Effects of Flupyradifurone and Two Reference Insecticides Commonly Used in Toxicological Studies on the Larval Proteome of the Honey bee Apis mellifera
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Honey Bees
2.2. Honey Bee Brood Test
2.3. Protein Analysis
2.3.1. Preparation of Protein Extracts and Two-Dimensional PAGE (2D-PAGE)
2.3.2. Protein Picking and Identification
2.4. Statistical Analysis
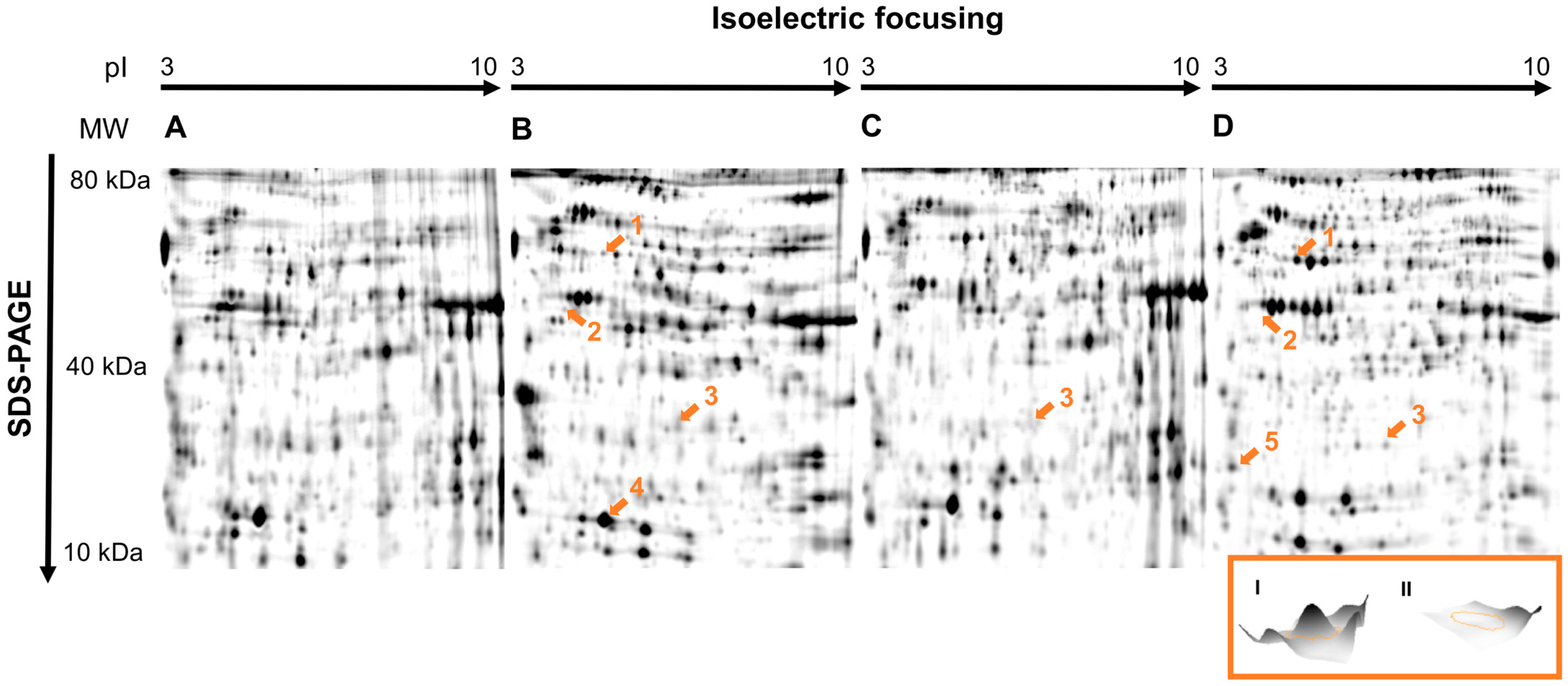
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Klein, A.-M.; Vaissiere, B.E.; Cane, J.H.; Steffan-Dewenter, I.; Cunningham, S.A.; Kremen, C.; Tscharntke, T. Importance of pollinators in changing landscapes for world crops. Proc. R. Soc. B 2006, 274, 303–313. [Google Scholar] [CrossRef]
- Hung, K.-L.J.; Kingston, J.M.; Albrecht, M.; Holway, D.A.; Kohn, J.R. The worldwide importance of honey bees as pollinators in natural habitats. Proc. R. Soc. B 2018, 285, 20172140. [Google Scholar] [CrossRef]
- Moritz, R.F.A.; Erler, S. Lost colonies found in a data mine: Global honey trade but not pests or pesticides as a major cause of regional honeybee colony declines. Agric. Ecosyst. Environ. 2016, 216, 44–50. [Google Scholar] [CrossRef]
- Thomann, M.; Imbert, E.; Devaux, C.; Cheptou, P.-O. Flowering plants under global pollinator decline. Trends Plant Sci. 2013, 18, 353–359. [Google Scholar] [CrossRef]
- van der Zee, R.; Pisa, L.; Andonov, S.; Brodschneider, R.; Charrière, J.-D.; Chlebo, R.; Coffey, M.F.; Crailsheim, K.; Dahle, B.; Gaja, A.; et al. Managed honey bee colony losses in Canada, China, Europe, Israel and Turkey, for the winters of 2008–9 and 2009–10. J. Apic. Res. 2012, 51, 100–114. [Google Scholar] [CrossRef]
- van der Zee, R.; Gray, A.; Pisa, L.; de Rijk, T. An observational study of honey bee colony winter losses and their association with Varroa destructor, neonicotinoids and other risk factors. PLoS ONE 2015, 10, e0131611. [Google Scholar] [CrossRef]
- Genersch, E.; von der Ohe, W.; Kaatz, H.; Schroeder, A.; Otten, C.; Büchler, R.; Berg, S.; Ritter, W.; Mühlen, W.; Gisder, S.; et al. The German bee monitoring project: A long term study to understand periodically high winter losses of honey bee colonies. Apidologie 2010, 41, 332–352. [Google Scholar] [CrossRef]
- van Dooremalen, C.; Cornelissen, B.; Poleij-Hok-Ahin, C.; Blacquière, T. Single and interactive effects of Varroa destructor, Nosema spp., and imidacloprid on honey bee colonies (Apis mellifera). Ecosphere 2018, 9, e02378. [Google Scholar] [CrossRef]
- Sur, R.; Stork, A. Uptake, translocation and metabolism of imidacloprid in plants. Bull. Insectol. 2003, 56, 35–40. [Google Scholar]
- Jeschke, P.; Nauen, R. Neonicotinoids—From zero to hero in insecticide chemistry. Pest Manag. Sci. 2008, 64, 1084–1098. [Google Scholar] [CrossRef]
- EFSA. EFSA Guidance Document on the risk assessment of plant protection products on bees (Apis mellifera, Bombus spp. and solitary bees). EFSA J. 2013, 11, 3295. [Google Scholar] [CrossRef]
- Bass, C.; Denholm, I.; Williamson, M.S.; Nauen, R. The global status of insect resistance to neonicotinoid insecticides. Pestic. Biochem. Physiol. 2015, 121, 78–87. [Google Scholar] [CrossRef]
- Carnesecchi, E.; Svendsen, C.; Lasagni, S.; Grech, A.; Quignot, N.; Amzal, B.; Toma, C.; Tosi, S.; Rortais, A.; Cortinas-Abrahantes, J.; et al. Investigating combined toxicity of binary mixtures in bees: Meta-analysis of laboratory tests, modelling, mechanistic basis and implications for risk assessment. Environ. Int. 2019, 133, 105256. [Google Scholar] [CrossRef]
- Raby, M.; Nowierski, M.; Perlov, D.; Zhao, X.; Hao, C.; Poirier, D.G.; Sibley, P.K. Acute toxicity of 6 neonicotinoid insecticides to freshwater invertebrates. Environ. Toxicol. Chem. 2018, 37, 1430–1445. [Google Scholar] [CrossRef]
- EFSA. Evaluation of the data on clothianidin, imidacloprid and thiamethoxam for the updated risk assessment to bees for seed treatments and granules in the EU. EFSA Support. Publ. 2018, 15, 1378E. [Google Scholar] [CrossRef]
- Nauen, R.; Jeschke, P.; Velten, R.; Beck, M.E.; Ebbinghaus-Kintscher, U.; Thielert, W.; Wölfel, K.; Haas, M.; Kunz, K.; Raupach, G. Flupyradifurone: A brief profile of a new butenolide insecticide. Pest Manag. Sci. 2015, 71, 850–862. [Google Scholar] [CrossRef]
- Hesselbach, H.; Scheiner, R. The novel pesticide flupyradifurone (Sivanto) affects honeybee motor abilities. Ecotoxicology 2019, 28, 354–366. [Google Scholar] [CrossRef]
- Tan, K.; Wang, C.; Dong, S.; Li, X.; Nieh, J.C. The pesticide flupyradifurone impairs olfactory learning in Asian honey bees (Apis cerana) exposed as larvae or as adults. Sci. Rep. 2017, 7, 17772. [Google Scholar] [CrossRef]
- Al Naggar, Y.; Baer, B. Consequences of a short time exposure to a sublethal dose of Flupyradifurone (Sivanto) pesticide early in life on survival and immunity in the honeybee (Apis mellifera). Sci. Rep. 2019, 9, 19753. [Google Scholar] [CrossRef]
- Wu, J.Y.; Anelli, C.M.; Sheppard, W.S. Sub-lethal effects of pesticide residues in brood comb on worker honey bee (Apis mellifera) development and longevity. PLoS ONE 2011, 6, e14720. [Google Scholar] [CrossRef]
- Wu-Smart, J.; Spivak, M. Sub-lethal effects of dietary neonicotinoid insecticide exposure on honey bee queen fecundity and colony development. Sci. Rep. 2016, 6, 32108. [Google Scholar] [CrossRef]
- Yang, E.-C.; Chang, H.-C.; Wu, W.-Y.; Chen, Y.-W. Impaired olfactory associative behavior of honeybee workers due to contamination of imidacloprid in the larval stage. PLoS ONE 2012, 7, e49472. [Google Scholar] [CrossRef]
- Aupinel, P.; Fortini, D.; Dufour, H.; Tasei, J.; Michaud, B.; Odoux, J.; Pham-Delègue, M. Improvement of artificial feeding in a standard in vitro method for rearing Apis mellifera larvae. Bull. Insectology 2005, 58, 107–111. [Google Scholar]
- OECD. Guidance Document on Honey Bee (Apis mellifera) Larval Toxicity Test Following Repeated Exposure. Series on Testing and Assessment. No. 239. In OECD Environment, Health and Safety Publications; ENV/JM/MONO 34; IOMC: Paris, France, 2016. [Google Scholar]
- Thompson, H.M.; Maus, C. The relevance of sublethal effects in honey bee testing for pesticide risk assessment. Pest Manag. Sci. 2007, 63, 1058–1061. [Google Scholar] [CrossRef]
- Kablau, A.; Eckert, J.; Pistorius, J.; Sharbati, S.; Einspanier, R. Effects of selected insecticidal substances on mRNA transcriptome in larvae of Apis mellifera. Pestic. Biochem. Physiol. 2020, 170, 104703. [Google Scholar] [CrossRef]
- EFSA. Conclusion on the peer review of the pesticide risk assessment of the active substance flupyradifurone. EFSA J. 2015, 13, 4020. [Google Scholar] [CrossRef]
- McAfee, A.; Metz, B.N.; Milone, J.P.; Foster, L.J.; Tarpy, D.R. Drone honey bees are disproportionately sensitive to abiotic stressors despite expressing high levels of stress response proteins. Commun. Biol. 2022, 5, 141. [Google Scholar] [CrossRef]
- Lukaszewicz-Hussain, A. Role of oxidative stress in organophosphate insecticide toxicity–Short review. Pestic. Biochem. Physiol. 2010, 98, 145–150. [Google Scholar] [CrossRef]
- Mărgăoan, R.; Mărghitaş, L.A.; Dezmirean, D.S.; Dulf, F.V.; Bunea, A.; Socaci, S.A.; Bobiş, O. Predominant and secondary pollen botanical origins influence the carotenoid and fatty acid profile in fresh honeybee-collected pollen. J. Agric. Food Chem. 2014, 62, 6306–6316. [Google Scholar] [CrossRef]
- Helmer, S.H.; Kerbaol, A.; Aras, P.; Jumarie, C.; Boily, M. Effects of realistic doses of atrazine, metolachlor, and glyphosate on lipid peroxidation and diet-derived antioxidants in caged honey bees (Apis mellifera). Environ. Sci. Pollut. Res. 2015, 22, 8010–8021. [Google Scholar] [CrossRef]
- Carlson, S.D.; Steeves, H.R.; Vandeberg, J.S.; Robbins, W.E. Vitamin A deficiency: Effect on retinal structure of the moth Manduca sexta. Science 1967, 158, 268–270. [Google Scholar] [CrossRef] [PubMed]
- Němec, V.; Kodrík, D.; Matolín, S.; Laufer, H. Juvenile hormone-like effects of retinoic acid in insect metamorphosis, embryogenesis and reproduction. J. Insect Physiol. 1993, 39, 1083–1093. [Google Scholar] [CrossRef]
- Halme, A.; Cheng, M.; Hariharan, I.K. Retinoids regulate a developmental checkpoint for tissue regeneration in Drosophila. Curr. Biol. 2010, 20, 458–463. [Google Scholar] [CrossRef] [PubMed]
- Maden, M.; Hind, M. Retinoic acid, a regeneration-inducing molecule. Dev. Dyn. 2003, 226, 237–244. [Google Scholar] [CrossRef]
- Chakrabarti, P.; Carlson, E.A.; Lucas, H.M.; Melathopoulos, A.P.; Sagili, R.R. Field rates of Sivanto™ (flupyradifurone) and Transform® (sulfoxaflor) increase oxidative stress and induce apoptosis in honey bees (Apis mellifera L.). PLoS ONE 2020, 15, e0233033. [Google Scholar] [CrossRef]
- Uchida, Y.; Izai, K.; Orii, T.; Hashimoto, T. Novel fatty acid beta-oxidation enzymes in rat liver mitochondria. II. Purification and properties of enoyl-coenzyme A (CoA) hydratase/3-hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase trifunctional protein. J. Biol. Chem. 1992, 267, 1034–1041. [Google Scholar] [CrossRef]
- Słowińska, M.; Nynca, J.; Bąk, B.; Wilde, J.; Siuda, M.; Ciereszko, A. (2019). 2D-DIGE proteomic analysis reveals changes in haemolymph proteome of 1-day-old honey bee (Apis mellifera) workers in response to infection with Varroa destructor mites. Apidologie 2019, 50, 632–656. [Google Scholar] [CrossRef]
- Akram, M. Citric acid cycle and role of its intermediates in metabolism. Cell Biochem. Biophys. 2014, 68, 475–478. [Google Scholar] [CrossRef]
- Pearce, E.L.; Pearce, E.J. Metabolic pathways in immune cell activation and quiescence. Immunity 2013, 38, 633–643. [Google Scholar] [CrossRef]
- Feyereisen, R. Insect P450 enzymes. Annu. Rev. Entomol. 1999, 44, 507–533. [Google Scholar] [CrossRef]
- Eaton, S.; Bartlett, K.; Pourfarzam, M. Mammalian mitochondrial beta-oxidation. Biochem J 1996, 320, 345–357. [Google Scholar] [CrossRef] [PubMed]
- Moore, B. First description of 14-3-3 proteins in mammalian brains. In Physiological and Biochemical Aspects of Nervous Integration; Carlson, F.D., Ed.; Prentice-Hall: Hoboken, NJ, USA, 1967; pp. 343–359. [Google Scholar]
- van Heusden, G.P.H.; Wenzel, T.J.; Lagendijk, E.L.; de Steensma, H.; van den Berg, J.A. Characterization of the yeast BMH1 gene encoding a putative protein homologous to mammalian protein kinase II activators and protein kinase C inhibitors. FEBS Lett. 1992, 302, 145–150. [Google Scholar] [CrossRef] [PubMed]
- van Heusden, G.P.H.; Griffiths, D.J.; Ford, J.C.; Chin-A-Woeng, T.F.; Schrader, P.A.; Carr, A.M.; Steensma, H.Y. The 14-3-3 proteins encoded by the BMH1 and BMH2 genes are essential in the yeast Saccharomyces cerevisiae and can be replaced by a plant homologue. Eur. J. Biochem. 1995, 229, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Acevedo, S.F.; Tsigkari, K.K.; Grammenoudi, S.; Skoulakis, E.M. In vivo functional specificity and homeostasis of Drosophila 14-3-3 proteins. Genetics 2007, 177, 239–253. [Google Scholar] [CrossRef]
- Chang, H.C.; Rubin, G.M. 14-3-3 epsilon positively regulates Ras-mediated signaling in Drosophila. Genes Dev. 1997, 11, 1132–1139. [Google Scholar] [CrossRef]
- Kockel, L.; Vorbrüggen, G.; Jäckle, H.; Mlodzik, M.; Bohmann, D. Requirement for Drosophila 14-3-3 zeta in Raf-dependent photoreceptor development. Genes Dev. 1997, 11, 1140–1147. [Google Scholar] [CrossRef][Green Version]
- Dong, S.; Kang, S.; Gu, T.-L.; Kardar, S.; Fu, H.; Lonial, S.; Khoury, H.J.; Khuri, F.; Chen, J. 14–3-3 integrates prosurvival signals mediated by the AKT and MAPK pathways in ZNF198-FGFR1–transformed hematopoietic cells. Blood 2007, 110, 360–369. [Google Scholar] [CrossRef]
- Le, T.P.; Vuong, L.T.; Kim, A.R.; Hsu, Y.C.; Choi, K.W. 14-3-3 proteins regulate Tctp–Rheb interaction for organ growth in Drosophila. Nat. Commun. 2016, 7, 11501. [Google Scholar] [CrossRef]
- Shandala, T.; Woodcock, J.M.; Ng, Y.; Biggs, L.; Skoulakis, E.M.; Brooks, D.A.; Lopez, A.F. Drosophila 14-3-3ε has a crucial role in anti-microbial peptide secretion and innate immunity. J. Cell Sci. 2011, 124, 2165–2174. [Google Scholar] [CrossRef]
- Ulvila, J.; Vanha-Aho, L.M.; Kleino, A.; Vähä-Mäkilä, M.; Vuoksio, M.; Eskelinen, S.; Hultmark, D.; Kocks, C.; Hallman, M.; Parikka, M.; et al. Cofilin regulator 14-3-3ζ is an evolutionarily conserved protein required for phagocytosis and microbial resistance. J. Leukoc. Biol. 2011, 89, 649–659. [Google Scholar] [CrossRef]
- Zaluski, R.; Bittarello, A.C.; Souza Vieira, J.C.; Pereira Braga, C.; de Magalhaes Padilha, P.; da Silva Fernandes, M.; de Souza Bovi, T.; de Oliveira Orsi, R. Modification of the head proteome of nurse honeybees (Apis mellifera) exposed to field-relevant doses of pesticides. Sci. Rep. 2020, 10, 2190. [Google Scholar] [CrossRef] [PubMed]
- Snodgrass, R.E. Anatomy and Physiology of the Honeybee, 1st ed.; McGraw-Hill Book Company: New York, NY, USA, 1925. [Google Scholar]
- Buttstedt, A.; Moritz, R.F.A.; Erler, S. Origin and function of the major royal jelly proteins of the honeybee (Apis mellifera) as members of the yellow gene family. Biol. Rev. 2014, 89, 255–269. [Google Scholar] [CrossRef] [PubMed]
- Kubo, T.; Sasaki, M.; Nakamura, J.; Sasagawa, H.; Ohashi, K.; Takeuchi, H.; Natori, S. Change in the expression of hypopharyngeal-gland proteins of the worker honeybees (Apis mellifera L.) with age and/or role. J. Biochem. 1996, 119, 291–295. [Google Scholar] [CrossRef] [PubMed]
- Maori, E.; Navarro, I.C.; Boncristiani, H.; Seilly, D.J.; Rudolph, K.L.M.; Sapetschnig, A.; Lin, C.-C.; Ladbury, J.E.; Evans, J.D.; Heeney, J.L.; et al. A secreted RNA binding protein forms RNA-stabilizing granules in the honeybee royal jelly. Mol. Cell 2019, 74, 598–608. [Google Scholar] [CrossRef]
- Park, M.J.; Kim, B.Y.; Park, H.G.; Deng, Y.; Yoon, H.J.; Choi, Y.S.; Lee, K.S.; Jin, B.R. Major royal jelly protein 2 acts as an antimicrobial agent and antioxidant in royal jelly. J. Asia Pac. Entomol. 2019, 22, 684–689. [Google Scholar] [CrossRef]
- du Rand, E.E.; Smit, S.; Beukes, M.; Apostolides, Z.; Pirk, C.W.W.; Nicolson, S. Detoxification mechanisms of honey bees (Apis mellifera) resulting in tolerance of dietary nicotine. Sci. Rep. 2015, 5, 11779. [Google Scholar] [CrossRef]
- Baron, G.L.; Raine, N.E.; Brown, M.J.F. General and species-specific impacts of a neonicotinoid insecticide on the ovary development and feeding of wild bumblebee queens. Proc. R. Soc. B 2017, 284, 20170123. [Google Scholar] [CrossRef]
- Davis, A.R.; Solomon, K.; Shuel, R. Laboratory studies of honeybee larval growth and development as affected by systemic insecticides at adult-sublethal levels. J. Apic. Res. 1988, 27, 146–161. [Google Scholar] [CrossRef]
- Sgolastra, F.; Arnan, X.; Cabbri, R.; Isani, G.; Medrzycki, P.; Teper, D.; Bosch, J. Combined exposure to sublethal concentrations of an insecticide and a fungicide affect feeding, ovary development and longevity in a solitary bee. Proc. R. Soc. B 2018, 285, 20180887. [Google Scholar] [CrossRef]
- USEPA. Guidance on Exposure and Effects Testing for Assessing Risks to Bees; Office of Pesticide Programs, U.S. Environmental Protection Agency: Washington, DC, USA, 2016.
| Group | Treatment | Concentration (mg a.s./kg Diet) | Cumulative Doses (µg a.s./Larva) | Active Substance |
|---|---|---|---|---|
| C | Control | - | - | Water |
| CS | Solvent control | - | - | Water + acetone |
| T1 | Insecticide | 1.29 | 0.2 | Dimethoate |
| T2 | Insecticide | 0.32 | 0.05 | Fenoxycarb |
| T3 | Insecticide | 10 | 1.54 | Flupyradifurone |
| Protein | Dimethoate (Mean Fold Difference) | p-Value (t-Test) | Fenoxycarb (Mean Fold Difference) | p-Value (t-Test) | Flupyradifurone (Mean Fold Difference) | p-Value (t-Test) | No. of Matched Peptides |
|---|---|---|---|---|---|---|---|
| Glutathione S-transferase S1-like | −1.57 | 0.0005 | −1.02 | 0.86 | 1.17 | 0.37 | 633 |
| 3-Ketoacyl-CoA thiolase | −1.68 | 0.02 | −1.42 | 0.20 | −15.96 | 0.009 | 58 |
| Major royal jelly protein 2 | −2.11 | 0.02 | −4.10 | 0.0002 | −30.99 | 0.0007 | 43 |
| 14-3-3 Protein zeta | −1.17 | 0.35 | −1.21 | 0.44 | −1.60 | 0.01 | 66 |
| Retinal dehydrogenase 1 | −1.60 | 0.02 | 1.16 | 0.43 | 1.78 | 0.0002 | 120 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kablau, A.; Erler, S.; Eckert, J.H.; Pistorius, J.; Sharbati, S.; Einspanier, R. Effects of Flupyradifurone and Two Reference Insecticides Commonly Used in Toxicological Studies on the Larval Proteome of the Honey bee Apis mellifera. Insects 2023, 14, 77. https://doi.org/10.3390/insects14010077
Kablau A, Erler S, Eckert JH, Pistorius J, Sharbati S, Einspanier R. Effects of Flupyradifurone and Two Reference Insecticides Commonly Used in Toxicological Studies on the Larval Proteome of the Honey bee Apis mellifera. Insects. 2023; 14(1):77. https://doi.org/10.3390/insects14010077
Chicago/Turabian StyleKablau, Arne, Silvio Erler, Jakob H. Eckert, Jens Pistorius, Soroush Sharbati, and Ralf Einspanier. 2023. "Effects of Flupyradifurone and Two Reference Insecticides Commonly Used in Toxicological Studies on the Larval Proteome of the Honey bee Apis mellifera" Insects 14, no. 1: 77. https://doi.org/10.3390/insects14010077
APA StyleKablau, A., Erler, S., Eckert, J. H., Pistorius, J., Sharbati, S., & Einspanier, R. (2023). Effects of Flupyradifurone and Two Reference Insecticides Commonly Used in Toxicological Studies on the Larval Proteome of the Honey bee Apis mellifera. Insects, 14(1), 77. https://doi.org/10.3390/insects14010077








