Pyrethroids in an AlphaFold2 Model of the Insect Sodium Channel
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. AlphaFold2 Models
2.2. Modeling Channel States with the Open PM and Resting VSMs
2.3. Residue Designations and 3D Alignment of Channel Structures
2.4. Energy Optimizations, Ligand Docking, and Visualization
3. Results and Discussions
3.1. The AF2 Model of AaNav1-1
3.2. Docking of PMT in the PyR1 Site in Model iAaNav1-1
3.3. Docking of DMT in the PyR1 and PyR2 Sites in Model oAaNav1-1
3.4. In Silico Inactivating oAaNav1-1a with Two DMT Molecules in the PyR1 and PyR2 Sites
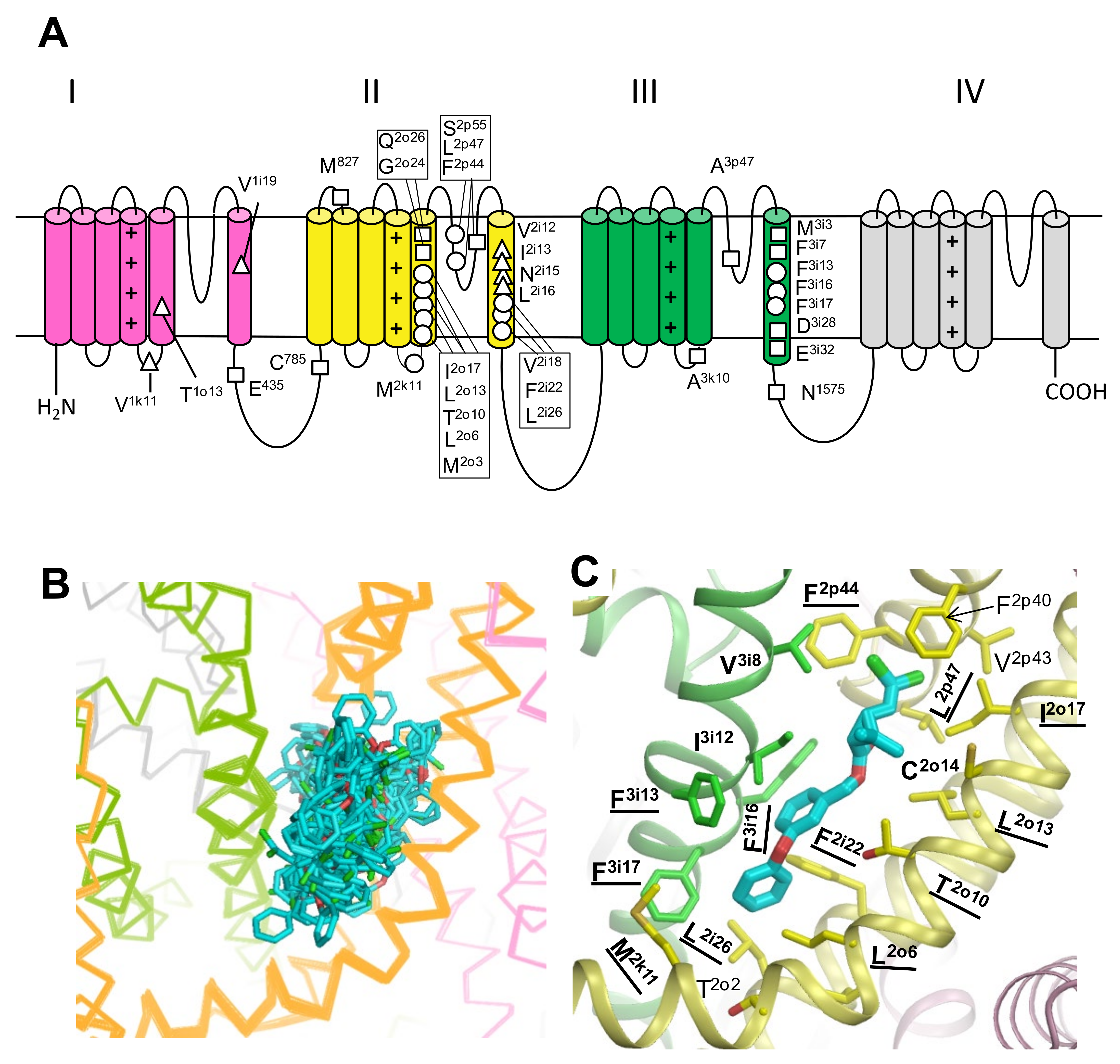

3.5. In Silico Deactivating VSM-I and VSM-II in oAaNav1-1 with DMT in the PyR1 and PyR2 Sites
3.6. Allosteric Effects on Pyrethroid Action of kdr Mutations beyond the PyR1 and PyR2 Sites
3.6.1. M827I in the VSM-II Linker IIS1-S2
3.6.2. G943A and Q945R in IIS6
3.6.3. S989/2p55P and D1763Y in P-Loops
3.6.4. F1538/3i17L, D1549/3i28V, and E1553/3i32G in IIIS6
3.7. Engineered Substitutions Not Reported as kdr Mutations
3.8. Cryo-EM Structure of NavPaS vs. AF2 Model of AaNav1-1
3.9. Limitations of Our Modeling Approach
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Narahashi, T. Mechanisms of action of pyretrhoids on sodium and calcium channel gating. Neuropharmacol. Pestic. Action 1986, 36–60. [Google Scholar]
- Narahashi, T. Neuronal ion channels as the target sites of insecticides. Pharmacol. Toxicol. 1996, 79, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Bloomquist, J.R. Ion channels as targets for insecticides. Ann. Rev. Entomol. 1996, 41, 163–190. [Google Scholar] [CrossRef] [PubMed]
- Voth, A.R.; Ho, P.S. The role of halogen bonding in inhibitor recognition and binding by protein kinases. Curr. Top. Med. Chem. 2007, 7, 1336–1348. [Google Scholar] [CrossRef] [PubMed]
- Dong, K.; Du, Y.; Rinkevich, F.; Nomura, Y.; Xu, P.; Wang, L.; Silver, K.; Zhorov, B.S. Molecular biology of insect sodium channels and pyrethroid resistance. Insect Biochem. Mol. Biol. 2014, 50, 1–17. [Google Scholar] [CrossRef]
- Soderlund, D. Sodium channels. In Comprehensive Molecular Insect Science; Gilbert, L.I., Latrou, K., Gill, S.S., Eds.; Elsevier: New York, NY, USA, 2005; Volume 5, pp. 1–24. [Google Scholar]
- Davies, T.G.; Field, L.M.; Usherwood, P.N.; Williamson, M.S. DDT, pyrethrins, pyrethroids and insect sodium channels. IUBMB Life 2007, 59, 151–162. [Google Scholar] [CrossRef]
- Rinkevich, F.D.; Du, Y.; Dong, K. Diversity and Convergence of Sodium Channel Mutations Involved in Resistance to Pyrethroids. Pestic. Biochem. Physiol. 2013, 106, 93–100. [Google Scholar] [CrossRef]
- O’Reilly, A.O.; Khambay, B.P.; Williamson, M.S.; Field, L.M.; Wallace, B.A.; Davies, T.G. Modelling insecticide-binding sites in the voltage-gated sodium channel. Biochem. J. 2006, 396, 255–263. [Google Scholar] [CrossRef]
- Long, S.B.; Campbell, E.B.; Mackinnon, R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science 2005, 309, 897–903. [Google Scholar] [CrossRef]
- Du, Y.; Nomura, Y.; Satar, G.; Hu, Z.; Nauen, R.; He, S.Y.; Zhorov, B.S.; Dong, K. Molecular evidence for dual pyrethroid-receptor sites on a mosquito sodium channel. Proc. Natl. Acad. Sci. USA 2013, 110, 11785–11790. [Google Scholar] [CrossRef]
- Du, Y.; Nomura, Y.; Zhorov, B.S.; Dong, K. Rotational Symmetry of Two Pyrethroid Receptor Sites in the Mosquito Sodium Channel. Mol. Pharmacol. 2015, 88, 273–280. [Google Scholar] [CrossRef]
- O’Reilly, A.O.; Williamson, M.S.; Gonzalez-Cabrera, J.; Turberg, A.; Field, L.M.; Wallace, B.A.; Davies, T.G. Predictive 3D modelling of the interactions of pyrethroids with the voltage-gated sodium channels of ticks and mites. Pest Manag. Sci. 2014, 70, 369–377. [Google Scholar] [CrossRef]
- Gosselin-Badaroudine, P.; Moreau, A.; Delemotte, L.; Cens, T.; Collet, C.; Rousset, M.; Charnet, P.; Klein, M.L.; Chahine, M. Characterization of the honeybee AmNaV1 channel and tools to assess the toxicity of insecticides. Sci. Rep. 2015, 5, 12475. [Google Scholar] [CrossRef]
- Wu, S.; Nomura, Y.; Du, Y.; Zhorov, B.S.; Dong, K. Molecular basis of selective resistance of the bumblebee BiNav1 sodium channel to tau-fluvalinate. Proc. Natl. Acad. Sci. USA 2017, 114, 12922–12927. [Google Scholar] [CrossRef]
- Itokawa, K.; Furutani, S.; Takaoka, A.; Maekawa, Y.; Sawabe, K.; Komagata, O.; Tomita, T.; de Lima Filho, J.L.; Alves, L.C.; Kasai, S. A first, naturally occurring substitution at the second pyrethroid receptor of voltage-gated sodium channel of Aedes aegypti. Pest Manag. Sci. 2021, 77, 2887–2893. [Google Scholar] [CrossRef]
- Sun, H.; Nomura, Y.; Du, Y.; Liu, Z.; Zhorov, B.S.; Dong, K. Two kdr mutations at the predicted second pyrethroid receptor site in sodium channels of Aedes aegypti and Nilaparvata lugens. Insect Biochem. Mol. Biol. 2022; in press. [Google Scholar] [CrossRef]
- Jiang, D.; Banh, R.; Gamal El-Din, T.M.; Tonggu, L.; Lenaeus, M.J.; Pomes, R.; Zheng, N.; Catterall, W.A. Open-state structure and pore gating mechanism of the cardiac sodium channel. Cell 2021, 184, 5151–5162.e11. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Callaway, E. ‘It will change everything’: DeepMind’s AI makes gigantic leap in solving protein structures. Nature 2020, 588, 203–204. [Google Scholar] [CrossRef]
- Korkosh, V.S.; Kiselev, A.M.; Mikhaylov, E.N.; Kostareva, A.A.; Zhorov, B.S. Atomic Mechanisms of Timothy Syndrome-Associated Mutations in Calcium Channel Cav1.2. Front. Physiol. 2019, 10, 335. [Google Scholar] [CrossRef]
- Zhorov, B.S.; Du, Y.; Song, W.; Luo, N.; Gordon, D.; Gurevitz, M.; Dong, K. Mapping the interaction surface of scorpion beta-toxins with an insect sodium channel. Biochem. J. 2021, 478, 2843–2869. [Google Scholar] [CrossRef]
- Zhorov, B.S.; Tikhonov, D.B. Potassium, sodium, calcium and glutamate-gated channels: Pore architecture and ligand action. J. Neurochem. 2004, 88, 782–799. [Google Scholar] [CrossRef]
- Zhorov, B.S. Vector method for calculating derivatives of energy of atom-atom interactions of complex molecules according to generalized coordiantes. J. Struct. Chem. 1981, 22, 4–8. [Google Scholar] [CrossRef]
- Garden, D.P.; Zhorov, B.S. Docking flexible ligands in proteins with a solvent exposure- and distance-dependent dielectric function. J. Comput. Aided Mol. Des. 2010, 24, 91–105. [Google Scholar] [CrossRef]
- Li, Z.; Scheraga, H.A. Monte Carlo-minimization approach to the multiple-minima problem in protein folding. Proc. Natl. Acad. Sci. USA 1987, 84, 6611–6615. [Google Scholar] [CrossRef]
- Tikhonov, D.B.; Zhorov, B.S. Mechanism of sodium channel block by local anesthetics, antiarrhythmics, and anticonvulsants. J. Gen. Physiol. 2017, 149, 465–481. [Google Scholar] [CrossRef]
- Shen, H.; Zhou, Q.; Pan, X.; Li, Z.; Wu, J.; Yan, N. Structure of a eukaryotic voltage-gated sodium channel at near-atomic resolution. Science 2017, 355. [Google Scholar] [CrossRef]
- Jiang, D.; Shi, H.; Tonggu, L.; Gamal El-Din, T.M.; Lenaeus, M.J.; Zhao, Y.; Yoshioka, C.; Zheng, N.; Catterall, W.A. Structure of the Cardiac Sodium Channel. Cell 2020, 180, 122–134.e10. [Google Scholar] [CrossRef]
- Auffinger, P.; Hays, F.A.; Westhof, E.; Ho, P.S. Halogen bonds in biological molecules. Proc. Natl. Acad. Sci. USA 2004, 101, 16789–16794. [Google Scholar] [CrossRef]
- Shen, H.; Liu, D.; Wu, K.; Lei, J.; Yan, N. Structures of human Nav1.7 channel in complex with auxiliary subunits and animal toxins. Science 2019, 363, 1303–1308. [Google Scholar] [CrossRef]
- Hopkins, B.W.; Pietrantonio, P.V. The Helicoverpa zea (Boddie) (Lepidoptera: Noctuidae) voltage-gated sodium channel and mutations associated with pyrethroid resistance in field-collected adult males. Insect Biochem. Mol. Biol. 2010, 40, 385–393. [Google Scholar] [CrossRef]
- Yoon, K.S.; Kwon, D.H.; Strycharz, J.P.; Hollingsworth, C.S.; Lee, S.H.; Clark, J.M. Biochemical and molecular analysis of deltamethrin resistance in the common bed bug (Hemiptera: Cimicidae). J. Med. Entomol. 2008, 45, 1092–1101. [Google Scholar] [CrossRef]
- Singh, O.P.; Dykes, C.L.; Das, M.K.; Pradhan, S.; Bhatt, R.M.; Agrawal, O.P.; Adak, T. Presence of two alternative kdr-like mutations, L1014F and L1014S, and a novel mutation, V1010L, in the voltage gated Na+ channel of Anopheles culicifacies from Orissa, India. Malar. J. 2010, 9, 146. [Google Scholar] [CrossRef] [PubMed]
- Brengues, C.; Hawkes, N.J.; Chandre, F.; McCarroll, L.; Duchon, S.; Guillet, P.; Manguin, S.; Morgan, J.C.; Hemingway, J. Pyrethroid and DDT cross-resistance in Aedes aegypti is correlated with novel mutations in the voltage-gated sodium channel gene. Med. Vet. Entomol. 2003, 17, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Saavedra-Rodriguez, K.; Urdaneta-Marquez, L.; Rajatileka, S.; Moulton, M.; Flores, A.E.; Fernandez-Salas, I.; Bisset, J.; Rodriguez, M.; McCall, P.J.; Donnelly, M.J.; et al. A mutation in the voltage-gated sodium channel gene associated with pyrethroid resistance in Latin American Aedes aegypti. Insect Mol. Biol. 2007, 16, 785–798. [Google Scholar] [CrossRef] [PubMed]
- Tan, W.L.; Li, C.X.; Wang, Z.M.; Liu, M.D.; Dong, Y.D.; Feng, X.Y.; Wu, Z.M.; Guo, X.X.; Xing, D.; Zhang, Y.M.; et al. First detection of multiple knockdown resistance (kdr)-like mutations in voltage-gated sodium channel using three new genotyping methods in Anopheles sinensis from Guangxi Province, China. J. Med. Entomol. 2012, 49, 1012–1020. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Torres, D.; Chandre, F.; Williamson, M.S.; Darriet, F.; Berge, J.B.; Devonshire, A.L.; Guillet, P.; Pasteur, N.; Pauron, D. Molecular characterization of pyrethroid knockdown resistance (kdr) in the major malaria vector Anopheles gambiae s.s. Insect Mol. Biol. 1998, 7, 179–184. [Google Scholar] [CrossRef] [PubMed]
- Stump, A.D.; Atieli, F.K.; Vulule, J.M.; Besansky, N.J. Dynamics of the pyrethroid knockdown resistance allele in western Kenyan populations of Anopheles gambiae in response to insecticide-treated bed net trials. Am. J. Trop. Med. Hyg. 2004, 70, 591–596. [Google Scholar] [CrossRef]
- Kim, H.; Baek, J.H.; Lee, W.-J.; Lee, S.-H. Frequency detection of pyrethroid resistance allele in Anopheles sinensis populations by real-time PCR amplification of specific allele (rtPASA). Pestic. Biochem. Physiol. 2007, 87, 54–61. [Google Scholar] [CrossRef]
- Cassanelli, S.; Cerchiari, B.; Giannini, S.; Bizzaro, D.; Mazzoni, E.; Manicardi, G.C. Use of the RFLP-PCR diagnostic test for characterizing MACE and kdr insecticide resistance in the peach potato aphid Myzus persicae. Pest Manag. Sci. 2005, 61, 91–96. [Google Scholar] [CrossRef]
- Jones, C.M.; Liyanapathirana, M.; Agossa, F.R.; Weetman, D.; Ranson, H.; Donnelly, M.J.; Wilding, C.S. Footprints of positive selection associated with a mutation (N1575Y) in the voltage-gated sodium channel of Anopheles gambiae. Proc. Natl. Acad. Sci. USA 2012, 109, 6614–6619. [Google Scholar] [CrossRef]
- Liu, Z.; Valles, S.M.; Dong, K. Novel point mutations in the German cockroach para sodium channel gene are associated with knockdown resistance (kdr) to pyrethroid insecticides. Insect Biochem. Mol. Biol. 2000, 30, 991–997. [Google Scholar] [CrossRef]
- Guerrero, F.D.; Jamroz, R.C.; Kammlah, D.; Kunz, S.E. Toxicological and molecular characterization of pyrethroid-resistant horn flies, Haematobia irritans: Identification of kdr and super-kdr point mutations. Insect Biochem. Mol. Biol. 1997, 27, 745–755. [Google Scholar] [CrossRef]
- Wu, M.; Gotoh, H.; Waters, T.; Walsh, D.B.; Lavine, L.C. Identification of an alternative knockdown resistance (kdr)-like mutation, M918L, and a novel mutation, V1010A, in the Thrips tabaci voltage-gated sodium channel gene. Pest Manag. Sci. 2013. [Google Scholar] [CrossRef]
- Sonoda, S.; Tsukahara, Y.; Ashfaq, M.; Tsumuki, H. Genomic organization of the para-sodium channel alpha-subunit genes from the pyrethroid-resistant and -susceptible strains of the diamondback moth. Arch. Insect Biochem. Physiol. 2008, 69, 1–12. [Google Scholar] [CrossRef]
- Forcioli, D.; Frey, B.; Frey, J.E. High nucleotide diversity in the para-like voltage-sensitive sodium channel gene sequence in the western flower thrips (Thysanoptera: Thripidae). J. Econ. Entomol. 2002, 95, 838–848. [Google Scholar] [CrossRef]
- Sonoda, S. Molecular analysis of pyrethroid resistance conferred by target insensitivity and increased metabolic detoxification in Plutella xylostella. Pest Manag. Sci. 2010, 66, 572–575. [Google Scholar] [CrossRef]
- Bass, C.; Schroeder, I.; Turberg, A.; Field, L.M.; Williamson, M.S. Identification of mutations associated with pyrethroid resistance in the para-type sodium channel of the cat flea, Ctenocephalides felis. Insect Biochem. Mol. Biol. 2004, 34, 1305–1313. [Google Scholar] [CrossRef]
- Rinkevich, F.D.; Su, C.; Lazo, T.; Hawthorne, D.; Tingey, W.; Naimov, S.; Scott, J.G. Multiple evolutionary origins of knockdown resistance (kdr) in pyrethroid-resistant Colorado potato beetle, Leptinotarsa decemlineata. Pestic. Biochem. Physiol. 2012, 104, 192–200. [Google Scholar] [CrossRef]
- Marshall, K.L.; Moran, C.; Chen, Y.; Herron, G.A. Detection of kdr pyrethroid resistance in the cotton aphid, Aphis gossypii (Hemiptera: Aphididae), using a PCR-RFLP assay. J. Pestic. Sci. 2012, 37, 169–172. [Google Scholar] [CrossRef]
- Carletto, J.; Martin, T.; Vanlerberghe-Masutti, F.; Brevault, T. Insecticide resistance traits differ among and within host races in Aphis gossypii. Pest Manag. Sci. 2009, 66, 301–307. [Google Scholar] [CrossRef]
- Karatolos, N.; Gorman, K.; Williamson, M.S.; Denholm, I. Mutations in the sodium channel associated with pyrethroid resistance in the greenhouse whitefly, Trialeurodes vaporariorum. Pest Manag. Sci. 2012, 68, 834–838. [Google Scholar] [CrossRef]
- Morin, S.; Williamson, M.S.; Goodson, S.J.; Brown, J.K.; Tabashnik, B.E.; Dennehy, T.J. Mutations in the Bemisia tabaci para sodium channel gene associated with resistance to a pyrethroid plus organophosphate mixture. Insect Biochem. Mol. Biol. 2002, 32, 1781–1791. [Google Scholar] [CrossRef]
- Gonzalez-Cabrera, J.; Davies, T.G.; Field, L.M.; Kennedy, P.J.; Williamson, M.S. An Amino Acid Substitution (L925V) Associated with Resistance to Pyrethroids in Varroa destructor. PLoS ONE 2013, 8, e82941. [Google Scholar] [CrossRef]
- Toda, S.; Morishita, M. Identification of three point mutations on the sodium channel gene in pyrethroid-resistant Thrips tabaci (Thysanoptera: Thripidae). J. Econ. Entomol. 2009, 102, 2296–2300. [Google Scholar] [CrossRef]
- Lee, S.H.; Smith, T.J.; Ingles, P.J.; Soderlund, D.M. Cloning and functional characterization of a putative sodium channel auxiliary subunit gene from the house fly (Musca domestica). Insect Biochem. Mol. Biol. 2000, 30, 479–487. [Google Scholar] [CrossRef]
- Hodgdon, H.E.; Yoon, K.S.; Previte, D.J.; Kim, H.J.; Aboelghar, G.E.; Lee, S.H.; Clark, J.M. Determination of knockdown resistance allele frequencies in global human head louse populations using the serial invasive signal amplification reaction. Pest Manag. Sci. 2010, 66, 1031–1040. [Google Scholar] [CrossRef]
- Roditakis, E.; Tsagkarakou, A.; Vontas, J. Identification of mutations in the para sodium channel of Bemisia tabaci from Crete, associated with resistance to pyrethroids. Pestic. Biochem. Physiol. 2006, 85, 161–166. [Google Scholar] [CrossRef]
- Srisawat, R.; Komalamisra, N.; Eshita, Y.; Zheng, M.; Ono, K.; Itoh, T.Q.; Matsumoto, A.; Petmitr, S.; Rongsriyam, Y. Point mutations in domain II of the voltage-gated sodium channel gene in deltamethrin-resistant Aedes aegypti (Diptera: Culicidae). Appl. Entomol. Zool. 2010, 45, 275–282. [Google Scholar] [CrossRef]
- Chang, C.; Shen, W.K.; Wang, T.T.; Lin, Y.H.; Hsu, E.L.; Dai, S.M. A novel amino acid substitution in a voltage-gated sodium channel is associated with knockdown resistance to permethrin in Aedes aegypti. Insect Biochem. Mol. Biol. 2009, 39, 272–278. [Google Scholar] [CrossRef]
- Pridgeon, J.W.; Appel, A.G.; Moar, W.J.; Liu, N. Variability of resistance mechanisms in pyrethroid resistant German cockroaches (Dictyoptera: Blattellidae). Pestic. Biochem. Physiol. 2002, 73, 149–156. [Google Scholar] [CrossRef]
- Kwon, D.H.; Clark, J.M.; Lee, S.H. Cloning of a sodium channel gene and identification of mutations putatively associated with fenpropathrin resistance in Tetranychus urticae. Pestic. Biochem. Physiol. 2010, 97, 93–100. [Google Scholar] [CrossRef]
- Kawada, H.; Higa, Y.; Komagata, O.; Kasai, S.; Tomita, T.; Thi Yen, N.; Loan, L.L.; Sanchez, R.A.; Takagi, M. Widespread distribution of a newly found point mutation in voltage-gated sodium channel in pyrethroid-resistant Aedes aegypti populations in Vietnam. PLoS Negl. Trop. Dis. 2009, 3, e527. [Google Scholar] [CrossRef] [PubMed]
- Kushwah, R.B.S.; Kaur, T.; Dykes, C.L.; Ravi Kumar, H.; Kapoor, N.; Singh, O.P. A new knockdown resistance (kdr) mutation, F1534L, in the voltage-gated sodium channel of Aedes aegypti, co-occurring with F1534C, S989P and V1016G. Parasit. Vectors 2020, 13, 327. [Google Scholar] [CrossRef] [PubMed]
- Katsavou, E.; Vlogiannitis, S.; Karp-Tatham, E.; Blake, D.P.; Ilias, A.; Strube, C.; Kioulos, I.; Dermauw, W.; Van Leeuwen, T.; Vontas, J. Identification and geographical distribution of pyrethroid resistance mutations in the poultry red mite Dermanyssus gallinae. Pest Manag. Sci. 2020, 76, 125–133. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Chen, A.C.; Davey, R.B.; Ivie, G.W.; George, J.E. Identification of a point mutation in the para-type sodium channel gene from a pyrethroid-resistant cattle tick. Biochem. Biophys. Res. Commun. 1999, 261, 558–561. [Google Scholar] [CrossRef]
- Usherwood, P.N.; Davies, T.G.; Mellor, I.R.; O’Reilly, A.O.; Peng, F.; Vais, H.; Khambay, B.P.; Field, L.M.; Williamson, M.S. Mutations in DIIS5 and the DIIS4-S5 linker of Drosophila melanogaster sodium channel define binding domains for pyrethroids and DDT. FEBS Lett. 2007, 581, 5485–5492. [Google Scholar] [CrossRef]
- Du, Y.; Lee, J.E.; Nomura, Y.; Zhang, T.; Zhorov, B.S.; Dong, K. Identification of a cluster of residues in transmembrane segment 6 of domain III of the cockroach sodium channel essential for the action of pyrethroid insecticides. Biochem. J. 2009, 419, 377–385. [Google Scholar] [CrossRef]
- Soderlund, D.M. State-Dependent Modification of Voltage-Gated Sodium Channels by Pyrethroids. Pestic. Biochem. Physiol. 2010, 97, 78–86. [Google Scholar] [CrossRef]
- Vais, H.; Williamson, M.S.; Goodson, S.J.; Devonshire, A.L.; Warmke, J.W.; Usherwood, P.N.; Cohen, C.J. Activation of Drosophila sodium channels promotes modification by deltamethrin. Reductions in affinity caused by knock-down resistance mutations. J. Gen. Physiol. 2000, 115, 305–318. [Google Scholar] [CrossRef]
- Ford, M.C.; Saxton, M.; Ho, P.S. Sulfur as an Acceptor to Bromine in Biomolecular Halogen Bonds. J. Phys. Chem. Lett. 2017, 8, 4246–4252. [Google Scholar] [CrossRef]
- Lee, S.H.; Smith, T.J.; Knipple, D.C.; Soderlund, D.M. Mutations in the house fly Vssc1 sodium channel gene associated with super-kdr resistance abolish the pyrethroid sensitivity of Vssc1/tipE sodium channels expressed in Xenopus oocytes. Insect Biochem. Mol. Biol. 1999, 29, 185–194. [Google Scholar] [CrossRef]
- Park, Y.; Taylor, M.F.; Feyereisen, R. A valine421 to methionine mutation in IS6 of the hscp voltage-gated sodium channel associated with pyrethroid resistance in Heliothis virescens F. Biochem. Biophys. Res. Commun. 1997, 239, 688–691. [Google Scholar] [CrossRef]
- Xu, Q.; Zhang, L.; Li, T.; Zhang, L.; He, L.; Dong, K.; Liu, N. Evolutionary adaptation of the amino acid and codon usage of the mosquito sodium channel following insecticide selection in the field mosquitoes. PLoS ONE 2012, 7, e47609. [Google Scholar] [CrossRef]
- Pittendrigh, B.; Reenan, R.; ffrench-Constant, R.H.; Ganetzky, B. Point mutations in the Drosophila sodium channel gene para associated with resistance to DDT and pyrethroid insecticides. Mol. Gen. Genet. 1997, 256, 602–610. [Google Scholar] [CrossRef]
- Kristensen, M. Identification of sodium channel mutations in human head louse (Anoplura: Pediculidae) from Denmark. J. Med. Entomol. 2005, 42, 826–829. [Google Scholar] [CrossRef]
- Fallang, A.; Denholm, I.; Horsberg, T.E.; Williamson, M.S. Novel point mutation in the sodium channel gene of pyrethroid-resistant sea lice Lepeophtheirus salmonis (Crustacea: Copepoda). Dis. Aquat. Organ. 2005, 65, 129–136. [Google Scholar] [CrossRef]
- Wang, R.; Liu, Z.; Dong, K.; Elzen, P.J.; Pettis, J.; Huang, Z. Association of novel mutations in a sodium channel gene with fluvinate resistance in the mite, Varroa destructor. J. Apic. Res. 2002, 41, 17–25. [Google Scholar] [CrossRef]
- Head, D.J.; McCaffery, A.R.; Callaghan, A. Novel mutations in the para-homologous sodium channel gene associated with phenotypic expression of nerve insensitivity resistance to pyrethroids in Heliothine lepidoptera. Insect Mol. Biol. 1998, 7, 191–196. [Google Scholar] [CrossRef]
- Tsagkarakou, A.; Van Leeuwen, T.; Khajehali, J.; Ilias, A.; Grispou, M.; Williamson, M.S.; Tirry, L.; Vontas, J. Identification of pyrethroid resistance associated mutations in the para sodium channel of the two-spotted spider mite Tetranychus urticae (Acari: Tetranychidae). Insect Mol. Biol. 2009, 18, 583–593. [Google Scholar] [CrossRef]
- Du, Y.; Song, W.; Groome, J.R.; Nomura, Y.; Luo, N.; Dong, K. A negative charge in transmembrane segment 1 of domain II of the cockroach sodium channel is critical for channel gating and action of pyrethroid insecticides. Toxicol. Appl. Pharmacol. 2010, 247, 53–59. [Google Scholar] [CrossRef]
- Du, Y.; Nomura, Y.; Zhorov, B.S.; Dong, K. Sodium Channel Mutations and Pyrethroid Resistance in Aedes aegypti. Insects 2016, 7, 60. [Google Scholar] [CrossRef]
- Zhorov, B.S. Possible Mechanism of Ion Selectivity in Eukaryotic Voltage-Gated Sodium Channels. J. Phys. Chem. B 2021, 125, 2074–2088. [Google Scholar] [CrossRef]
- Chen, M.; Du, Y.; Nomura, Y.; Zhu, G.; Zhorov, B.S.; Dong, K. Alanine to valine substitutions in the pore helix IIIP1 and linker-helix IIIL45 confer cockroach sodium channel resistance to DDT and pyrethroids. Neurotoxicology 2017, 60, 197–206. [Google Scholar] [CrossRef]
- Chen, M.; Du, Y.; Nomura, Y.; Zhu, G.; Zhorov, B.S.; Dong, K. Mutations of two acidic residues at the cytoplasmic end of segment IIIS6 of an insect sodium channel have distinct effects on pyrethroid resistance. Insect Biochem. Mol. Biol. 2017, 82, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Du, Y.; Wu, S.; Nomura, Y.; Zhu, G.; Zhorov, B.S.; Dong, K. Molecular evidence of sequential evolution of DDT- and pyrethroid-resistant sodium channel in Aedes aegypti. PLoS Negl. Trop. Dis. 2019, 13, e0007432. [Google Scholar] [CrossRef]
- Nguyen, P.T.; DeMarco, K.R.; Vorobyov, I.; Clancy, C.E.; Yarov-Yarovoy, V. Structural basis for antiarrhythmic drug interactions with the human cardiac sodium channel. Proc. Natl. Acad. Sci. USA 2019, 116, 2945–2954. [Google Scholar] [CrossRef]
- Jiang, D.; Tonggu, L.; Gamal El-Din, T.M.; Banh, R.; Pomes, R.; Zheng, N.; Catterall, W.A. Structural basis for voltage-sensor trapping of the cardiac sodium channel by a deathstalker scorpion toxin. Nat. Commun. 2021, 12, 128. [Google Scholar] [CrossRef] [PubMed]
- Buyan, A.; Sun, D.; Corry, B. Protonation state of inhibitors determines interaction sites within voltage-gated sodium channels. Proc. Natl. Acad. Sci. USA 2018, 115, E3135–E3144. [Google Scholar] [CrossRef] [PubMed]
- Mazola, Y.; Marquez Montesinos, J.C.E.; Ramirez, D.; Zuniga, L.; Decher, N.; Ravens, U.; Yarov-Yarovoy, V.; Gonzalez, W. Common Structural Pattern for Flecainide Binding in Atrial-Selective Kv1.5 and Nav1.5 Channels: A Computational Approach. Pharmaceutics 2022, 14, 1356. [Google Scholar] [CrossRef] [PubMed]




| Mutation 1 | Location | Species | Reference | Figure |
|---|---|---|---|---|
| V253/1k11F | PyR2 | Aedes aegypti | [16] | 2b,d |
| T267/1o13A | PyR2 | Plant hopper | [17] | 2b,d |
| V410/1i19M | PyR2 | Helicoverpa zea | [32] | 2b,d |
| V410/1i19L | PyR2 | Cimex luctularis | [33] | 2b,d |
| V410/1i19G/A | PyR2 | Helicoverpa zea | [32] | 2b,d |
| V1010/2i12L + L1014/2i16S | PyR2 + PyR2 | Anopheles culicifacies | [34] | 2b,d |
| I1011/2i13M | PyR2 | Aedes aegypti | [35] | 2b,d |
| I1011/2i13V | PyR2 | Aedes aegypti | [36] | 2b,d |
| N1013/2i15S | PyR2 | Anopheles sinensis | [37] | 2b,d |
| L1014/2i16F | PyR2 | Anopheles gambiae | [38] | 2b,d |
| L1014/2i16S | PyR2 | Anopheles arabiensis | [39] | 2b,d |
| L1014/2i16H | PyR2 | Helicoverpa zea | [32] | 2b,d |
| L1014/2i16C | PyR2 | Anopheles sinensis | [40] | 2b,d |
| L1014/2i16W | PyR2 | Anopheles sinensis | [37] | 2b,d |
| L1014/2i16F + F979S | PyR2 + IIP1 | Myzus persicae | [41] | 2b,d |
| L1014/2i16F + N1575Y | PyR2 + III-IV | Anopheles gambiae | [42] | 2b,d |
| L1014/2i16F+E435K+C785R | PyR2 | Blattella germanica | [43] | 2b,d |
| M918/2k11T + L1014/2i16F | PyR1 + PyR2 | Haematobia i. irritans | [44] | 1c, 4a,b,d |
| M918/2k11L + V1010/2i12A | PyR1 + PyR2 | Thrips tabaci | [45] | 1c, 2a |
| M918/2k11I + L1014/2i16F | PyR1 + PyR2 | Plutella xylostella | [46] | 1c, 2a,b,d |
| T929/2o10I + L1014/2i16F | PyR1 + PyR2 | Frankliniella occidentalis | [47] | 1c, 2a,b,d |
| T929/2o10I + L1014/2i16F | PyR1 + PyR2 | Plutella xylostella | [48] | 1c, 2a,b,d |
| T929/2o10C + L1014/2i16F | PyR1 + PyR2 | Frankliniella occidentalis | [47] | 1c, 2a |
| T929/2o10V + L1014/2i16F | PyR1 + PyR2 | Ctenocephalides felis | [49] | 1c, 2a |
| T929/2o10N + L1014/2i16F | PyR1 + PyR2 | L. decemlineata | [50] | 1c, 2a,b,d |
| F979/2p44S + L1014/2i16F | PyR1 + PyR2 | Myzus persicae | [41] | 1c, 2a,b,d |
| M918/2k11T | PyR1 | Aphis gossypii | [51] | 1c, 2a |
| M918/2k11L | PyR1 | Aphis gossypii | [52] | 1c, 2a |
| M918/2k11L + L925/2o6I | PyR1 + PyR1 | Trialeurodesvaporariorum | [53] | 1c, 2a |
| M918/2k11V | PyR1 | Bemisia tabaci | [54] | 1c, 2a |
| L925/2o6I | PyR1 | Bemisia tabaci | [54] | 1c, 2a |
| L925/2o6V | PyR1 | Varroa destructor | [55] | 1c, 2a |
| T929/2o10I | PyR1 | Thrips tabaci | [56] | 1c, 2a |
| T929/2o10I + L932/2o13F | PyR1 + PyR1 | P. humanus capitis | [57] | 1c, 2a |
| T929/2o10I + M827I | PyR1 + VSM2 | P. humanus capitis | [58] | 1c, 2a |
| T929/2o10C | PyR1 | Frankliniella occidentalis | [47] | 1c, 2a |
| T929/2o10V | PyR1 | Bemisia tabaci | [59] | 1c, 2a |
| L932/2o13F + M827I | PyR1 + VSM2 | P. humanus capitis | [58] | 1c, 2a |
| I936/2o17V | PyR1 | Helicoverpa zea | [32] | 1c, 2a |
| L982/2p47W | PyR1 | Aedes aegypti | [35] | 1c |
| V1016/2i18G | PyR1 | Aedes aegypti | [35] | 2a,c |
| V1016/2i18G + S989/2p55P | PyR1 | Aedes aegypti | [60] | 2a,c |
| V1016/2i18G + D1763Y | PyR1 | Aedes aegypti | [61] | 2a,c |
| V1016/2i18I | PyR1 | Aedes aegypti | [36] | 2a,c |
| F1020/2i22S | PyR1 | Blattella germanica | [62] | 2a |
| L1024/2i26V | PyR1 | Tetranychus urticae | [63] | 1c, S5b |
| F1534/3i13C | PyR1 | Aedes aegypti | [64] | 1c, 2a |
| F1534/3i13L | PyR1 | Aedes aegypti | [65] | 1c, 2a |
| F1537/3i16L | PyR1 | Dermanyssus gallinae | [66] | 1c |
| F1538/3i17I | PyR1 | Rhipicephalus microplus | [67] | 1c, 2a |
| Mutation 1 | Location | Channel | Reference | Figure |
|---|---|---|---|---|
| L260/1o6A | PyR2 | AaNav1-1 | [12] | 2b,d |
| I264/1o10C | PyR2 | AaNav1-1 | [11] | 2b,d |
| I413/1i22A | PyR2 | AaNav1-1 | [12] | 2b,d |
| M918/2k11T | PyR1 | DmNav | [68] | 4c,2a,c |
| V922/2o3I a | PyR1 | BiNav1-1 | [15] | 6c |
| L925/2o6I | PyR1 | DmNav | [68] | 1c |
| T929/2o10 I | PyR1 | DmNav | [68] | 1c,2a |
| L932/2o13F | PyR1 | DmNav | [68] | 1c,2a |
| C933/2o14A | PyR1 | DmNav | [68] | 1c,2a |
| I936/2o17V | PyR1 | DmNav | [68] | 1c,2a |
| N1013/2i15S | PyR2 | AaNav1-1 | [12] | 2b,d |
| F1020/2i22S | PyR1 | AaNav1-1 | [12] | 1c,2a |
| L1023/2i25A | PyR1 | AaNav1-1 | [12] | 2a |
| L1024/2i26A | PyR1 | AaNav1-1 | [12] | 1c |
| F1526/3i5L | PyR1 (τFVL) | BiNav1-1 | [15] | 6c |
| V1529/3i8A | PyR1 (τFVL) | BiNav1-1 | [15] | 1c,2a |
| I1533/3i12A | PyR1 | BgNav1-1a | [69] | 1c,2a |
| F1537/3i16A | PyR1 | BgNav1-1a | [69] | 2a |
| Mutation 1 | Species | Reference | Impact | Figure |
|---|---|---|---|---|
| D59G | Blattella germanica | [43] | ||
| A99S | Culex quinquefasciatus | [75] | ||
| I254/1k12N | Drosophila melanogaster | [76] | ||
| E435/1i45K | Blattella germanica | [43] | ||
| C785R | Blattella germanica | [43] | ||
| M827/IIS1-S2I | P. humanus capitis | [58] | PyR1 | 5a |
| G943/2o24A | P. humanus capitis | [77] | PyR1, PyR2 | 5b |
| Q945/2o26R | L. salmonis | [78] | PyR2 | 5b |
| S989/2p55P + V1016/2i18G | Aedes aegypti | [60] | IIP1-P2 + PyR1 | 5c, 2a,c |
| A1101T + P1879/CTDS | Plutella xylostella | [46] | II/III + III/IV | |
| A1410/3k10V | Drosophila melanogaster | [76] | PyR1 | 6a |
| A1494/3p47V | Drosophila melanogaster | [76] | PyR1 | 6b |
| M1524/3i3I | Drosophila melanogaster | [76] | PyR1 | 6c |
| F1528/3i7L + M1823/4i3I | Varroa destructor | [79] | PyR1 + IVS6 | 6d |
| D1549/3i28V + E1553/3i32G | Helicoverpa armigera | [80] | PyR1, PyR2 | 6e |
| A1215D + F1538/3i17I | Tetranychus urticae | [81] | II/III + PyR1 | |
| W1594R | Culex quinquefasciatus | [75] | N-end of IVS0 |
| Mutation a | Species | Reference | Impact | Figure |
|---|---|---|---|---|
| I249/1k7A | Aedes aegypti | [11] | PyR2, PyR1 | 5d |
| S420/1i29A | Aedes aegypti | [12] | PyR1 | 5e |
| D823G/A/K | Blattella germanica | [82] | PyR1 | 5a |
| L914/2k7F/I | Drosophila melanogaster | [68] | PyR1 | 6f |
| N927/2o8I | Drosophila melanogaster | [68] | PyR1, PyR2 | 5f |
| A1410/3k10 V | Blattella germanica | [85] | PyR1 | 6a |
| A1494/3p47V | Blattella germanica | [85] | PyR1 | 6b |
| G1535/3i14A b | Blattella germanica | [69] | PyR1 | |
| N1541/3i20A | Blattella germanica | [69] | PyR1 | |
| D1549/3i28V | Blattella germanica | [86] | 6e |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhorov, B.S.; Dong, K. Pyrethroids in an AlphaFold2 Model of the Insect Sodium Channel. Insects 2022, 13, 745. https://doi.org/10.3390/insects13080745
Zhorov BS, Dong K. Pyrethroids in an AlphaFold2 Model of the Insect Sodium Channel. Insects. 2022; 13(8):745. https://doi.org/10.3390/insects13080745
Chicago/Turabian StyleZhorov, Boris S., and Ke Dong. 2022. "Pyrethroids in an AlphaFold2 Model of the Insect Sodium Channel" Insects 13, no. 8: 745. https://doi.org/10.3390/insects13080745
APA StyleZhorov, B. S., & Dong, K. (2022). Pyrethroids in an AlphaFold2 Model of the Insect Sodium Channel. Insects, 13(8), 745. https://doi.org/10.3390/insects13080745







