, 60, 371–377. [Google Scholar] [CrossRef] [Green Version]
- Goergen, G.; Vayssières, J.-F.; Gnanvossou, D.; Tindo, M. Bactrocera invadens (Diptera: Tephritidae), a New Invasive Fruit Fly Pest for the Afrotropical Region: Host Plant Range and Distribution in West and Central Africa. Env. Entomol. 2011, 40, 844–854. [Google Scholar] [CrossRef] [PubMed]
- EPPO Global Database. Available online: https://gd.eppo.int (accessed on 21 June 2022).
- Crop Protection Compendium. Available online: https://www.cabi.org/cpc (accessed on 21 June 2022).
- Mwatawala, M.; Makundi, R.; Maerere, A.P.; De Meyer, M. Occurrence of the Solanum Fruit Fly Bactrocera latifrons (Hendel) (Diptera: Tephritidae) in Tanzania. J. Afrotrop. Zool. 2010, 6, 83–89. [Google Scholar]
- Mziray, H.A.; Makundi, R.H.; Mwatawala, M.; Maerere, A.; De Meyer, M. Host Use of Bactrocera latifrons, a New Invasive Tephritid Species in Tanzania. J. Econ. Entomol. 2010, 103, 70–76. [Google Scholar] [CrossRef]
- Nugnes, F.; Russo, E.; Viggiani, G.; Bernardo, U. First Record of an Invasive Fruit Fly Belonging to Bactrocera dorsalis Complex (Diptera: Tephritidae) in Europe. Insects 2018, 9, 182. [Google Scholar] [CrossRef] [Green Version]
- Gargiulo, S.; Nugnes, F.; Benedetta, F.; Bernardo, U. Bactrocera latifrons in Europe: The Importance of the Right Attractant for Detection. Bull. Insectol. 2021, 74, 311–320. [Google Scholar]
- Doorenweerd, C.; Leblanc, L.; Norrbom, A.L.; San Jose, M.; Rubinoff, D. A Global Checklist of the 932 Fruit Fly Species in the Tribe Dacini (Diptera, Tephritidae). ZooKeys 2018, 730, 19–56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vargas, R.I.; Piñero, J.C.; Leblanc, L. An Overview of Pest Species of Bactrocera Fruit Flies (Diptera: Tephritidae) and the Integration of Biopesticides with Other Biological Approaches for Their Management with a Focus on the Pacific Region. Insects 2015, 6, 297–318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, I.M.; Elson-Harris, M.M. Fruit Flies of Economic Significance: Their Identification and Bionomics; CAB International: Wallingford, UK, 1992; ISBN 0-85198-790-7. [Google Scholar]
- Ometto, L.; Cestaro, A.; Ramasamy, S.; Grassi, A.; Revadi, S.; Siozios, S.; Moretto, M.; Fontana, P.; Varotto, C.; Pisani, D.; et al. Linking Genomics and Ecology to Investigate the Complex Evolution of an Invasive Drosophila Pest. Genome Biol. Evol. 2013, 5, 745–757. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clarke, A.R.; Armstrong, K.F.; Carmichael, A.E.; Milne, J.R.; Raghu, S.; Roderick, G.K.; Yeates, D.K. Invasive Phytophagous Pests Arising through a Recent Tropical Evolutionary Radiation: The Bactrocera dorsalis Complex of Fruit Flies. Annu. Rev. Entomol. 2004, 50, 293–319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drew, R.A.I. The Tropical Fruit Flies (Diptera: Tephritidae: Dacinae) of the Australasian Region and Oceanic Regions. Mem. Qld. Mus. 1989, 26, 1–521. [Google Scholar]
- Drew, R.A.I.; Hancock, D.L. Phylogeny of the Tribe Dacini (Dacinae) Based on Morphological, Distributional, and Biological Data; CRC Press: Boca Raton, FL, USA, 1999; pp. 509–522. ISBN 978-0-429-12467-9. [Google Scholar]
- Drew, R.A.I.; Romig, M.C. Keys to the Tropical Fruit Flies of South-East Asia; CABI: Wallingford, UK, 2016. [Google Scholar]
- Krosch, M.N.; Schutze, M.K.; Armstrong, K.F.; Graham, G.C.; Yeates, D.K.; Clarke, A.R. A Molecular Phylogeny for the Tribe Dacini (Diptera: Tephritidae): Systematic and Biogeographic Implications. Mol. Phylogenet. Evol. 2012, 64, 513–523. [Google Scholar] [CrossRef] [Green Version]
- San Jose, M.; Doorenweerd, C.; Leblanc, L.; Barr, N.; Geib, S.; Rubinoff, D. Incongruence between Molecules and Morphology: A Seven-Gene Phylogeny of Dacini Fruit Flies Paves the Way for Reclassification (Diptera: Tephritidae). Mol. Phylogenet. Evol. 2018, 121, 139–149. [Google Scholar] [CrossRef]
- Virgilio, M.; Jordaens, K.; Verwimp, C.; White, I.M.; De Meyer, M. Higher Phylogeny of Frugivorous Flies (Diptera, Tephritidae, Dacini): Localised Partition Conflicts and a Novel Generic Classification. Mol. Phylogenet. Evol. 2015, 85, 171–179. [Google Scholar] [CrossRef]
- Dupuis, J.R.; Bremer, F.T.; Kauwe, A.; San Jose, M.; Leblanc, L.; Rubinoff, D.; Geib, S.M. HiMAP: Robust Phylogenomics from Highly Multiplexed Amplicon Sequencing. Mol. Ecol. Resour. 2018, 18, 1000–1019. [Google Scholar] [CrossRef]
- Muraji, M.; Nakahara, S. Phylogenetic Relationships among Fruit Flies, Bactrocera (Diptera, Tephritidae), Based on the Mitochondrial rDNA Sequences. Insect Mol. Biol. 2001, 10, 549–559. [Google Scholar] [CrossRef] [PubMed]
- Nakahara, S.; Muraji, M. Phylogenetic Analyses of Bactrocera Fruit Flies (Diptera: Tephritidae) Based on Nucleotide Sequences of the Mitochondrial COI and COII Genes. Res. Bull. Plant Prot. Jpn. 2007, 44, 1–12. [Google Scholar]
- Smith, P.T.; Kambhampati, S.; Armstrong, K.A. Phylogenetic Relationships among Bactrocera Species (Diptera: Tephritidae) Inferred from Mitochondrial DNA Sequences. Mol. Phylogenet. Evol. 2003, 26, 8–17. [Google Scholar] [CrossRef]
- Yong, H.-S.; Song, S.-L.; Lim, P.-E.; Eamsobhana, P.; Suana, I.W. Complete Mitochondrial Genome of Three Bactrocera Fruit Flies of Subgenus Bactrocera (Diptera: Tephritidae) and Their Phylogenetic Implications. PLoS ONE 2016, 11, e0148201. [Google Scholar] [CrossRef]
- Zhang, B.; Liu, Y.H.; Wu, W.X.; Wang, Z.L. Molecular Phylogeny of Bactrocera Species (Diptera: Tephritidae: Dacini) Inferred from Mitochondrial Sequences of 16S RDNA and COI Sequences. Fla. Entomol. 2010, 93, 369–377. [Google Scholar] [CrossRef]
- Choo, A.; Nguyen, T.N.M.; Ward, C.M.; Chen, I.Y.; Sved, J.; Shearman, D.; Gilchrist, A.S.; Crisp, P.; Baxter, S.W. Identification of Y-Chromosome Scaffolds of the Queensland Fruit Fly Reveals a Duplicated Gyf Gene Paralogue Common to Many Bactrocera Pest Species. Insect Mol. Biol. 2019, 28, 873–886. [Google Scholar] [CrossRef]
- Papanicolaou, A.; Schetelig, M.F.; Arensburger, P.; Atkinson, P.W.; Benoit, J.B.; Bourtzis, K.; Castañera, P.; Cavanaugh, J.P.; Chao, H.; Childers, C.; et al. The Whole Genome Sequence of the Mediterranean Fruit Fly, Ceratitis capitata (Wiedemann), Reveals Insights into the Biology and Adaptive Evolution of a Highly Invasive Pest Species. Genome Biol. 2016, 17, 192. [Google Scholar] [CrossRef] [Green Version]
- Sim, S.B.; Geib, S.M. A Chromosome-Scale Assembly of the Bactrocera cucurbitae Genome Provides Insight to the Genetic Basis of White Pupae. G3 Genes Genomes Genet. 2017, 7, 1927–1940. [Google Scholar] [CrossRef] [Green Version]
- Geib, S.M.; Calla, B.; Hall, B.; Hou, S.; Manoukis, N.C. Characterizing the Developmental Transcriptome of the Oriental Fruit Fly, Bactrocera dorsalis (Diptera: Tephritidae) through Comparative Genomic Analysis with Drosophila melanogaster Utilizing ModENCODE Datasets. BMC Genom. 2014, 15, 924. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Xiong, K.-C.; Liu, Y.-H. De Novo Transcriptome Analysis of Chinese Citrus Fly, Bactrocera minax (Diptera: Tephritidae), by High-Throughput Illumina Sequencing. PLoS ONE 2016, 11, e0157656. [Google Scholar] [CrossRef]
- Bayega, A.; Djambazian, H.; Tsoumani, K.T.; Gregoriou, M.E.; Sagri, E.; Drosopoulou, E.; Mavragani-Tsipidou, P.; Giorda, K.; Tsiamis, G.; Bourtzis, K.; et al. De Novo Assembly of the Olive Fruit Fly (Bactrocera oleae) Genome with Linked-Reads and Long-Read Technologies Minimizes Gaps and Provides Exceptional y Chromosome Assembly. BMC Genom. 2020, 21, 259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-Length Transcriptome Assembly from RNA-Seq Data without a Reference Genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and Applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [Green Version]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Löytynoja, A.; Goldman, N. A Model of Evolution and Structure for Multiple Sequence Alignment. Philos. Trans. R. Soc. B Biol. Sci. 2008, 363, 3913–3919. [Google Scholar] [CrossRef]
- Abascal, F.; Zardoya, R.; Telford, M.J. TranslatorX: Multiple Alignment of Nucleotide Sequences Guided by Amino Acid Translations. Nucleic Acids Res. 2010, 38, W7–W13. [Google Scholar] [CrossRef] [Green Version]
- Ramasamy, S.; Ometto, L.; Crava, C.M.; Revadi, S.; Kaur, R.; Horner, D.S.; Pisani, D.; Dekker, T.; Anfora, G.; Rota-Stabelli, O. The Evolution of Olfactory Gene Families in Drosophila and the Genomic Basis of Chemical-Ecological Adaptation in Drosophila suzukii. Genome Biol. Evol. 2016, 8, 2297–2311. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Stamatakis, A. RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Zhang, C.; Rabiee, M.; Sayyari, E.; Mirarab, S. ASTRAL-III: Polynomial Time Species Tree Reconstruction from Partially Resolved Gene Trees. BMC Bioinform. 2018, 19, 153. [Google Scholar] [CrossRef] [Green Version]
- Bouckaert, R.; Vaughan, T.G.; Barido-Sottani, J.; Duchêne, S.; Fourment, M.; Gavryushkina, A.; Heled, J.; Jones, G.; Kühnert, D.; De Maio, N.; et al. BEAST 2.5: An Advanced Software Platform for Bayesian Evolutionary Analysis. PLoS Comput. Biol. 2019, 15, e1006650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lartillot, N.; Philippe, H. A Bayesian Mixture Model for Across-Site Heterogeneities in the Amino-Acid Replacement Process. Mol. Biol. Evol. 2004, 21, 1095–1109. [Google Scholar] [CrossRef] [PubMed]
- Ogilvie, H.A.; Bouckaert, R.R.; Drummond, A.J. StarBEAST2 Brings Faster Species Tree Inference and Accurate Estimates of Substitution Rates. Mol. Biol. Evol. 2017, 34, 2101–2114. [Google Scholar] [CrossRef]
- Barido-Sottani, J.; Bošková, V.; Du Plessis, L.; Kühnert, D.; Magnus, C.; Mitov, V.; Müller, N.F.; Pečerska, J.; Rasmussen, D.A.; Zhang, C.; et al. Taming the BEAST—A Community Teaching Material Resource for BEAST 2. Syst. Biol. 2018, 67, 170–174. [Google Scholar] [CrossRef] [PubMed]
- Bouckaert, R.R.; Heled, J. DensiTree 2: Seeing Trees Through the Forest. bioRxiv 2014, 012401. [Google Scholar] [CrossRef] [Green Version]
- Norrbom, A. New Genera of Tephritidae (Diptera) from Brazil and Dominican Amber, with Phylogenetic Analysis of the Tribe Ortalotrypetini. Insecta Mundi 1994, 8, 1–15. [Google Scholar]
- Wiegmann, B.M.; Trautwein, M.D.; Winkler, I.S.; Barr, N.B.; Kim, J.-W.; Lambkin, C.; Bertone, M.A.; Cassel, B.K.; Bayless, K.M.; Heimberg, A.M.; et al. Episodic Radiations in the Fly Tree of Life. Proc. Natl. Acad. Sci. USA 2011, 108, 5690–5695. [Google Scholar] [CrossRef] [Green Version]
- Junqueira, A.C.M.; Azeredo-Espin, A.M.L.; Paulo, D.F.; Marinho, M.A.T.; Tomsho, L.P.; Drautz-Moses, D.I.; Purbojati, R.W.; Ratan, A.; Schuster, S.C. Large-Scale Mitogenomics Enables Insights into Schizophora (Diptera) Radiation and Population Diversity. Sci. Rep. 2016, 6, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Keightley, P.D.; Trivedi, U.; Thomson, M.; Oliver, F.; Kumar, S.; Blaxter, M.L. Analysis of the Genome Sequences of Three Drosophila melanogaster Spontaneous Mutation Accumulation Lines. Genome Res. 2009, 19, 1195–1201. [Google Scholar] [CrossRef] [Green Version]
- Obbard, D.J.; Maclennan, J.; Kim, K.-W.; Rambaut, A.; O’Grady, P.M.; Jiggins, F.M. Estimating Divergence Dates and Substitution Rates in the Drosophila Phylogeny. Mol. Biol. Evol. 2012, 29, 3459–3473. [Google Scholar] [CrossRef] [Green Version]
- Keightley, P.D.; Pinharanda, A.; Ness, R.W.; Simpson, F.; Dasmahapatra, K.K.; Mallet, J.; Davey, J.W.; Jiggins, C.D. Estimation of the Spontaneous Mutation Rate in Heliconius melpomene. Mol. Biol. Evol. 2015, 32, 239–243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keightley, P.D.; Ness, R.W.; Halligan, D.L.; Haddrill, P.R. Estimation of the Spontaneous Mutation Rate per Nucleotide Site in a Drosophila melanogaster Full-Sib Family. Genetics 2014, 196, 313–320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vargas, R.I.; Walsh, W.A.; Kanehisa, D.; Jang, E.B.; Armstrong, J.W. Demography of Four Hawaiian Fruit Flies (Diptera: Tephritidae) Reared at Five Constant Temperatures. Ann. Entomol. Soc. Am. 1997, 90, 162–168. [Google Scholar] [CrossRef]
- Stephens, A.E.A.; Kriticos, D.J.; Leriche, A. The Current and Future Potential Geographical Distribution of the Oriental Fruit Fly, Bactrocera dorsalis (Diptera: Tephritidae). Bull. Entomol. Res. 2007, 97, 369–378. [Google Scholar] [CrossRef]
- Theron, C.D.; Manrakhan, A.; Weldon, C.W. Host Use of the Oriental Fruit Fly, Bactrocera dorsalis (Hendel) (Diptera: Tephritidae), in South Africa. J. Appl. Entomol. 2017, 141, 810–816. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Yang, H.; Wang, T.; Wang, J.; Wei, H. Life History and Adult Dynamics of Bactrocera dorsalis in the Citrus Orchard of Nanchang, a Subtropical Area from China: Implications for a Control Timeline. ScienceAsia 2019, 45, 212–220. [Google Scholar] [CrossRef] [Green Version]
- Russel, P.M.; Brewer, B.J.; Klaere, S.; Bouckaert, R.R. Model Selection and Parameter Inference in Phylogenetics Using Nested Sampling. Syst. Biol. 2019, 68, 219–233. [Google Scholar] [CrossRef]
- Kass, R.E.; Raftery, A.E. Bayes Factors. J. Am. Stat. Assoc. 1995, 90, 774–795. [Google Scholar] [CrossRef]
- Schrempf, D.; Minh, B.Q.; De Maio, N.; von Haeseler, A.; Kosiol, C. Reversible Polymorphism-Aware Phylogenetic Models and Their Application to Tree Inference. J. Theor. Biol. 2016, 407, 362–370. [Google Scholar] [CrossRef] [Green Version]
- Drew, R.A.I.; Lambert, D.M. On the Specific Status of Dacus (Bactrocera) aquilonis and D. (Bactrocera) tryoni (Diptera: Tephritidae). Ann. Entomol. Soc. Am. 1986, 79, 870–878. [Google Scholar] [CrossRef]
- Cruickshank, L.; Jessup, A.J.; Cruickshank, D.J. Interspecific Crosses of Bactrocera tryoni (Froggatt) and Bactrocera jarvisi (Tryon) (Diptera: Tephritidae) in the Laboratory. Aust. J. Entomol. 2001, 40, 278–280. [Google Scholar] [CrossRef]
- Pike, N.; Wang, W.Y.S.; Meats, A. The Likely Fate of Hybrids of Bactrocera tryoni and Bactrocera neohumeralis. Heredity 2003, 90, 365–370. [Google Scholar] [CrossRef] [PubMed]
- Wee, S.-L.; Tan, K.-H. Evidence of Natural Hybridization between Two Sympatric Sibling Species of Bactrocera dorsalis Complex Based on Pheromone Analysis. J. Chem. Ecol. 2005, 31, 845–858. [Google Scholar] [CrossRef] [PubMed]
- Ebina, T.; Ohto, K. Morphological Characters and PCR-RFLP Markers in the Interspecific Hybrids between Bactrocera carambolae and B. papayae of the B. dorsalis Species Complex (Diptera: Tephritidae). Res. Bull. Plant Prot. Serv. Jpn. 2006, 42, 23–34. [Google Scholar]
- Shearman, D.C.A.; Frommer, M.; Morrow, J.L.; Raphael, K.A.; Gilchrist, A.S. Interspecific Hybridization as a Source of Novel Genetic Markers for the Sterile Insect Technique in Bactrocera tryoni (Diptera: Tephritidae). J. Econ. Entomol. 2010, 103, 1071–1079. [Google Scholar] [CrossRef]
- Schutze, M.K.; Jessup, A.; Ul-Haq, I.; Vreysen, M.J.B.; Wornoayporn, V.; Vera, M.T.; Clarke, A.R. Mating Compatibility Among Four Pest Members of the Bactrocera dorsalis Fruit Fly Species Complex (Diptera: Tephritidae). J. Econ. Entomol. 2013, 106, 695–707. [Google Scholar] [CrossRef]
- Augustinos, A.A.; Drosopoulou, E.; Gariou-Papalexiou, A.; Bourtzis, K.; Mavragani-Tsipidou, P.; Zacharopoulou, A. The Bactrocera dorsalis Species Complex: Comparative Cytogenetic Analysis in Support of Sterile Insect Technique Applications. BMC Genet. 2014, 15, S16. [Google Scholar] [CrossRef] [Green Version]
- Bo, W.; Ahmad, S.; Dammalage, T.; Tomas, U.S.; Wornoayporn, V.; Ul Haq, I.; Cáceres, C.; Vreysen, M.J.B.; Schutze, M.K. Mating Compatibility between Bactrocera invadens and Bactrocera dorsalis (Diptera: Tephritidae). J. Econ. Entomol. 2014, 107, 623–629. [Google Scholar] [CrossRef]
- Gilchrist, A.S.; Shearman, D.C.; Frommer, M.; Raphael, K.A.; Deshpande, N.P.; Wilkins, M.R.; Sherwin, W.B.; Sved, J.A. The Draft Genome of the Pest Tephritid Fruit Fly Bactrocera tryoni: Resources for the Genomic Analysis of Hybridising Species. BMC Genom. 2014, 15, 1153. [Google Scholar] [CrossRef] [Green Version]
- Yeap, H.L.; Lee, S.F.; Robinson, F.; Mourant, R.G.; Sved, J.A.; Frommer, M.; Papanicolaou, A.; Edwards, O.R.; Oakeshott, J.G. Separating Two Tightly Linked Species-Defining Phenotypes in Bactrocera with Hybrid Recombinant Analysis. BMC Genet. 2020, 21, 132. [Google Scholar] [CrossRef]
- Pollard, D.A.; Iyer, V.N.; Moses, A.M.; Eisen, M.B. Widespread Discordance of Gene Trees with Species Tree in Drosophila: Evidence for Incomplete Lineage Sorting. PLoS Genet. 2006, 2, e173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yaakop, S.; Ibrahim, N.J.; Shariff, S.; Zain, B.M.M. Molecular Clock Analysis on Five Bactrocera Species Flies (Diptera: Tephritidae) Based on Combination of COI and NADH Sequences. Orient. Insects 2015, 49, 150–164. [Google Scholar] [CrossRef]
- Nardi, F.; Carapelli, A.; Boore, J.L.; Roderick, G.K.; Dallai, R.; Frati, F. Domestication of Olive Fly through a Multi-Regional Host Shift to Cultivated Olives: Comparative Dating Using Complete Mitochondrial Genomes. Mol. Phylogenet. Evol. 2010, 57, 678–686. [Google Scholar] [CrossRef] [PubMed]
- Zhong, G.; Geng, J.; Wong, H.K.; Ma, Z.; Wu, N. A Semi-Quantitative Method for the Reconstruction of Eustatic Sea Level History from Seismic Profiles and Its Application to the Southern South China Sea. Earth Planet. Sci. Lett. 2004, 223, 443–459. [Google Scholar] [CrossRef]
- DeSalle, R.; Giddings, L.V. Discordance of Nuclear and Mitochondrial DNA Phylogenies in Hawaiian Drosophila. Proc. Natl. Acad. Sci. USA 1986, 83, 6902–6906. [Google Scholar] [CrossRef] [Green Version]
- Beltrán, M.; Jiggins, C.D.; Bull, V.; Linares, M.; Mallet, J.; McMillan, W.O.; Bermingham, E. Phylogenetic Discordance at the Species Boundary: Comparative Gene Genealogies Among Rapidly Radiating Heliconius Butterflies. Mol. Biol. Evol. 2002, 19, 2176–2190. [Google Scholar] [CrossRef] [Green Version]
- Putnam, A.S.; Scriber, J.M.; Andolfatto, P. Discordant Divergence Times among Z-Chromosome Regions between Two Ecologically Distinct Swallowtail Butterfly Species. Evolution 2007, 61, 912–927. [Google Scholar] [CrossRef]
- Toews, D.P.L.; Brelsford, A. The Biogeography of Mitochondrial and Nuclear Discordance in Animals. Mol. Ecol. 2012, 21, 3907–3930. [Google Scholar] [CrossRef]
- Drummond, A.J.; Ho, S.Y.W.; Phillips, M.J.; Rambaut, A. Relaxed Phylogenetics and Dating with Confidence. PLoS Biol. 2006, 4, e88. [Google Scholar] [CrossRef]
- Ho, S.Y.W.; Lanfear, R.; Bromham, L.; Phillips, M.J.; Soubrier, J.; Rodrigo, A.G.; Cooper, A. Time-Dependent Rates of Molecular Evolution. Mol. Ecol. 2011, 20, 3087–3101. [Google Scholar] [CrossRef]
- Ho, S.Y.W.; Lo, N. The Insect Molecular Clock. Aust. J. Entomol. 2013, 52, 101–105. [Google Scholar] [CrossRef]
- Misof, B.; Liu, S.; Meusemann, K.; Peters, R.S.; Donath, A.; Mayer, C.; Frandsen, P.B.; Ware, J.; Flouri, T.; Beutel, R.G.; et al. Phylogenomics Resolves the Timing and Pattern of Insect Evolution. Science 2014, 346, 763–767. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Valerio, F.; Zadra, N.; Rota-Stabelli, O.; Ometto, L. The Impact of Fast Radiation on the Phylogeny of Bactrocera Fruit Flies as Revealed by Multiple Evolutionary Models and Mutation Rate-Calibrated Clock. Insects 2022, 13, 603. https://doi.org/10.3390/insects13070603
Valerio F, Zadra N, Rota-Stabelli O, Ometto L. The Impact of Fast Radiation on the Phylogeny of Bactrocera Fruit Flies as Revealed by Multiple Evolutionary Models and Mutation Rate-Calibrated Clock. Insects. 2022; 13(7):603. https://doi.org/10.3390/insects13070603
Chicago/Turabian StyleValerio, Federica, Nicola Zadra, Omar Rota-Stabelli, and Lino Ometto. 2022. "The Impact of Fast Radiation on the Phylogeny of Bactrocera Fruit Flies as Revealed by Multiple Evolutionary Models and Mutation Rate-Calibrated Clock" Insects 13, no. 7: 603. https://doi.org/10.3390/insects13070603
APA StyleValerio, F., Zadra, N., Rota-Stabelli, O., & Ometto, L. (2022). The Impact of Fast Radiation on the Phylogeny of Bactrocera Fruit Flies as Revealed by Multiple Evolutionary Models and Mutation Rate-Calibrated Clock. Insects, 13(7), 603. https://doi.org/10.3390/insects13070603






