Immunodetection of Truncated Forms of the α6 Subunit of the nAChR in the Brain of Spinosad Resistant Ceratitis capitata Phenotypes
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ceratitis capitata and Drosophila melanogaster Strains
2.2. Heterologous Expression of Truncated Isoform Ccα63aQ68*
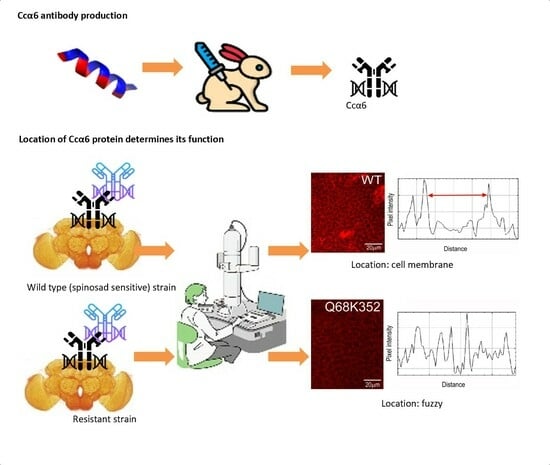
2.3. Generation and Validation of the Anti-Ccα6 Antibody
2.4. Immunofluorescence Assays
3. Results
3.1. Evaluation of the Anti-Ccα6 Antibody
3.2. Presence of Ccα6 in the Brains of Ceratitis capitata
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sparks, T.C.; Storer, N.; Porter, A.; Slater, R.; Nauen, R. Insecticide resistance management and industry: The origins and evolution of the Insecticide Resistance Action Committee (IRAC) and the mode of action classification scheme. Pest Manag. Sci. 2021, 77, 2609–2619. [Google Scholar] [CrossRef] [PubMed]
- Magaña, C.; Hernandez-Crespo, P.; Ortego, F.; Castañera, P. Resistance to malathion in field populations of Ceratitis capitata. J. Econ. Entomol. 2007, 100, 1836–1843. [Google Scholar] [CrossRef] [PubMed]
- Arouri, R.; Le Goff, G.; Hemden, H.; Navarro-Llopis, V.; M’Saad, M.; Castañera, P.; Feyereisen, R.; Hernández-Crespo, P.; Ortego, F. Resistance to lambda-cyhalothrin in Spanish field populations of Ceratitis capitata and metabolic resistance mediated by P450 in a resistant strain. Pest Manag. Sci. 2015, 71, 1281–1291. [Google Scholar] [CrossRef] [PubMed]
- Guillem-Amat, A.; Sánchez, L.; López-Errasquín, E.; Ureña, E.; Hernández-Crespo, P.; Ortego, F. Field detection and predicted evolution of spinosad resistance in Ceratitis capitata. Pest Manag. Sci. 2020, 76, 3702–3710. [Google Scholar] [CrossRef] [PubMed]
- Van Leeuwen, T.; Dermauw, W.; Mavridis, K.; Vontas, J. Significance and interpretation of molecular diagnosis for insecticide resistance management of agricultural pests. Curr. Opin. Insect Sci. 2020, 39, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Ureña, E.; Guillem-Amat, A.; Couso-Ferrer, F.; Beroiz, B.; Perera, N.; López-Errasquín, E.; Castañera, P.; Ortego, F.; Hernández-Crespo, P. Multiple mutations in the nicotinic acetylcholine receptor Ccα6 gene associated with resistance to spinosad in medfly. Sci. Rep. 2019, 9, 2961. [Google Scholar] [CrossRef] [PubMed]
- Guillem-Amat, A.; Ureña, E.; López-Errasquín, E.; Navarro-Llopis, V.; Batterham, P.; Sánchez, L.; Perry, T.; Hernández-Crespo, P.; Ortego, F. Functional characterization and fitness cost of spinosad resistance in Ceratitis capitata. J. Pest Sci. 2020, 93, 1043–1058. [Google Scholar] [CrossRef]
- Papanicolaou, A.; Schetelig, M.F.; Arensburger, P.; Atkinson, P.W.; Benoit, J.B.; Bourtzis, K.; Castañera, P.; Cavanaugh, J.P.; Chao, H.; Childers, C.; et al. The whole genome sequence of the Mediterranean fruit fly, Ceratitis capitata (Wiedemann), reveals insights into the biology and adaptive evolution of a highly invasive pest species. Genome Biol. 2016, 17, 192. [Google Scholar]
- Thany, S.H.; Lenaers, G.; Raymond-Delpech, V.; Sattelle, D.B.; Lapied, B. Exploring the pharmacological properties of insect nicotinic acetylcholine receptors. Trends Pharmacol. Sci. 2007, 28, 14–22. [Google Scholar] [CrossRef]
- Jones, A.K.; Sattelle, D.B. Diversity of Insect Nicotinic Acetylcholine Receptor Subunits. In Insect Nicotinic Acetylcholine Receptors, Advances in Experimental Medicine and Biology; Thany, S.H., Ed.; Springer: New York, NY, USA, 2010; Volume 683, pp. 24–44. [Google Scholar]
- Ihara, M.; Buckingham, S.D.; Matsuda, K.; Sattelle, D.B. Modes of action, resistance and toxicity of insecticides targeting nicotinic acetylcholine receptors. Curr. Med. Chem. 2017, 24, 2925–2934. [Google Scholar] [CrossRef]
- Baxter, S.W.; Chen, M.; Dawson, A.; Zhao, J.-Z.; Vogel, H.; Shelton, A.M.; Heckel, D.G.; Jiggins, C.D. Mis-spliced transcripts of nicotinic acetylcholine receptor α6 are associated with field evolved spinosad resistance in Plutella xylostella (L.). PLoS Genet. 2010, 6, e1000802. [Google Scholar] [CrossRef] [PubMed]
- Rinkevich, F.D.; Chen, M.; Shelton, A.M.; Scott, J.G. Transcripts of the nicotinic acetylcholine receptor subunit gene Pxyla6 with premature stop codons are associated with spinosad resistance in diamondback moth, Plutella xylostella. Invertebr. Neurosci. 2010, 10, 25–33. [Google Scholar] [CrossRef]
- Hsu, J.C.; Feng, H.T.; Wu, W.J.; Geib, S.M.; Mao, C.-H.; Vontas, J. Truncated transcripts of nicotinic acetylcholine subunit gene Bdα6 are associated with spinosad resistance in Bactrocera dorsalis. Insect Biochem. Mol. Biol. 2012, 42, 806–815. [Google Scholar] [CrossRef]
- Perry, T.; McKenzie, J.A.; Batterham, P. A Dalpha6 knockout strain of Drosophila melanogaster confers a high level of resistance to spinosad. Insect Biochem. Mol. Biol. 2007, 37, 184–188. [Google Scholar] [CrossRef] [PubMed]
- Watson, G.B.; Chouinard, S.W.; Cook, K.R.; Geng, C.; Gifford, J.M.; Gustafson, G.D.; Hasler, J.M.; Larrinua, I.M.; Letherer, T.J.; Mitchell, J.C.; et al. A spinosyn-sensitive Drosophila melanogaster nicotinic acetylcholine receptor identified through chemically induced target site resistance, resistance gene identification, and heterologous expression. Insect Biochem. Mol. Biol. 2010, 40, 376–384. [Google Scholar] [CrossRef] [PubMed]
- Perry, T.; Chen, W.; Ghazali, R.; Yang, Y.T.; Christesen, D.; Martelli, F.; Lumb, C.; Luong, H.N.B.; Mitchell, J.; Holien, J.K.; et al. Role of nicotinic acetylcholine receptor subunits in the mode of action of neonicotinoid, sulfoximine and spinosyn insecticides in Drosophila melanogaster. Insect Biochem. Mol. Biol. 2021, 131, 103547. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Liu, Z.; Fan, X.; Zhang, X.; Qiao, X.; Huang, J. Nicotinic acetylcholine receptor modulator insecticides act on diverse receptor subtypes with distinct subunit compositions. PLoS Genet. 2022, 18, e1009920. [Google Scholar] [CrossRef] [PubMed]
- Rosenthal, J.S.; Yin, J.; Lei, J.; Sathyamurthy, A.; Short, J.; Long, C.; Spillman, E.; Sheng, C.; Yuan, Q. Temporal regulation of nicotinic acetylcholine receptor subunits supports central cholinergic synapse development in Drosophila. Proc. Natl. Acad. Sci. USA 2021, 118, e2004685118. [Google Scholar] [CrossRef]
- Zhang, G.; Zhang, W. Direct protein-protein interaction network for insecticide resistance based on subcellular localization analysis in Drosophila melanogaster. J. Environ. Sci. Health 2020, 55, 732–748. [Google Scholar] [CrossRef]
- García-Guzmán, M.; Sala, F.; Sala, S.; Campos-Caro, A.; Stuhmer, W.; Gutiérrez, L.M.; Criado, M. α-Bungarotoxin-sensitive nicotinic receptors on bovine chromaffin cells: Molecular cloning, functional expression and alternative splicing of the α7 subunit. Eur. J. Neurol. 1995, 7, 647–655. [Google Scholar] [CrossRef]
- Saragoza, P.A.; Modir, J.G.; Goel, N.; French, K.L.; Li, L.; Nowak, M.W.; A Stitzel, J. Identification of an alternatively processed nicotinic receptor α7 subunit RNA in mouse brain. Mol. Brain Res. 2003, 117, 15–26. [Google Scholar] [PubMed]
- Somers, J.; Luong, H.N.B.; Batterham, P.; Perry, T. Deletion of the nicotinic acetylcholine receptor subunit gene Dα1 confers insecticide resistance, but at what cost? Fly 2018, 12, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Smit, A.B.; Brejc, K.; Syed, N.; Sixma, T.K. Structure and Function of AChBP, Homologue of the Ligand-Binding Domain of the Nicotinic Acetylcholine Receptor. Ann. N. Y. Acad. Sci. 2003, 998, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Thany, S.H.; Tricoire-Leignel, H.; Lapied, B. Identification of cholinergic synaptic transmission in the insect nervous system. Adv. Exp. Med. Biol. 2010, 683, 1–10. [Google Scholar] [PubMed]
- Chamaon, K.; Schulz, R.; Smalla, K.H.; Seidel, B.; Gundelfinger, E.D. Neuronal nicotinic acetylcholine receptors of Drosophila melanogaster: The alpha-subunit Da3 and the beta-type subunit ARD co-assemble within the same receptor complex. FEBS Lett. 2000, 482, 189–192. [Google Scholar] [CrossRef]
- Chamaon, K.; Smalla, K.H.; Thomas, U.; Gundelfinger, E.D. Nicotinic acetylcholine receptors of Drosophila: Three subunits encoded by genomically linked genes can co-assemble into the same receptor complex. J. Neurochem. 2002, 80, 149–157. [Google Scholar] [CrossRef]
- Fayyazuddin, A.; Zaheer, M.A.; Hiesinger, P.R.; Bellen, H.J. The nicotinic acetylcholine receptor Dα7 is required for an escape behavior in Drosophila. PLoS Biol. 2006, 4, e63. [Google Scholar] [CrossRef]
- Schuster, R.; Phannavong, B.; Schroder, C.; Gundelfinger, E.D. Immunohistochemical localization of a ligand-binding and a structural subunit of nicotinic acetylcholine receptors in the central nervous system of Drosophila melanogaster. J. Comp. Neurol. 1993, 335, 149–162. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guillem-Amat, A.; López-Errasquín, E.; García-Ricote, I.; Barbero, J.L.; Sánchez, L.; Casas-Tintó, S.; Ortego, F. Immunodetection of Truncated Forms of the α6 Subunit of the nAChR in the Brain of Spinosad Resistant Ceratitis capitata Phenotypes. Insects 2023, 14, 857. https://doi.org/10.3390/insects14110857
Guillem-Amat A, López-Errasquín E, García-Ricote I, Barbero JL, Sánchez L, Casas-Tintó S, Ortego F. Immunodetection of Truncated Forms of the α6 Subunit of the nAChR in the Brain of Spinosad Resistant Ceratitis capitata Phenotypes. Insects. 2023; 14(11):857. https://doi.org/10.3390/insects14110857
Chicago/Turabian StyleGuillem-Amat, Ana, Elena López-Errasquín, Irene García-Ricote, José Luis Barbero, Lucas Sánchez, Sergio Casas-Tintó, and Félix Ortego. 2023. "Immunodetection of Truncated Forms of the α6 Subunit of the nAChR in the Brain of Spinosad Resistant Ceratitis capitata Phenotypes" Insects 14, no. 11: 857. https://doi.org/10.3390/insects14110857
APA StyleGuillem-Amat, A., López-Errasquín, E., García-Ricote, I., Barbero, J. L., Sánchez, L., Casas-Tintó, S., & Ortego, F. (2023). Immunodetection of Truncated Forms of the α6 Subunit of the nAChR in the Brain of Spinosad Resistant Ceratitis capitata Phenotypes. Insects, 14(11), 857. https://doi.org/10.3390/insects14110857