Resistance to Extreme Stresses by a Newly Discovered Japanese Tardigrade Species, Macrobiotus kyoukenus (Eutardigrada, Macrobiotidae)
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
Stress Resistance
3. Results
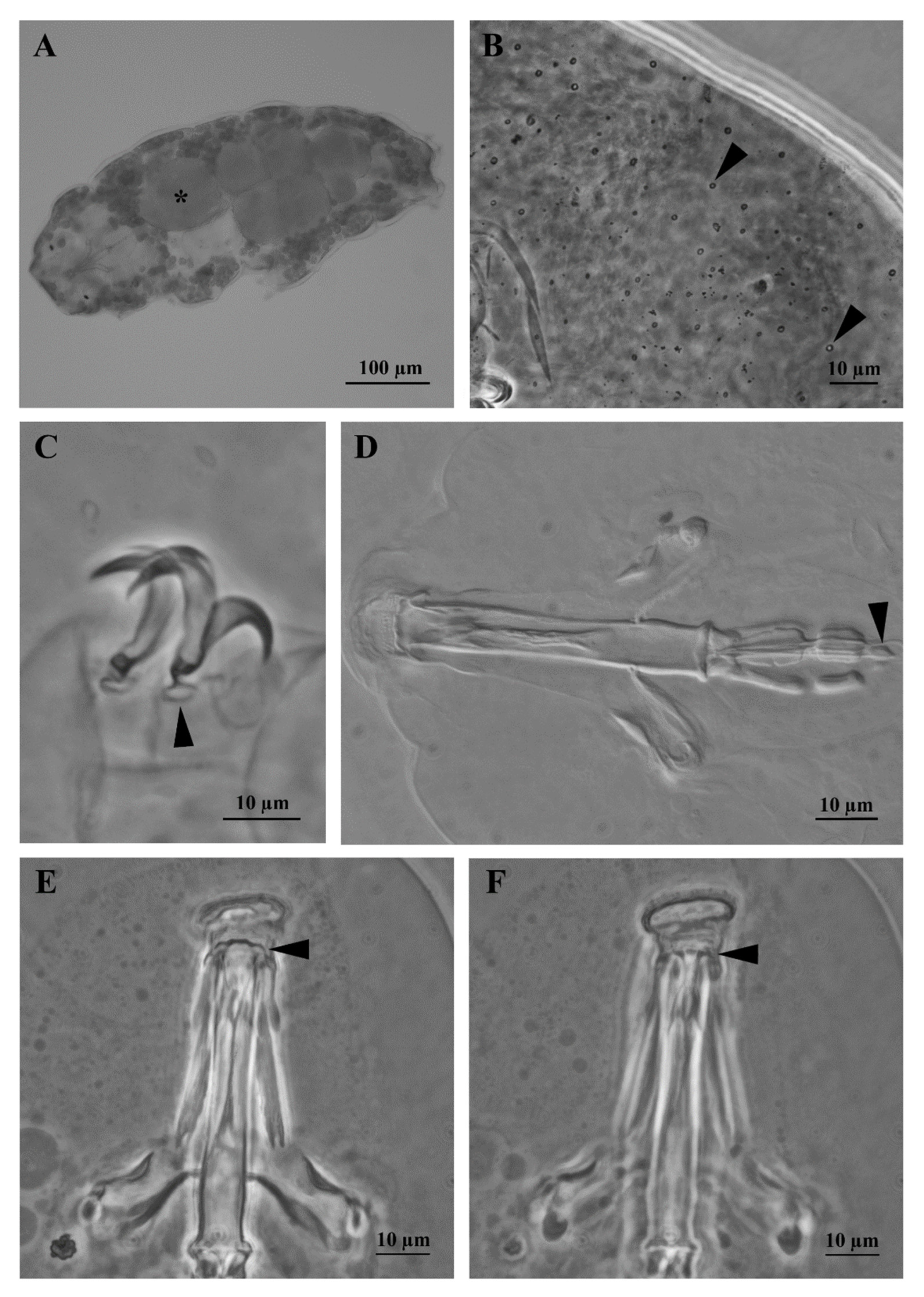
3.1. Morphological Analyses
3.1.1. Reproduction
3.1.2. Differential Diagnosis
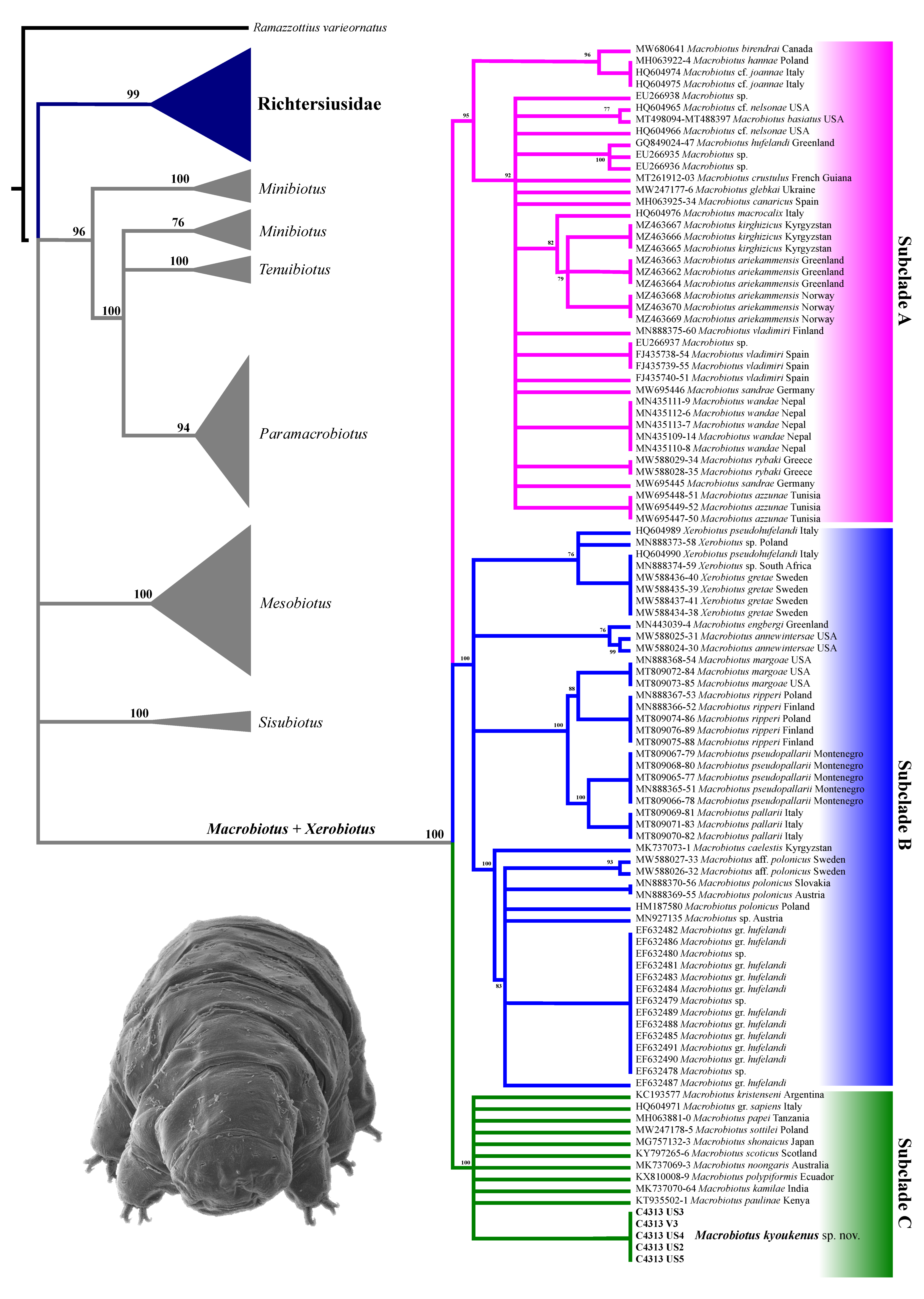
3.2. Molecular Analysis
3.3. Stress Resistance
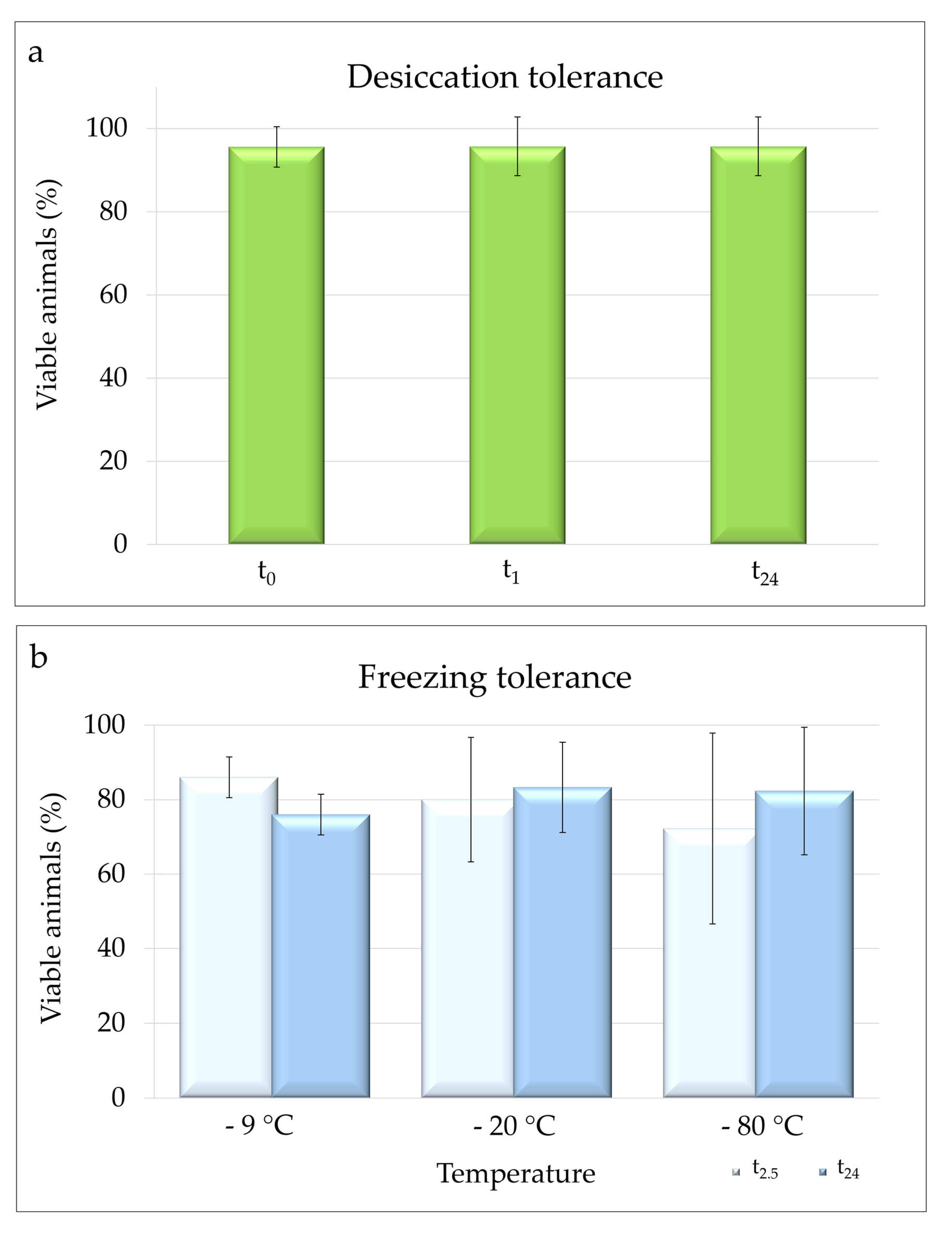
3.3.1. Desiccation Tolerance
3.3.2. Freezing Tolerance
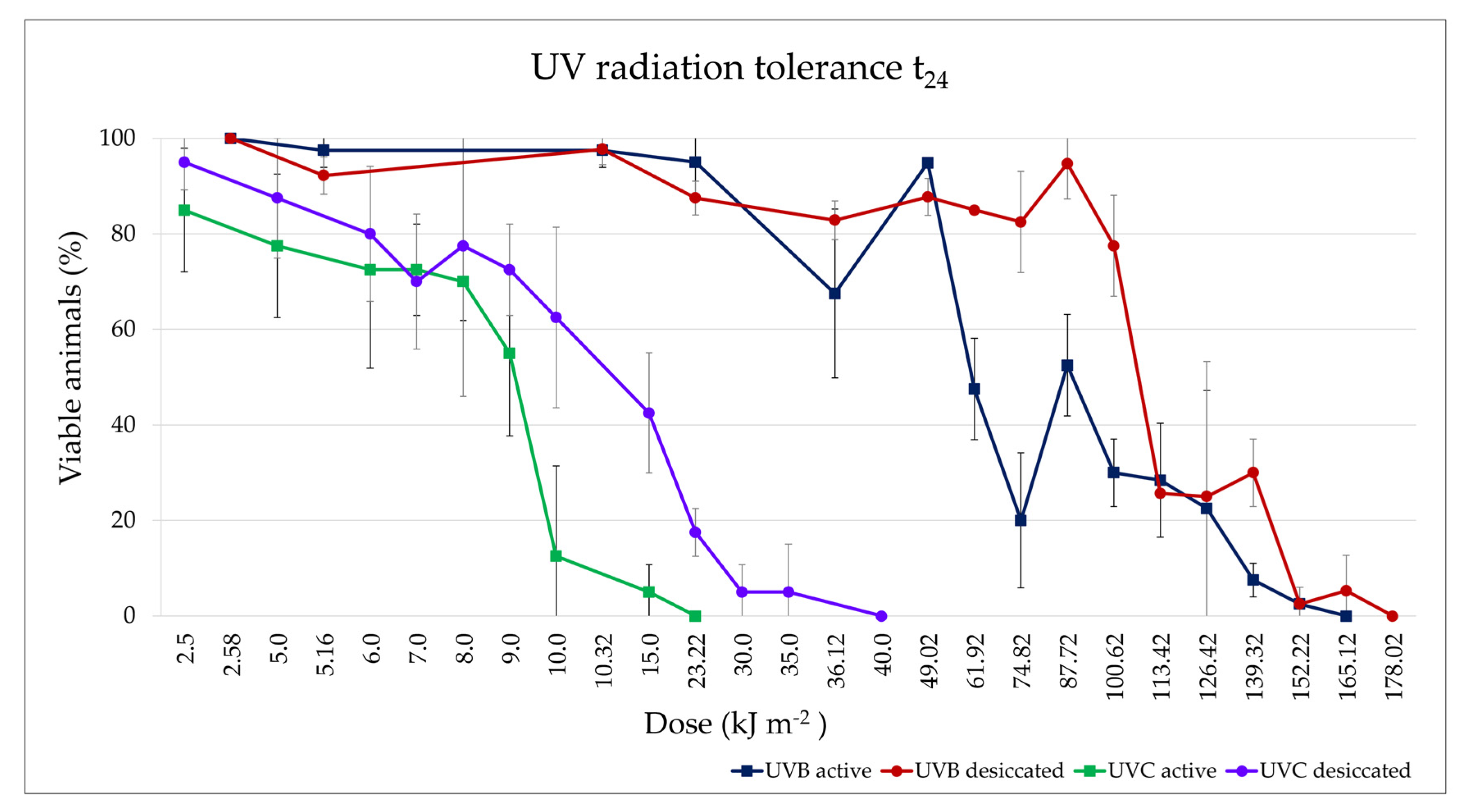
3.3.3. UV Radiation Tolerance
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Guidetti, R.; Altiero, T.; Rebecchi, L. On dormancy strategies in tardigrades. J. Insect Physiol. 2011, 57, 567–576. [Google Scholar] [CrossRef] [PubMed]
- Rebecchi, L.; Altiero, T.; Guidetti, R. Anhydrobiosis: The extreme limit of desiccation tolerance. ISJ Invert. Surviv. J. 2007, 4, 65–81. [Google Scholar]
- Møbjerg, N.; Halberg, K.A.; Jørgensen, A.; Persson, D.; Bjørn, M.; Ramløv, H.; Kristensen, R.M. Survival in extreme environments—On the current knowledge of adaptations in tardigrades. Acta Physiol. 2011, 202, 409–420. [Google Scholar] [CrossRef] [PubMed]
- Horikawa, D.D.; Kunieda, T.; Abe, W.; Watanabe, M.; Nakahara, Y.; Yukuhiro, F.; Sakashita, T.; Hamada, N.; Wada, S.; Funayama, T.; et al. Establishment of a rearing system of the extremotolerant tardigrade Ramazzottius varieornatus: A new model animal for astrobiology. Astrobiology 2008, 8, 549–556. [Google Scholar] [CrossRef]
- Jönsson, K.I.; Rabbow, E.; Schill, R.O.; Harms-Ringdahl, M.; Rettberg, P. Tardigrades survive exposure to space in low Earth orbit. Curr. Biol. 2008, 18, R729–R731. [Google Scholar] [CrossRef] [Green Version]
- Rebecchi, L.; Cesari, M.; Altiero, T.; Frigieri, A.; Guidetti, R. Survival and DNA degradation in anhydrobiotic tardigrades. J. Exp. Biol. 2009, 212, 4033–4039. [Google Scholar] [CrossRef] [Green Version]
- Neves, R.C.; Hvidepil, L.K.B.; Sørensen-Hygum, T.L.; Stuart, R.M.; Møbjerg, N. Thermotolerance experiments on active and desiccated states of Ramazzottius varieornatus emphasize that tardigrades are sensitive to high temperatures. Sci. Rep. UK 2020, 10, 94. [Google Scholar] [CrossRef] [Green Version]
- Møbjerg, N.; Neves, R.C. New insights into survival strategies of tardigrades. Comp. Biochem. Phys. A 2021, 254, 110890. [Google Scholar] [CrossRef]
- Altiero, T.; Guidetti, R.; Caselli, V.; Cesari, M.; Rebecchi, L. Ultraviolet radiation tolerance in hydrated and desiccated eutardigrades. J. Zool. Syst. Evol. Res. 2011, 49, 104–110. [Google Scholar] [CrossRef]
- Horikawa, D.D.; Cumbers, J.; Sakakibara, I.; Rogo, D.; Leuko, S.; Harnoto, R.; Arakawa, K.; Katayama, T.; Kunieda, T.; Toyoda, A.; et al. Analysis of DNA repair and protection in the Tardigrade Ramazzottius varieornatus and Hypsibius dujardini after exposure to UVC radiation. PLoS ONE 2013, 8, e64793. [Google Scholar] [CrossRef] [Green Version]
- Hashimoto, T.; Horikawa, D.D.; Saito, Y.; Kuwahara, H.; Kozuka-Hata, H.; Shin, I.T.; Minakuchi, Y.; Ohishi, K.; Motoyama, A.; Aizu, T.; et al. Extremotolerant tardigrade genome and improved radiotolerance of human cultured cells by tardigrade-unique protein. Nat. Commun. 2016, 7, 12808. [Google Scholar] [CrossRef] [PubMed]
- Giovannini, I.; Altiero, T.; Guidetti, R.; Rebecchi, L. Will the Antarctic tardigrade Acutuncus antarcticus be able to withstand environmental stresses related to global climate change? J. Exp. Biol. 2018, 221, jeb160622. [Google Scholar] [PubMed] [Green Version]
- Jönsson, K.I. Radiation tolerance in tardigrades: Current knowledge and potential applications in medicine. Cancers 2019, 11, 1333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guidetti, R.; Bertolani, R. Tardigrade taxonomy: An updated checklist of the taxa and a list of characters for their identification. Zootaxa 2005, 845, 1–46. [Google Scholar] [CrossRef]
- Degma, P.; Guidetti, R. Notes to the current checklist of Tardigrada. Zootaxa 2007, 1579, 41–53. [Google Scholar] [CrossRef]
- Mathews, G. Tardigrada from Japan. Peking Nat. Hist. Bull. 1937, 11, 411–412. [Google Scholar]
- Utsugi, K. Urban tardigrades in Kyushu. Zool. Sci. 1986, 3, 1110. [Google Scholar]
- Utsugi, K. Tardigrades in Hokkaido area. Zool. Sci. 1988, 5, 1335. [Google Scholar]
- Hatai, S. On the Japanese Tardigrada. Sci. Rep. Yokosuka City Mus. 1956, 1, 1–13. [Google Scholar]
- Utsugi, K. Study on terrestrial tardigrades in Japan. II. Summary of urban tardigrades in Japan. Nat. Environ. Sci. Res. 1996, 9, 33–46. (In Japanese) [Google Scholar]
- Suzuki, A.C.; Heard, L.; Sugiura, K. Terrestrial tardigrades from Mikurajima Island (the first report). Mikurensis 2018, 7, 3–8. [Google Scholar]
- Utsugi, K. Urban tardigrades in Hokuriku area. Zool. Sci. 1987, 4, 1111. [Google Scholar]
- Biserov, V.I.; Dudichev, A.L.; Biserova, N.M. Preliminary data on tardigrades of Lake Biwa (Japan). Arthropoda Sel. 2001, 10, 307–310. [Google Scholar]
- Ishida, M.; Matsui, T. Noteworthy tardigrades from Kochi Prefecture. Nat. Environ. Sci. Res. 2007, 9, 45–58. (In Japanese) [Google Scholar]
- Stec, D.; Arakawa, K.; Michalczyk, Ł. An integrative description of Macrobiotus shonaicus sp. nov. (Tardigrada: Macrobiotidae) from Japan with notes on its phylogenetic position within the hufelandi group. PLoS ONE 2018, 13, e0192210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugiura, K.; Arakawa, K.; Matsumoto, M. Distribution of Macrobiotus shonaicus Stec, Arakawa & Michalczyk, 2018 (Tardigrada: Eutardigrada: Macrobiotidae) in Japan. Zootaxa 2020, 4767, 56–70. [Google Scholar]
- Bertolani, R.; Rebecchi, L. A revision of the Macrobiotus hufelandi group (Tardigrada, Macrobiotidae), with some observations on the taxonomic characters of eutardigrades. Zool. Scr. 1993, 22, 127–152. [Google Scholar] [CrossRef]
- Guidetti, R.; Peluffo, J.R.; Rocha, A.M.; Cesari, M.; Moly de Peluffo, M.C. The morphological and molecular analyses of a new South American urban tardigrade offer new insights on the biological meaning of the Macrobiotus hufelandi group of species (Tardigrada: Macrobiotidae). J. Nat. Hist. 2013, 47, 2409–2426. [Google Scholar] [CrossRef]
- Stec, D.; Vecchi, M.; Calhim, S.; Michalczyk, Ł. New multilocus phylogeny reorganises the family Macrobiotidae (Eutardigrada) and unveils complex morphological evolution of the Macrobiotus hufelandi group. Mol. Phylogenet. Evol. 2021, 160, 106987. [Google Scholar] [CrossRef]
- Bertolani, R.; Biserov, V.; Rebecchi, L.; Cesari, M. Taxonomy and biogeography of tardigrades using an integrated approach: New results on species of the Macrobiotus hufelandi group. Invertebr. Zool. 2011, 8, 23–36. [Google Scholar] [CrossRef]
- Kaczmarek, Ł.; Michalczyk, Ł. The Macrobiotus hufelandi group (Tardigrada) revisited. Zootaxa 2017, 4363, 101–123. [Google Scholar] [CrossRef] [PubMed]
- Michalczyk, Ł.; Kaczmarek, Ł. The Tardigrada Register: A comprehensive online data repository for tardigrade taxonomy. J. Limnol. 2013, 201372, 175–181. [Google Scholar] [CrossRef]
- Massa, E.; Guidetti, R.; Cesari, M.; Rebecchi, L.; Jönsson, K.I. Tardigrades of Kristianstads Vattenrike Biosphere Reserve with description of four new species from Sweden. Sci. Rep. 2021, 11, 4861. [Google Scholar] [CrossRef]
- Bartels, P.J.; Nelson, D.R.; Exline, R.P. Allometry and the removal of body size effects in the morphometric analysis of tardigrades. J. Zool. Syst. Evol. Res. 2011, 49, 17–25. [Google Scholar] [CrossRef]
- Bertolani, R.; Guidetti, R.; Marchioro, T.; Altiero, T.; Rebecchi, L.; Cesari, M. Phylogeny of Eutardigrada: New molecular data and their morphological support lead to the identification of new evolutionary lineages. Mol. Phylogenet. Evol. 2014, 76, 110–126. [Google Scholar] [CrossRef] [PubMed]
- Cesari, M.; Vecchi, M.; Palmer, A.; Bertolani, R.; Pilato, G.; Rebecchi, L.; Guidetti, R. What if the claws are reduced? Morphological and molecular phylogenetic relationships of the genus Haplomacrobiotus May, 1948 (Eutardigrada, Parachela). Zool. J. Linn. Soc. 2016, 178, 819–827. [Google Scholar] [CrossRef] [Green Version]
- Stec, D.; Morek, W.; Gąsiorek, P.; Michalczyk, Ł. Unmasking hidden species diversity within the Ramazzottius oberhaeuseri complex, with an integrative redescription of the nominal species for the family Ramazzottiidae (Tardigrada: Eutardigrada: Parachela). Syst. Biodivers. 2018, 16, 357–376. [Google Scholar] [CrossRef]
- Bertolani, R.; Rebecchi, L.; Giovannini, I.; Cesari, M. DNA barcoding and integrative taxonomy of Macrobiotus hufelandi C.A.S. Schultze 1834, the first tardigrade species to be described, and some related species. Zootaxa 2011, 2997, 19–36. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.I.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [Green Version]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2017, 20, 1160–1166. [Google Scholar] [CrossRef] [Green Version]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Gascuel, O. A simple, fast and accurate method to estimate large phylogenies by maximum-likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z. Molecular Evolution: A Statistical Approach; Oxford University Press: Oxford, UK, 2014; p. 512. [Google Scholar]
- Stamatakis, A.; Hoover, P.; Rougemont, J. A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 2008, 57, 758–771. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Templeton, A.R.; Crandall, K.A.; Sing, C.F. A cladistic analysis of phenotypic association with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram estimation. Genetics 1992, 132, 619–633. [Google Scholar] [CrossRef]
- Clement, M.; Posada, D.; Crandall, K. TCS: A computer program to estimate gene genealogies. Mol. Ecol. 2000, 9, 1657–1660. [Google Scholar] [CrossRef] [Green Version]
- Santos, A.M.; Cabezas, M.P.; Tavares, A.I.; Xavier, R.; Branco, M. tcsBU: A tool to extend TCS network layout and visualization. Bioinformatics 2016, 32, 627–628. [Google Scholar] [CrossRef] [Green Version]
- Hart, M.W.; Sunday, J. Things fall apart: Biological species form unconnected parsimony networks. Biol. Lett. 2007, 3, 509–512. [Google Scholar] [CrossRef] [Green Version]
- Puillandre, N.; Brouillet, S.; Achaz, G. ASAP: Assemble species by automatic partitioning. Mol. Ecol. Resour. 2021, 21, 609–620. [Google Scholar] [CrossRef]
- Zhang, J.; Kapli, P.; Pavlidis, P.; Stamatakis, A. A general species delimitation method with applications to phylogenetic placements. Bioinformatics 2013, 29, 2869–2876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rebecchi, L.; Altiero, T.; Guidetti, R.; Cesari, M.; Bertolani, R.; Negroni, M.; Rizzo, A.M. Tardigrade resistance to space effects: First results of experiments on the LIFE-TARSE mission on FOTON-M3 (September 2007). Astrobiology 2009, 9, 581–591. [Google Scholar] [CrossRef] [PubMed]
- Guidetti, R.; Altiero, T.; Bertolani, R.; Grazioso, P.; Rebecchi, L. Survival of freezing by hydrated tardigrades inhabiting terrestrial and freshwater habitats. Zoology 2011, 114, 123–128. [Google Scholar] [CrossRef]
- Stec, D.; Vončina, K.; Kristensen, R.M.; Michalczyk, Ł. The Macrobiotus ariekammensis species complex provides evidence for parallel evolution of claw elongation in macrobiotid tardigrades. Zool. J. Linn. Soc. 2022, zlab101. [Google Scholar] [CrossRef]
- Walz, B. Electron microscopic investigation of cephalic sense organs of the tardigrade Macrobiotus hufelandi C.A.S. Schulze. Zoomorphologie 1978, 89, 1–19. [Google Scholar] [CrossRef]
- Wiederhöft, H.; Greven, H. The cerebral ganglia of Milnesium tardigradum Doyère (Apochela, Tardigrada): Three dimensional reconstruction and notes on their ultrastructure. Zool. J. Linn. Soc. 1996, 116, 71–84. [Google Scholar] [CrossRef]
- Wiederhöft, H.; Greven, H. Notes on head sensory organs of Milnesium tardigradum Doyère, 1840 (Apochela, Eutardigrada). Zoologischer Anzeiger 1999, 238, 338–346. [Google Scholar]
- Biserova, N.M.; Kuznetsova, K.G. Head sensory organs of Halobiotus stenostomus (Eutardigrada, Hypsibiidae). Biol. Bull. 2012, 39, 579–589. [Google Scholar] [CrossRef]
- Guidetti, R.; Cesari, M.; Giovannini, I.; Ebel, C.; Förschler, M.I.; Rebecchi, L.; Schill, R.O. Morphology and taxonomy of the genus Ramazzottius (Eutardigrada; Ramazzottidae) with the integrative description of Ramazzottius kretschmanni sp. nov. Eur. Zool. J. 2022, 89, 339–363. [Google Scholar] [CrossRef]
- Dastych, H. Ramazzottius agannae sp. nov., a new tardigrade species from the nival zone of the Austrian Central Alps (Tardigrada). Ent. Mitt. Zool. Mus. Hambg. 2011, 15, 237–253. [Google Scholar]
- Kaczmarek, L.; Michalczyk, L.; Diduszko, D. Ramazzottius bunikowskae, a new species of Tardigrada (Eutardigrada, Hypsibiidae) from Russia. Zootaxa 2006, 1229, 49–57. [Google Scholar] [CrossRef] [Green Version]
- Tumanov, D.V. Integrative description of Mesobiotus anastasiae sp. nov. (Eutardigrada, Macrobiotoidea) and first record of Lobohalacarus (Chelicerata, Trombidiformes) from the Republic of South Africa. Eur. J. Taxon. 2020, 726, 102–131. [Google Scholar] [CrossRef]
- Jönsson, K.I.; Borsari, S.; Rebecchi, L. Anhydrobiotic survival in populations of the tardigrades Richtersius coronifer and Ramazzottius oberhaeuseri from Italy and Sweden. Zool. Anz. 2001, 240, 419–423. [Google Scholar] [CrossRef]
- Horikawa, D.D.; Higashi, S. Desiccation tolerance of the tardigrade Milnesium tardigradum collected in Sapporo, Japan, and Bogor, Indonesia. Zool. Sci. 2004, 21, 813–816. [Google Scholar] [CrossRef] [Green Version]
- Hengherr, S.; Worland, M.R.; Reuner, A.; Brümmer, F.; Schill, R.O. Freeze tolerance, supercooling points and ice formation: Comparative studies on the subzero temperature survival of limno-terrestrial tardigrades. J. Exp. Biol. 2009, 212, 802–807. [Google Scholar] [CrossRef] [Green Version]
- Zang, L.; Shimada, Y.; Miyake, H.; Nishimura, N. Transcriptome analysis of molecular response to UVC irradiation in zebrafish embryos. Ecotox. Environ. Saf. 2022, 231, 113211. [Google Scholar] [CrossRef]
- Jönsson, K.I.; Levine, E.B.; Wojcik, A.; Haghdoost, S.; Harms-Ringdahl, M. Environmental adaptations: Radiation tolerance. In Water Bears: The Biology of Tardigrades; Schill, R.O., Ed.; Springer: Cham, Switzerland, 2018; pp. 311–330. [Google Scholar]
- Hashimoto, T.; Kunieda, T. DNA protection protein, a novel mechanism of radiation tolerance: Lessons from Tardigrades. Life 2017, 7, 26. [Google Scholar] [CrossRef] [Green Version]
- May, R.M.; Maria, M.; Guimard, J. Actions différentielles des rayons X et Ultraviolets sur le tardigrade Macrobiotus areolatus, à l´état actif et desséché. Bull. Biol. Fr. Belg. 1964, 98, 349–367. [Google Scholar]
- United Nations Environment Programme. Environmental Effects and Interactions of Stratospheric Ozone Depletion, UV Radiation, and Climate Change: 2018 Assessment Report; Ozone Secretariat—The Secretariat for the Vienna Convention for the Protection of the Ozone Layer and for the Montreal Protocol on Substances that Deplete the Ozone Layer—United Nations Environment Programme: Nairobi, Kenya, 2018; p. 394. [Google Scholar]





| Specimen | Voucher Specimen Type | 18S | 28S | ITS-2 | cox1 |
| C4313 V2 | hologenophore | N/A | ON818306 | N/A | ON809460 |
| C4313 V3 | hologenophore | ON818312 | ON818307 | ON818300 | ON809461 |
| C4313 US2 | hologenophore | ON818313 | ON818308 | N/A | N/A |
| C4313 US3 | hologenophore | ON818314 | ON818309 | ON818301 | ON809462 |
| C4313 US4 | hologenophore | ON818315 | ON818310 | ON818302 | ON809463 |
| C4313 US5 | hologenophore | ON818316 | ON818311 | ON818303 | ON809464 |
| C4313 US6 | hologenophore | N/A | N/A | ON818304 | N/A |
| C4313 US7 | hologenophore | N/A | N/A | ON818305 | N/A |
| C4313 US8 | hologenophore | N/A | N/A | N/A | ON809465 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cesari, M.; Giovannini, I.; Altiero, T.; Guidetti, R.; Cornette, R.; Kikawada, T.; Rebecchi, L. Resistance to Extreme Stresses by a Newly Discovered Japanese Tardigrade Species, Macrobiotus kyoukenus (Eutardigrada, Macrobiotidae). Insects 2022, 13, 634. https://doi.org/10.3390/insects13070634
Cesari M, Giovannini I, Altiero T, Guidetti R, Cornette R, Kikawada T, Rebecchi L. Resistance to Extreme Stresses by a Newly Discovered Japanese Tardigrade Species, Macrobiotus kyoukenus (Eutardigrada, Macrobiotidae). Insects. 2022; 13(7):634. https://doi.org/10.3390/insects13070634
Chicago/Turabian StyleCesari, Michele, Ilaria Giovannini, Tiziana Altiero, Roberto Guidetti, Richard Cornette, Takahiro Kikawada, and Lorena Rebecchi. 2022. "Resistance to Extreme Stresses by a Newly Discovered Japanese Tardigrade Species, Macrobiotus kyoukenus (Eutardigrada, Macrobiotidae)" Insects 13, no. 7: 634. https://doi.org/10.3390/insects13070634
APA StyleCesari, M., Giovannini, I., Altiero, T., Guidetti, R., Cornette, R., Kikawada, T., & Rebecchi, L. (2022). Resistance to Extreme Stresses by a Newly Discovered Japanese Tardigrade Species, Macrobiotus kyoukenus (Eutardigrada, Macrobiotidae). Insects, 13(7), 634. https://doi.org/10.3390/insects13070634








