Species Identification of the Major Japanese Encephalitis Vectors within the Culex vishnui Subgroup (Diptera: Culicidae) in Thailand Using Geometric Morphometrics and DNA Barcoding
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
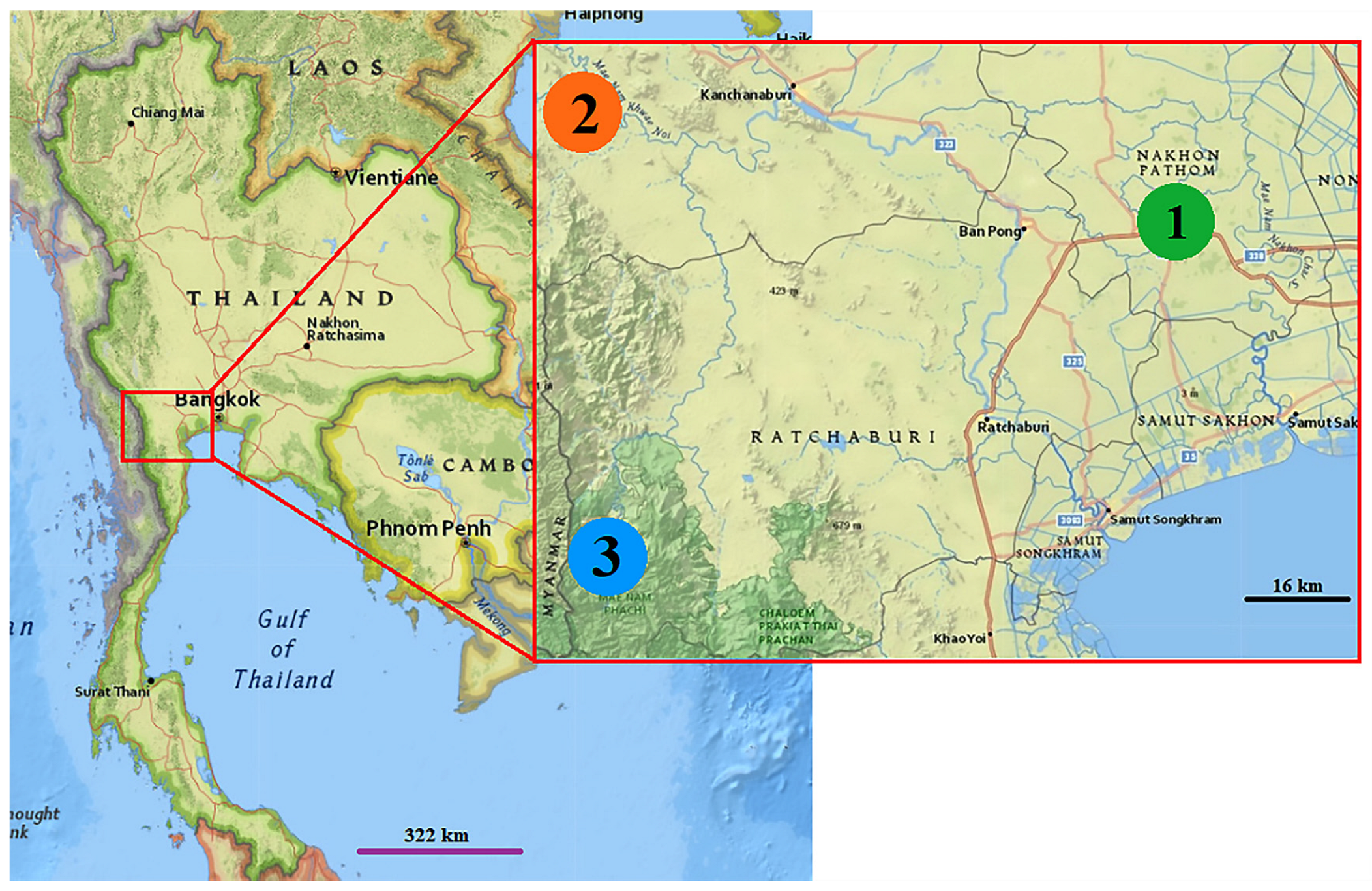
2.1. Mosquito Collections
2.2. Species Identification Based on the Morphology
2.3. Sample Preparation for GM and DNA Barcoding
2.4. Geometric Morphometrics
2.5. DNA Barcoding
3. Results
3.1. Geometric Morphometrics for Species Identification
3.2. Species Identification by DNA Barcoding
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Japanese Encephalitis 2019. Available online: https://www.who.int/news-room/fact-sheets/detail/japanese-encephalitis (accessed on 10 July 2022).
- Amicizia, D.; Zangrillo, F.; Lai, P.L.; Iovine, M.; Panatto, D. Overview of Japanese Encephalitis Disease and Its Prevention. Focus on IC51 Vaccine (IXIARO®). J. Prev. Med. Hyg. 2018, 59, E99–E107. [Google Scholar] [PubMed]
- World Health Organization. Japanese Encephalitis Reported Cases by WHO Region 2020. Available online: https://apps.who.int/gho/data/view.main.1520_42lang=en (accessed on 20 December 2022).
- Ministry of Public Health, Thailand. Annual Report. Bureau of Epidemiology: MOPH; Japanese B encephalitis. 2017–2019. Available online: http://www.boe.moph.go.th/boedb/surdata/index.php (accessed on 10 July 2022).
- Ministry of Public Health, Thailand. Epidemiological Report. Available online: http://www.boe.moph.go.th/boedb/surdata/index.php (accessed on 10 July 2022).
- World Health Organization. Mosquito Born Diseases. Available online: http://www.who.int/neglected_diseases/vector_ecology/mosquito-borne-diseases/en/ (accessed on 15 July 2022).
- Chaiphongpachara, T.; Laojun, S. Effectiveness of Ultraviolet (UV) Insect Light Traps for Mosquitoes Control in Coastal Areas of Samut Songkhram Province, Thailand. J. Anim. Behav. Biometeorol. 2019, 7, 25–30. [Google Scholar]
- Lee, H.; Halverson, S.; Ezinwa, N. Mosquito-Borne Diseases. Prim. Care-Clin. Off. Pract. 2018, 45, 393–407. [Google Scholar] [CrossRef] [PubMed]
- Harbach, R.E. Mosquito Taxonomic Inventory. Available online: https://mosquito-taxonomic-inventory.myspecies.info/simpletaxonomy/term/6192 (accessed on 1 October 2022).
- Rattanarithikul, R.; Harbach, R.E.; Harrison, B.A.; Panthusiri, P.; Jones, J.W.; Coleman, R.E. Illustrated Keys to the Mosquitoes of Thailand. II. Genera Culex and Lutzia. Southeast Asian J. Trop. Med. Public Health 2005, 36, 1–97. [Google Scholar] [PubMed]
- Gingrich, J.B.; Nisalak, A.; Latendresse, J.R.; Sattabongkot, J.; Hoke, C.H.; Pomsdhit, J.; Chantalakana, C.; Satayaphanta, C.; Uechiewcharnkit, K.; Innis, B.L. Japanese Encephalitis Virus in Bangkok: Factors Influencing Vector Infections in Three Suburban Communities. J. Med. Entomol. 1992, 29, 436–444. [Google Scholar] [CrossRef]
- Sumruayphol, S.; Chaiphongpachara, T.; Samung, Y.; Ruangsittichai, J.; Cui, L.; Zhong, D.; Sattabongkot, J.; Sriwichai, P. Seasonal Dynamics and Molecular Differentiation of Three Natural Anopheles Species (Diptera: Culicidae) of the Maculatus Group (Neocellia Series) in Malaria Hotspot Villages of Thailand. Parasit. Vectors 2020, 13, 574. [Google Scholar] [CrossRef]
- Kshitiz, S.; Sebastien, B.; Veronique, C.; Chalalai, R. Host Preference of Culex Mosquitoes in an Area with High Transmission Rate of Japanese Encephalitis of Kandal Province, Cambodia. J. Kasetsart Vet. 2008, 28, 126–137. [Google Scholar]
- Chaiphongpachara, T.; Laojun, S. Effectiveness of Landmark- and Semi-Landmark-Based Geometric Morphometric to Identify Four Species of Culex Mosquitoes in Thailand. J. Adv. Vet. Anim. Res. 2019, 6, 278–283. [Google Scholar] [CrossRef]
- Chaiphongpachara, T.; Sriwichai, P.; Samung, Y.; Ruangsittichai, J. Geometric Morphometrics Approach towards Discrimination of Three Member Species of Maculatus Group in Thailand. Acta Trop. 2019, 192, 66–74. [Google Scholar] [CrossRef]
- Champakaew, D.; Junkum, A.; Sontigun, N.; Sanit, S.; Limsopatham, K.; Saeung, A.; Somboon, P.; Pitasawat, B. Geometric Morphometric Wing Analysis as a Tool to Discriminate Female Mosquitoes from Different Suburban Areas of Chiang Mai Province, Thailand. PLoS ONE 2021, 16, e0260333. [Google Scholar] [CrossRef]
- Chaiphongpachara, T.; Changbunjong, T.; Sumruayphol, S.; Laojun, S.; Suwandittakul, N.; Kuntawong, K. Geometric Morphometrics versus DNA Barcoding for the Identification of Malaria Vectors Anopheles dirus and An. baimaii in the Thai-Cambodia Border. Sci. Rep. 2022, 12, 13236. [Google Scholar] [CrossRef]
- Chaiphongpachara, T.; Changbunjong, T.; Laojun, S. Geometric Morphometric and Molecular Techniques for Discriminating among Three Cryptic Species of the Anopheles barbirostris Complex (Diptera: Culicidae) in Thailand. Heliyon 2022, 8, e11261. [Google Scholar] [CrossRef]
- Oliveira-Christe, R.; Marrelli, M.T. Using Geometric Morphometric Analysis of Wings to Identify Mosquito Species from the Subgenus Microculex (Diptera: Culicidae). J. Vector Ecol. 2021, 46, 221–225. [Google Scholar] [CrossRef]
- Chatpiyaphat, K.; Sumruayphol, S.; Dujardin, J.P.; Samung, Y.; Phayakkaphon, A.; Cui, L.; Ruangsittichai, J.; Sungvornyothin, S.; Sattabongkot, J.; Sriwichai, P. Geometric Morphometrics to Distinguish the Cryptic Species Anopheles minimus and An. harrisoni in Malaria Hot Spot Villages, Western Thailand. Med. Vet. Entomol. 2021, 35, 293–301. [Google Scholar]
- Chaiphongpachara, T.; Changbunjong, T.; Laojun, S.; Nutepsu, T.; Suwandittakul, N.; Kuntawong, K.; Sumruayphol, S.; Ruangsittichai, J. Mitochondrial DNA Barcoding of Mosquito Species (Diptera: Culicidae) in Thailand. PLoS ONE 2022, 17, e0275090. [Google Scholar] [CrossRef]
- Ortega-Morales, A.I.; Hernandez-Triana, L.M.; Chan-Chable, R.J.; Garza-Hernandez, J.A.; Gonzalez-Alvarez, V.H.; Ruiz-Arrondo, I.; Nikolova, N.I.; Martinez-Arce, A.; Fooks, A.R.; Rodriguez-Perez, M.A. Dna Barcoding of Mosquitoes from the Pantanos de Centla Biosphere Reserve, Southeastern Mexico. J. Am. Mosq. Control Assoc. 2021, 37, 198–207. [Google Scholar] [CrossRef]
- Lorenz, C.; Almeida, F.; Almeida-Lopes, F.; Louise, C.; Pereira, S.N.; Petersen, V.; Vidal, P.O.; Virginio, F.; Suesdek, L. Geometric Morphometrics in Mosquitoes: What Has Been Measured? Infect. Genet. Evol. 2017, 54, 205–215. [Google Scholar] [CrossRef]
- Dujardin, J.P. Morphometrics Applied to Medical Entomology. Infect. Genet. Evol. 2008, 8, 875–890. [Google Scholar] [CrossRef]
- Suwandittakul, N.; Mungthin, M.; Kuntawong, K.; Laojun, S.; Pimsuka, S.; Chaiphongpachara, T. A Novel Use of a Geometric Morphometric Technique to Distinguish Human Parasite Eggs of Twelve Different Species. Exp. Parasitol. 2022, 238, 108281. [Google Scholar] [CrossRef]
- Simões, R.F.; Wilke, A.B.B.; Chagas, C.R.F.; de Menezes, R.M.T.; Suesdek, L.; Multini, L.C.; Silva, F.S.; Grech, M.G.; Marrelli, M.T.; Kirchgatter, K. Wing Geometric Morphometrics as a Tool for the Identification of Culex Subgenus Mosquitoes of Culex (Diptera: Culicidae). Insects 2020, 11, 567. [Google Scholar] [CrossRef]
- Sumruayphol, S.; Apiwathnasorn, C.; Ruangsittichai, J.; Sriwichai, P.; Attrapadung, S.; Samung, Y.; Dujardin, J.P. DNA Barcoding and Wing Morphometrics to Distinguish Three Aedes Vectors in Thailand. Acta Trop. 2016, 159, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Ruangsittichai, J.; Apiwathnasorn, C.; Dujardin, J.P. Interspecific and Sexual Shape Variation in the Filariasis Vectors Mansonia dives and Ma. bonneae. Infect. Genet. Evol. 2011, 11, 2089–2094. [Google Scholar] [CrossRef] [PubMed]
- Kress, W.J.; Erickson, D.L. DNA Barcodes: Genes, Genomics, and Bioinformatics. Proc. Natl. Acad. Sci. USA 2008, 105, 2761–2762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weeraratne, T.C.; Surendran, S.N.; Karunaratne, S.H.P.P. DNA Barcoding of Morphologically Characterized Mosquitoes Belonging to the Subfamily Culicinae from Sri Lanka. Parasit Vectors 2018, 11, 266. [Google Scholar] [CrossRef] [Green Version]
- Artur, T.; Anna, S.K.; James, J.B.; Neil, S.; Miroslawa, D. A New Method of Metabarcoding Microsporidia and Their Hosts Reveals High Levels of Microsporidian Infections in Mosquitoes (Culicidae). Mol. Ecol. Resour. 2020, 20, 1486–1504. [Google Scholar]
- Dujardin, S.; Dujardin, J.P. Geometric Morphometrics in the Cloud. Infect. Genet. Evol. 2019, 70, 189–196. [Google Scholar] [CrossRef]
- Demari-Silva, B.; Suesdek, L.; Sallum, M.A.M.; Marrelli, M.T. Wing Geometry of Culex Coronator (Diptera: Culicidae) from South and Southeast Brazil. Parasit. Vectors 2014, 7, 174. [Google Scholar] [CrossRef] [Green Version]
- Goodall, C.R. Procrustes Methods in the Statistical Analysis of Shape. J. R. Stat. Soc. Ser. B 1991, 53, 285–321. [Google Scholar] [CrossRef]
- Bookstein, F.L. Morphometric Tools for Landmark Data: Geometry and Biology; Cambridge University Press: Cambridge, UK, 1991. [Google Scholar]
- Rohlf, F.J. Rotational Fit (Procrustes) Methods. Proceedings of the Michigan Morphometrics Workshop; Rohlf, F.J., Bookstein, F.L., Eds.; University of Michigan, Museum of Zoology: Ann Arbor, MI, USA, 1990. [Google Scholar]
- Rohlf, F.J.; Slice, D.E. Extensions of de Procrustes Method for the Optimal Superimposition of Landmarks. Syst. Zool. 1990, 39, 40–59. [Google Scholar] [CrossRef] [Green Version]
- Dujardin, J.-P.; Dujardin, S.; Kaba, D.; Santillán-Guayasamín, S.; Villacís, A.G.; Piyaselakul, S.; Sumruayphol, S.; Samung, Y.; Morales Vargas, R. The Maximum Likelihood Identification Method Applied to Insect Morphometric Data. Zool. Syst. 2017, 42, 46–58. [Google Scholar]
- Dujardin, J.P.; Kaba, D.; Henry, A.B. The exchangeability of shape. BMC Res. Notes 2010, 3, 266. [Google Scholar] [CrossRef] [Green Version]
- Kumar, N.P.; Rajavel, A.R.; Natarajan, R.; Jambulingam, P. DNA Barcodes Can Distinguish Species of Indian Mosquitoes (Diptera: Culicidae). J. Med. Entomol. 2007, 44, 1–7. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; Mcgettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X Version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Kumari, R.; Kumar, K.; Rawat, A.; Singh, G.; Yadav, N.K.; Chauhan, L.S. First Indigenous Transmission of Japanese Encephalitis in Urban Areas of National Capital Territory of Delhi, India. Trop. Med. Int. Health 2013, 18, 743–749. [Google Scholar] [CrossRef]
- Faizah, A.N.; Kobayashi, D.; Isawa, H.; Amoa-Bosompem, M.; Murota, K.; Higa, Y.; Futami, K.; Shimada, S.; Kim, K.S.; Itokawa, K.; et al. Deciphering the Virome of Culex vishnui Subgroup Mosquitoes, the Major Vectors of Japanese Encephalitis, in Japan. Viruses 2020, 12, 264. [Google Scholar] [CrossRef] [Green Version]
- Chonephetsarath, S.; Raksakoon, C.; Sumruayphol, S.; Dujardin, J.P.; Potiwat, R. The Unequal Taxonomic Signal of Mosquito Wing Cells. Insects 2021, 12, 376. [Google Scholar] [CrossRef]
- Wang, G.; Li, C.; Guo, X.; Xing, D.; Dong, Y.; Wang, Z.; Zhang, Y.; Liu, M.; Zheng, Z.; Zhang, H.; et al. Identifying the Main Mosquito Species in China Based on DNA Barcoding. PLoS ONE 2012, 7, e47051. [Google Scholar] [CrossRef]
- Hernández-Triana, L.M.; Brugman, V.A.; Nikolova, N.I.; Ruiz-Arrondo, I.; Barrero, E.; Thorne, L.; de Marco, M.F.; Krüger, A.; Lumley, S.; Johnson, N.; et al. DNA Barcoding of British Mosquitoes (Diptera, Culicidae) to Support Species Identification, Discovery of Cryptic Genetic Diversity and Monitoring Invasive Species. Zookeys 2019, 832, 57–76. [Google Scholar] [CrossRef] [Green Version]
- Carvajal, T.M.; Amalin, D.M.; Watanabe, K. Wing Geometry and Genetic Analyses Reveal Contrasting Spatial Structures between Male and Female Aedes aegypti (L.) (Diptera: Culicidae) Populations in Metropolitan Manila, Philippines. Infect. Genet. Evol. 2021, 87, 104676. [Google Scholar] [CrossRef]







| Species | n | Mean (mm) | Min–Max | SD | SE |
|---|---|---|---|---|---|
| Cx. pseudovishnui | 40 | 2.45 a | 2.06–2.82 | 0.17 | 0.03 |
| Cx. tritaeniorhynchus | 58 | 2.23 b | 1.98–2.48 | 0.14 | 0.02 |
| Cx. vishnui | 65 | 2.58 c | 2.02–2.95 | 0.24 | 0.03 |
| Species | Cx. pseudovishnui | Cx. tritaeniorhynchus | Cx. vishnui |
|---|---|---|---|
| Cx. pseudovishnui | 0.000 | ||
| Cx. tritaeniorhynchus | 6.268 * | 0.000 | |
| Cx. vishnui | 3.498 * | 4.695 * | 0.000 |
| Species | Cross-Validated Reclassification Score (%) | |
|---|---|---|
| Wing Size (Assigned/Observed) | Wing Shape (Assigned/Observed) | |
| Cx. pseudovishnui | 35.00% (14/40) | 75.00% (30/40) |
| Cx. tritaeniorhynchus | 75.86% (44/58) | 98.28% (57/58) |
| Cx. vishnui | 56.92% (37/65) | 87.69% (57/65) |
| Total performance | 58.28% (95/163) | 88.34% (144/163) |
| Species | Average Percentage Genetic Divergence (Min–Max) | ||
|---|---|---|---|
| Cx. pseudovishnui | Cx. tritaeniorhynchus | Cx. vishnui | |
| Cx. pseudovishnui | 0.81% (0.00–1.43) | ||
| Cx. tritaeniorhynchus | 6.87% (6.33–7.89) | 0.90% (0.28–1.86) | |
| Cx. vishnui | 4.71% (3.91–5.90) | 5.40% (4.96–6.03) | 0.67% (0.00–1.57) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saiwichai, T.; Laojun, S.; Chaiphongpachara, T.; Sumruayphol, S. Species Identification of the Major Japanese Encephalitis Vectors within the Culex vishnui Subgroup (Diptera: Culicidae) in Thailand Using Geometric Morphometrics and DNA Barcoding. Insects 2023, 14, 131. https://doi.org/10.3390/insects14020131
Saiwichai T, Laojun S, Chaiphongpachara T, Sumruayphol S. Species Identification of the Major Japanese Encephalitis Vectors within the Culex vishnui Subgroup (Diptera: Culicidae) in Thailand Using Geometric Morphometrics and DNA Barcoding. Insects. 2023; 14(2):131. https://doi.org/10.3390/insects14020131
Chicago/Turabian StyleSaiwichai, Tawee, Sedthapong Laojun, Tanawat Chaiphongpachara, and Suchada Sumruayphol. 2023. "Species Identification of the Major Japanese Encephalitis Vectors within the Culex vishnui Subgroup (Diptera: Culicidae) in Thailand Using Geometric Morphometrics and DNA Barcoding" Insects 14, no. 2: 131. https://doi.org/10.3390/insects14020131
APA StyleSaiwichai, T., Laojun, S., Chaiphongpachara, T., & Sumruayphol, S. (2023). Species Identification of the Major Japanese Encephalitis Vectors within the Culex vishnui Subgroup (Diptera: Culicidae) in Thailand Using Geometric Morphometrics and DNA Barcoding. Insects, 14(2), 131. https://doi.org/10.3390/insects14020131





