What Are the Functional Roles of Piwi Proteins and piRNAs in Insects?
Abstract
:Simple Summary
Abstract
1. Introduction
2. Insights from Research on Piwi Proteins and piRNAs in Drosophila
2.1. Molecular Function of Argonaute Proteins of the Piwi Subfamily in Drosophila
2.1.1. Molecular Functions of Drosophila Piwi Proteins in the Ovary
2.1.2. Molecular Functions of Drosophila Piwi Proteins in the Testis
2.2. Piwi Subfamily Proteins and the Regulation of Stem Cell Function in the Gonads in Drosophila
2.2.1. The Gene piwi in Drosophila
The Gene piwi and the Regulation of Stem Cell Function in the Ovary in Drosophila
The Gene piwi and the Regulation of Stem Cell Function in the Testis in Drosophila
2.2.2. The Genes aub and ago3 in Drosophila
The Genes aub and ago3 and the Regulation of Stem Cell Function in the Ovary in Drosophila
The Genes aub and ago3 and the Regulation of Stem Cell Function in the Testis in Drosophila
2.3. Functional Roles of Piwi Proteins during Germ Cell Differentiation in Drosophila
2.3.1. Functional Roles of Drosophila Piwi Proteins during Germ Cell Differentiation in the Ovary
2.3.2. Functional Roles of Drosophila Piwi Proteins during Germ Cell Differentiation in the Testis
2.4. Functions of Drosophila Piwi Proteins and piRNAs Outside the Gonad
3. Insights from Research on Piwi Proteins and piRNAs in Mosquitoes
3.1. Diverisification of Piwi Proteins in Aedes Mosquitoes
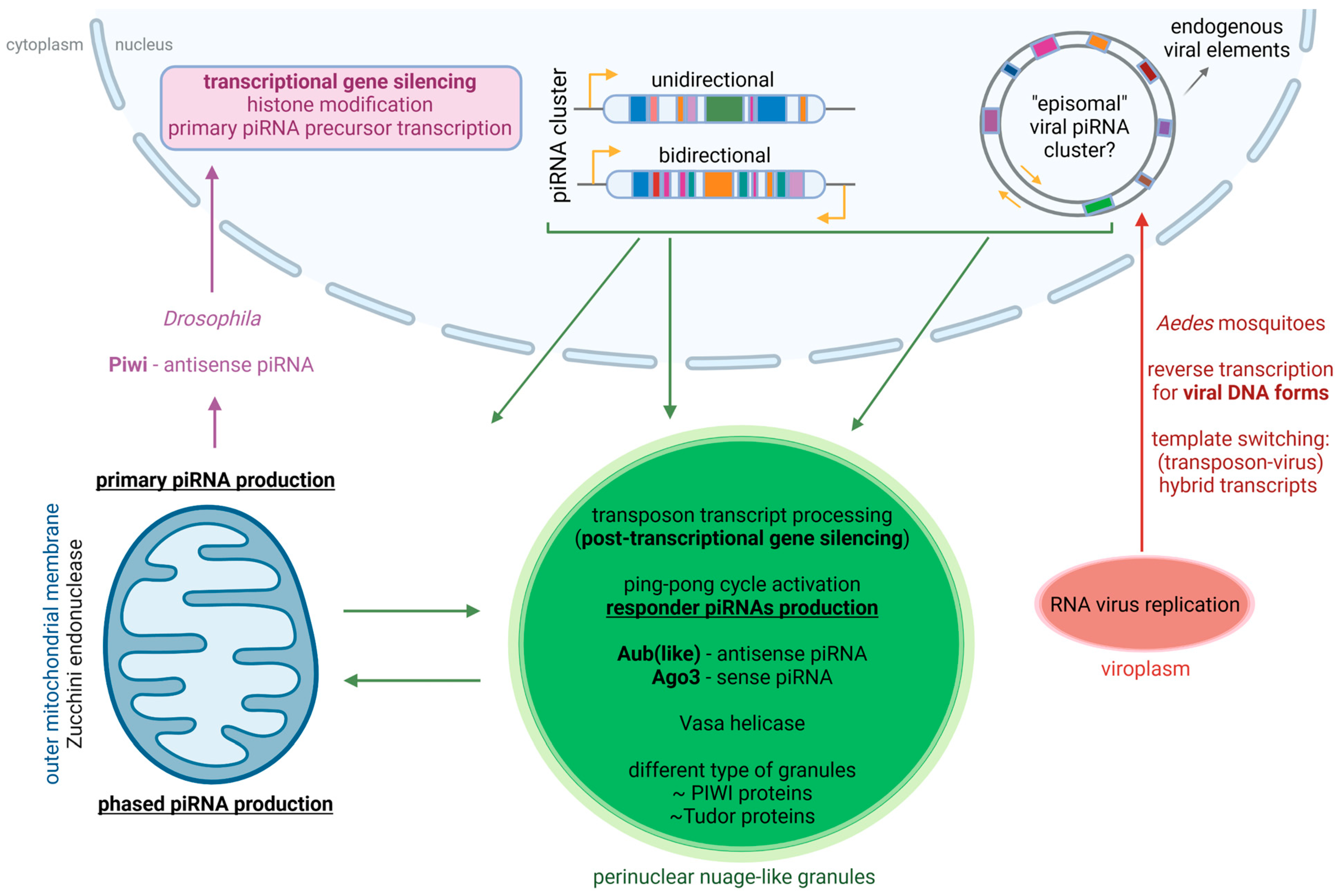
3.2. The piRNA Pathway as an Antiviral Defense Mechanism in Aedes Mosquitoes
3.3. Endogenous Viral Elements and Immunological Memory in Aedes Mosquitoes
3.4. Virus- and Mosquito-Dependent Variability of the piRNA Pathway as an Antiviral Defense Mechanism
3.5. Other (Non-Antiviral) Functions of piRNAs in Mosquitoes
4. Insights from Research on Piwi Proteins and piRNAs in the Silkworm
4.1. A Silkworm Cell Line as a Model for the Molecular Analysis of the piRNA Pathway
4.2. Control of Transposable Elements by the piRNA Pathway in the Silkworm
4.3. Sex Determination by the piRNA Pathway in the Silkworm
4.4. The piRNA Pathway as an Antiviral Defense Mechanism in Lepidoptera
4.5. piRNAs and the Stress Response in Silkworm Cells
5. Insights from Research on Piwi Proteins and piRNAs in Other Insects
5.1. piRNAs, piRNA Clusters and Piwi Genes in Cockroaches, Termites and Locusts
5.2. The Piwi Pathway in Hemiptera: Defense against Parasitic Elements in Whiteflies and Regulation of Reproductive Plasticity in Aphids
5.3. piRNAs, piRNA Clusters and Piwi Genes in Beetles (Coleoptera)
5.4. piRNAs, piRNA Clusters and Piwi Genes in Bees (Hymenoptera)
6. Discussion
6.1. A Universal piRNA Pathway
6.2. Rules for piRNA-Target Interactions
6.3. Expansion of the Function of piRNAs: From Transposon Control to Gene Regulation and Antiviral Defense
6.4. The piRNA Pathway as Guardian of the Genome: Implications for Stem Cell Function
7. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siomi, H.; Siomi, M.C. On the road to reading the RNA-interference code. Nature 2009, 457, 396–404. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Carthew, R.W.; Sontheimer, E.J. Origins and Mechanisms of miRNAs and siRNAs. Cell 2009, 136, 642–655. [Google Scholar] [CrossRef] [PubMed]
- Kingsolver, M.B.; Huang, Z.; Hardy, R.W. Insect antiviral innate immunity: Pathways, effectors, and connections. J. Mol. Biol. 2013, 425, 4921–4936. [Google Scholar] [CrossRef]
- Swevers, L.; Liu, J.; Smagghe, G. Defense mechanisms against viral infection in Drosophila: RNAi and non-RNAi. Viruses 2018, 10, 230. [Google Scholar] [CrossRef]
- Brennecke, J.; Aravin, A.A.; Stark, A.; Dus, M.; Kellis, M.; Sachidanandam, R.; Hannon, G.J. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 2007, 128, 1089–1103. [Google Scholar] [CrossRef]
- Gainetdinov, I.; Colpan, C.; Arif, A.; Cecchini, K.; Zamore, P.D. A single mechanism of biogenesis, initiated and directed by PIWI proteins, explains piRNA production in most Animals. Mol. Cell 2019, 71, 775–790. [Google Scholar] [CrossRef] [PubMed]
- Czech, B.; Hannon, G.J. One loop to rule them all: The ping-pong cycle and piRNA-guided silencing. Trends Biochem. Sci. 2016, 41, 324–337. [Google Scholar] [CrossRef]
- Robine, N.; Lau, N.C.; Balla, S.; Jin, Z.; Okamura, K.; Kuramochi-Miyagawa, S.; Blower, M.D.; Lai, E.C. A broadly conserved pathway generates 3′UTR-directed primary piRNAs. Curr. Biol. 2009, 19, 2066–2076. [Google Scholar] [CrossRef] [Green Version]
- Chen, P.; Kotov, A.A.; Godneeva, B.K.; Bazylev, S.S.; Olenina, L.V.; Aravin, A.A. piRNA-mediated gene regulation and adaptation to sex-specific transposon expression in D. melanogaster male germline. Genes Dev. 2021, 35, 914–935. [Google Scholar] [CrossRef]
- Blair, C.D. Deducing the role of virus genome-derived PIWI-associated RNAs in the mosquito–arbovirus arms race. Front. Genet. 2019, 10, 1114. [Google Scholar] [CrossRef]
- Liu, J.; Swevers, L.; Kolliopoulou, A.; Smagghe, G. Arboviruses and the challenge to establish systemic and persistent infections in competent mosquito vectors: The interaction with the RNAi mechanism. Front. Physiol. 2019, 10, 890. [Google Scholar] [CrossRef]
- Sakakibara, K.; Siomi, M.C. The PIWI-interacting RNA molecular pathway: Insights from cultured silkworm germline cells. BioEssays 2018, 40, 1700068. [Google Scholar] [CrossRef]
- Kiuchi, T.; Koga, H.; Kawamoto, M.; Shoji, K.; Sakai, H.; Arai, Y.; Ishihara, G.; Kawaoka, S.; Sugano, S.; Shimada, T.; et al. A single female-specific piRNA is the primary determiner of sex in the silkworm. Nature 2014, 509, 633–636. [Google Scholar] [CrossRef]
- Lewis, S.H.; Quarles, K.A.; Yang, Y.; Tanguy, M.; Frézal, L.; Smith, S.A.; Sharma, P.P.; Cordaux, R.; Gilbert, C.; Gitaud, I.; et al. Pan-arthropod analysis reveals somatic piRNAs as an ancestral defence against transposable elements. Nat. Ecol. Evol. 2018, 2, 174–181. [Google Scholar] [CrossRef]
- Malone, C.D.; Brennecke, J.; Dus, M.; Stark, A.; McCombie, W.R.; Sachidanandam, R.; Hannon, G.J. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 2009, 137, 522–535. [Google Scholar] [CrossRef]
- Nishida, K.M.; Saito, K.; Mori, T.; Kawamura, Y.; Nagami-Okada, T.; Inagaki, S.; Siomi, H.; Siomi, M.C. Gene silencing mechanisms mediated by Aubergine piRNA complexes in Drosophila male gonad. RNA 2007, 13, 1911–1922. [Google Scholar] [CrossRef]
- Czech, B.; Munafó, M.; Ciabrelli, F.; Eastwood, E.L.; Fabry, M.H.; Kneuss, E.; Hannon, G.J. piRNA-guided genome defense: From biogenesis to silencing. Annu. Rev. Genet. 2018, 52, 131–157. [Google Scholar] [CrossRef]
- Mohn, F.; Handler, D.; Brennecke, J. piRNA-guided slicing specifies transcripts for Zucchini dependent, phased piRNA biogenesis. Science 2015, 348, 812–817. [Google Scholar] [CrossRef] [Green Version]
- Gebert, D.; Neubert, L.K.; Lloyd, C.; Gui, J.; Lehmann, R.; Teixeira, F.K. Large Drosophila germline piRNA clusters are evolutionarily labile and dispensable for transposon regulation. Mol. Cell 2021, 81, 3965–3978. [Google Scholar] [CrossRef]
- Qi, H.; Watanabe, T.; Ku, H.-Y.; Liu, N.; Zhong, M.; Lin, H. The Yb body, a major site for Piwi-associated RNA biogenesis and a gateway for Piwi expression and transport to the nucleus in somatic cells. J. Biol. Chem. 2011, 286, 3789–3797. [Google Scholar] [CrossRef]
- Murota, Y.; Ishizu, H.; Nakagawa, S.; Iwasaki, Y.W.; Shibata, S.; Kamatani, M.K.; Saito, K.; Okano, H.; Siomi, H.; Siomi, M.C. Yb Integrates piRNA intermediates and processing factors into perinuclear bodies to enhance piRISC assembly. Cell Rep. 2014, 8, 103–113. [Google Scholar] [CrossRef]
- Klenov, M.S.; Sokolova, O.A.; Yakushev, E.Y.; Stolyarenko, A.D.; Mikhaleva, E.A.; Lavrov, S.A.; Gvozdev, V.A. Separation of stem cell maintenance and transposon silencing functions of Piwi protein. Proc. Natl. Acad. Sci. USA 2011, 108, 18760–18765. [Google Scholar] [CrossRef]
- Sienski, G.; Dönertas, D.; Brennecke, J. Transcriptional silencing of transposons by Piwi and maelstrom and its impact on chromatin state and gene expression. Cell 2012, 151, 964–980. [Google Scholar] [CrossRef]
- Schnabl, J.; Wang, J.; Hohmann, U.; Gehre, M.; Batki, J.; Andreev, V.I.; Purkhauser, K.; Fasching, N.; Duchek, P.; Novatchkova, M.; et al. Molecular principles of Piwi-mediated cotranscriptional silencing through the dimeric SFiNX complex. Genes Dev. 2021, 35, 392–409. [Google Scholar] [CrossRef]
- Stein, C.B.; Genzor, P.; Mitra, S.; Elchert, A.R.; Ipsaro, J.J.; Benner, L.; Sobti, S.; Su, Y.; Hammell, M.; Joshua-Tor, L.; et al. Decoding the 5′ nucleotide bias of PIWI-interacting RNAs. Nat. Commun. 2019, 10, 828. [Google Scholar] [CrossRef]
- Ozata, D.M.; Gainetdinov, I.; Zoch, A.; O’Carroll, D.; Zamore, P.D. PIWI-interacting RNAs: Small RNAs with big functions. Nat. Rev. Genet. 2019, 20, 89–108. [Google Scholar] [CrossRef]
- Handler, D.; Olivieri, D.; Novatchkova, M.; Gruber, F.S.; Meixner, K.; Mechtler, K.; Stark, A.; Sachidanandam, R.; Brennecke, J. A systematic analysis of Drosophila TUDOR domain-containing proteins identifies Vreteno and the Tdrd12 family as essential primary piRNA pathway factors. EMBO J. 2011, 30, 3977–3993. [Google Scholar] [CrossRef]
- Ge, D.T.; Wang, W.; Tipping, C.; Gainetdinov, I.; Weng, Z.; Zamore, P.D. The RNA-binding ATPase, Armitage, couples piRNA amplification in nuage to phased piRNA production on mitochondria. Mol. Cell 2019, 74, 982–995. [Google Scholar] [CrossRef]
- Reiss, D.; Josse, T.; Anxolabehere, D.; Ronsseray, S. Aubergine mutations in Drosophila melanogaster impair P cytotype determination by telomeric P elements inserted in heterochromatin. Mol. Genet. Genom. 2004, 272, 336–343. [Google Scholar] [CrossRef]
- Savitsky, M.; Kwon, D.; Georgiev, P.; Kalmykova, A.; Gvozdev, V. Telomere elongation is under the control of the RNAi-based mechanism in the Drosophila germline. Genes Dev. 2006, 20, 345–354. [Google Scholar] [CrossRef]
- Li, C.; Vagin, V.V.; Lee, S.; Xu, J.; Ma, S.; Xi, H.; Seitz, H.; Horwich, M.D.; Syrzycka, M.; Honda, B.M.; et al. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell 2009, 137, 509–521. [Google Scholar] [CrossRef]
- Senti, K.A.; Jurczak, D.; Sachidanandam, R.; Brennecke, J. piRNA guided slicing of transposon transcripts enforces their transcriptional silencing via specifying the nuclear piRNA repertoire. Genes Dev. 2015, 29, 1747–1762. [Google Scholar] [CrossRef] [PubMed]
- Duc, C.; Yoth, M.; Jensen, S.; Mouniée, N.; Bergman, C.M.; Vaury, C.; Brasset, E. Trapping a somatic endogenous retrovirus into a germline piRNA cluster immunizes the germline against further invasion. Genome Biol. 2019, 20, 127. [Google Scholar] [CrossRef]
- Luo, S.; Zhang, H.; Duan, Y.; Yao, X.; Clark, A.G.; Lu, J. The evolutionary arms race between transposable elements and piRNAs in Drosophila melanogaster. BMC Evol. Biol. 2020, 20, 14. [Google Scholar] [CrossRef]
- Mohn, F.; Sienski, G.; Handler, D.; Brennecke, J. The rhino-deadlock-cutoff complex licenses noncanonical transcription of dual-strand piRNA clusters in Drosophila. Cell 2014, 157, 1364–1379. [Google Scholar] [CrossRef]
- Andersen, P.R.; Tirian, L.; Vunjak, M.; Brennecke, J. A heterochromatin-dependent transcription machinery drives piRNA expression. Nature 2017, 549, 54–59. [Google Scholar] [CrossRef] [PubMed]
- Garavis, M.; Gonzalez, C.; Villasante, A. On the origin of the eukaryotic chromosome: The role of noncanonical DNA structures in telomere evolution. Genome Biol. Evol. 2013, 5, 1142–1150. [Google Scholar] [CrossRef]
- Radion, E.; Morgunova, V.; Ryazansky, S.; Akulenko, N.; Lavrov, S.; Abramov, Y.; Komarov, P.A.; Glukhov, S.I.; Olovnikov, I.; Kalmykova, A. Key role of piRNAs in telomeric chromatin maintenance and telomere nuclear positioning in Drosophila germline. Epigenetics Chromatin 2018, 11, 40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ilyin, A.A.; Stolyarenko, A.D.; Zenkin, N.; Klenov, M.S. Complex genetic interactions between Piwi and HP1a in the repression of transposable elements and tissue specific genes in the ovarian germline. Int. J. Mol. Sci. 2021, 22, 13430. [Google Scholar] [CrossRef]
- Ma, X.; Wang, S.; Do, T.; Song, X.; Inaba, M.; Nishimoto, Y.; Liu, L.P.; Gao, Y.; Mao, Y.; Li, H.; et al. Piwi is required in multiple cell types to control germline stem cell lineage development in the Drosophila ovary. PLoS ONE 2014, 9, e90267. [Google Scholar] [CrossRef] [PubMed]
- Klattenhoff, C.; Bratu, D.P.; McGinnis-Schultz, N.; Koppetsch, B.S.; Cook, H.A.; Theurkauf, W.E. Drosophila rasiRNA pathway mutations disrupt embryonic axis specification through activation of an ATR/Chk2 DNA damage response. Dev. Cell 2007, 12, 45–55. [Google Scholar] [CrossRef]
- Quénerch’du, E.; Anand, A.; Kai, T. The piRNA pathway is developmentally regulated during spermatogenesis in Drosophila. RNA 2016, 22, 1044–1054. [Google Scholar] [CrossRef] [PubMed]
- Saint-Leandre, B.; Capy, P.; Hua-Van, A.; Filée, J. piRNA and transposon dynamics in Drosophila: A female story. Genome Biol. Evol. 2020, 12, 931–947. [Google Scholar] [CrossRef]
- Nagao, A.; Mituyama, T.; Huang, H.; Chen, D.; Siomi, M.C.; Siomi, H. Biogenesis pathways of piRNAs loaded onto AGO3 in the Drosophila testis. RNA 2010, 16, 2503–2515. [Google Scholar] [CrossRef]
- Kibanov, M.V.; Egorova, K.S.; Ryazansky, S.S.; Sokolova, O.A.; Kotov, A.A.; Olenkina, O.M.; Stolyarenko, A.D.; Gvozdev, V.A.; Olenina, L.V. A novel organelle, the piNG-body, in the nuage of Drosophila male germ cells is associated with piRNA-mediated gene silencing. Mol. Biol. Cell 2011, 22, 3410–3419. [Google Scholar] [CrossRef]
- Malone, C.D.; Lehmann, R.; Teixeira, F.K. The cellular basis of hybrid dysgenesis and Stellate regulation in Drosophila. Curr. Opin. Genet. Dev. 2015, 34, 88–94. [Google Scholar] [CrossRef]
- Kotov, A.A.; Adashev, V.E.; Godneeva, B.K.; Ninova, M.; Shatskikh, A.S.; Bazylev, S.S.; Aravin, A.A.; Olenina, L.V. piRNA silencing contributes to interspecies hybrid sterility and reproductive isolation in Drosophila melanogaster. Nucleic Acids Res. 2019, 47, 4255–4271. [Google Scholar] [CrossRef]
- Molla Herman, A.; Brasset, E. Rhino breaks the deadlock in Drosophila testis. PLoS Genet. 2021, 17, e1009702. [Google Scholar] [CrossRef]
- Lin, H.; Spradling, A.C. A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development 1997, 124, 2463–2476. [Google Scholar] [CrossRef]
- Cox, D.N.; Chao, A.; Baker, J.; Chang, L.; Qiao, D.; Lin, H. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 1998, 12, 3715–3727. [Google Scholar] [CrossRef]
- Dansereau, D.A.; Lasko, P. The development of germline stem cells in Drosophila. Methods Mol. Biol. 2008, 450, 3–26. [Google Scholar] [PubMed]
- Cox, D.N.; Chao, A.; Lin, H. piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development 2000, 127, 503–514. [Google Scholar] [CrossRef] [PubMed]
- Szakmary, A.; Cox, D.N.; Wang, Z.; Lin, H. Regulatory relationship among piwi, pumilio, and bag-of-marbles in Drosophila germline stem cell self-renewal and differentiation. Curr. Biol. 2005, 15, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Klein, J.D.; Qu, C.; Yang, X.; Fan, Y.; Tang, C.; Peng, J.C. c-Fos repression by Piwi regulates Drosophila ovarian germline formation and tissue morphogenesis. PLoS Genet. 2016, 12, e1006281. [Google Scholar] [CrossRef]
- Peng, J.C.; Valouev, A.; Liu, N.; Lin, H. Piwi maintains germline stem cells and oogenesis in Drosophila through negative regulation of Polycomb group proteins. Nat. Genet. 2016, 48, 283–291. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, M.; Martino Cortez, Y.; Wong-Deyrup, S.; Tavares, L.; Schowalter, S.; Flora, P.; Hill, C.; Nasrallah, M.A.; Chittur, S.; Rangan, P. Transposon dysregulation modulates dWnt4 signaling to control germline stem cell differentiation in Drosophila. PLoS Genet. 2016, 12, e1005918. [Google Scholar] [CrossRef]
- Song, S.U.; Gerasimova, T.; Kurkulos, M.; Boeke, J.D.; Corces, V.G. An env-like protein encoded by a Drosophila retroelement: Evidence that gypsy is an infectious retrovirus. Genes Dev. 1994, 8, 2046–2057. [Google Scholar] [CrossRef]
- de Cuevas, M.; Matunis, E.L. The stem cell niche: Lessons from the Drosophila testis. Development 2011, 138, 2861–2869. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez, J.; Qi, H.; Liu, N.; Lin, H. Piwi is a key regulator of both somatic and germline stem cells in the Drosophila testis. Cell Rep. 2015, 12, 150–161. [Google Scholar] [CrossRef]
- Saito, K.; Inagaki, S.; Mituyama, T.; Kawamura, Y.; Ono, Y.; Sakota, E.; Kotani, H.; Asai, K.; Siomi, H.; Siomi, M.C. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature 2009, 461, 1296–1299. [Google Scholar] [CrossRef]
- Post, C.; Clark, J.P.; Sytnikova, Y.A.; Chirn, G.W.; Lau, N.C. The capacity of target silencing by Drosophila PIWI and piRNAs. RNA 2014, 20, 1977–1986. [Google Scholar] [CrossRef]
- Ishizu, H.; Iwasaki, Y.W.; Hirakata, S.; Ozaki, H.; Iwasaki, W.; Siomi, H.; Siomi, M.C. Somatic primary piRNA biogenesis driven by cis-acting RNA elements and trans-acting Yb. Cell Rep. 2015, 12, 429–440. [Google Scholar] [CrossRef]
- Ma, X.; Zhu, X.; Han, Y.; Story, B.; Do, T.; Song, X.; Wang, S.; Zhang, Y.; Blanchette, M.; Gogol, M.; et al. Aubergine controls germline stem cell self-renewal and progeny differentiation via distinct mechanisms. Dev. Cell 2017, 41, 157–169. [Google Scholar] [CrossRef]
- Rojas-Ríos, P.; Chartier, A.; Pierson, S.; Simonelig, M. Aubergine and piRNAs promote germline stem cell self-renewal by repressing the proto-oncogene Cbl. EMBO J. 2017, 36, 3194–3211. [Google Scholar] [CrossRef]
- Vagin, V.V.; Sigova, A.; Li, C.; Seitz, H.; Gvozdev, V.; Zamore, P.D. A distinct small RNA pathway silences selfish genetic elements in the germline. Science 2006, 313, 320–324. [Google Scholar] [CrossRef]
- Winata, C.L.; Khorz, V. The translational regulation of maternal mRNAs in time and space. FEBS Lett. 2018, 592, 3007–3023. [Google Scholar] [CrossRef]
- Barckmann, B.; Pierson, S.; Dufourt, J.; Papin, C.; Armenise, C.; Port, F.; Grentzinger, T.; Chambeyron, S.; Baronian, G.; Desvignes, J.-P.; et al. Aubergine iCLIP reveals piRNA-dependent decay of mRNAs involved in germ cell development in the early embryo. Cell Rep. 2015, 12, 1205–1216. [Google Scholar] [CrossRef]
- Vourekas, A.; Alexiou, P.; Vrettos, N.; Maragkakis, M.; Mourelatos, Z. Sequence-dependent but not sequence-specific piRNA adhesion traps mRNAs to the germ plasm. Nature 2016, 531, 390–394. [Google Scholar] [CrossRef] [Green Version]
- Rojas-Ríos, P.; Simonelig, M. piRNAs and PIWI proteins: Regulators of gene expression in development and stem cells. Development 2018, 145, dev161786. [Google Scholar] [CrossRef]
- Rouget, C.; Papin, C.; Boureux, A.; Meunier, A.-C.; Franco, B.; Robine, N.; Lai, E.C.; Pelisson, A.; Simonelig, M. Maternal mRNA deadenylation and decay by the piRNA pathway in the early Drosophila embryo. Nature 2010, 467, 1128–1132. [Google Scholar] [CrossRef]
- Harris, A.N.; Macdonald, P.M. Aubergine encodes a Drosophila polar granule component required for pole cell formation and related to eIF2C. Development 2001, 128, 2823–2832. [Google Scholar] [CrossRef]
- Mahowald, A.P. Assembly of the Drosophila germ plasm. Int. Rev. Cytol. 2001, 203, 187–213. [Google Scholar]
- Brennecke, J.; Malone, C.D.; Aravin, A.A.; Sachidanandam, R.; Stark, A.; Hannon, G.J. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science 2008, 322, 1387–1392. [Google Scholar] [CrossRef]
- Le Thomas, A.; Stuwe, E.; Li, S.; Du, J.; Marinov, G.; Rozhkov, N.; Chen, Y.C.; Luo, Y.; Sachidanandam, R.; Toth, K.F.; et al. Transgenerationally inherited piRNAs trigger piRNA biogenesis by changing the chromatin of piRNA clusters and inducing precursor processing. Genes Dev. 2014, 28, 1667–1680. [Google Scholar] [CrossRef] [PubMed]
- Dufourt, J.; Bontonou, G.; Chartier, A.; Jahan, C.; Meunier, A.-C.; Pierson, S.; Harrison, P.F.; Papin, C.; Beilharz, T.H.; Simonelig, M. piRNAs and Aubergine cooperate with Wispy poly(A) polymerase to stabilize mRNAs in the germ plasm. Nat. Commun. 2017, 8, 1305. [Google Scholar] [CrossRef]
- Mani, S.R.; Megosh, H.; Lin, H. Piwi proteins are essential for early Drosophila embryogenesis. Dev. Biol. 2014, 385, 340–349. [Google Scholar] [CrossRef]
- Gonzalez, L.E.; Tang, X.; Lin, H. Maternal Piwi regulates primordial germ cell development to ensure the fertility of female progeny in Drosophila. Genetics 2021, 219, iyab091. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Knowles, B.A.; Goldstein, L.S.; Hawley, R.S. A kinesin-like protein required for distributive chromosome segregation in Drosophila. Cell 1990, 62, 1053–1062. [Google Scholar] [CrossRef] [PubMed]
- Czech, B.; Malone, C.D.; Zhou, R.; Stark, A.; Schlingeheyde, C.; Dus, M.; Perrimon, N.; Kellis, M.; Wohlschlegel, J.A.; Sachidanandam, R.; et al. An endogenous small interfering RNA pathway in Drosophila. Nature 2008, 453, 798–802. [Google Scholar] [CrossRef] [Green Version]
- Ghildiyal, M.; Seitz, H.; Horwich, M.D.; Li, C.; Du, T.; Lee, S.; Xu, J.; Kittler, E.L.; Zapp, M.L.; Weng, Z.; et al. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science 2008, 320, 1077–1081. [Google Scholar] [CrossRef]
- Kawamura, Y.; Saito, K.; Kin, T.; Ono, Y.; Asai, K.; Sunohara, T.; Okada, T.N.; Siomi, M.C.; Siomi, H. Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature 2008, 453, 793–797. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Rand, T.A.; Kalidas, S.; Du, F.; Kim, H.E.; Smith, D.P.; Wang, X. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science 2003, 301, 1921–1925. [Google Scholar] [CrossRef]
- Ross, R.J.; Weiner, M.M.; Lin, H. PIWI proteins and PIWI-interacting RNAs in the soma. Nature 2014, 505, 353–359. [Google Scholar] [CrossRef] [PubMed]
- Brower-Toland, B.; Findley, S.D.; Jiang, L.; Liu, L.; Yin, H.; Dus, M.; Zhou, P.; Elgin, S.C.; Lin, H. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes Dev. 2007, 21, 2300–2311. [Google Scholar] [CrossRef] [PubMed]
- Jones, B.C.; Wood, J.G.; Chang, C.; Tam, A.D.; Franklin, M.J.; Siegel, E.R.; Helfand, S.L. A somatic piRNA pathway in the Drosophila fat body ensures metabolic homeostasis and normal lifespan. Nat. Commun. 2016, 7, 13856. [Google Scholar] [CrossRef] [PubMed]
- Perrat, P.N.; DasGupta, S.; Wang, J.; Theurkauf, W.; Weng, Z.; Rosbash, M.; Waddell, S. Transposition-driven genomic heterogeneity in the Drosophila brain. Science 2013, 340, 91–95. [Google Scholar] [CrossRef]
- Janic, A.; Mendizabal, L.; Llamazares, S.; Rossell, D.; Gonzalez, C. Ectopic expression of germline genes drives malignant brain tumor growth in Drosophila. Science 2010, 330, 1824–1827. [Google Scholar] [CrossRef]
- Lau, N.; Robine, N.; Martin, R.; Chung, W.J.; Niki, Y.; Berezikov, E.; Lai, E.C. Abundant primary piRNAs, endo-siRNAs and microRNAs in a Drosophila ovary cell line. Genome Res. 2009, 19, 1776–1785. [Google Scholar] [CrossRef]
- Wen, J.; Mohammed, J.; Bortolamiol-Becet, D.; Tsai, H.; Robine, N.; Westholm, J.O.; Ladewig, E.; Dai, Q.; Okamura, K.; Flynt, A.S.; et al. Diversity of miRNAs, siRNAs, and piRNAs across 25 Drosophila cell lines. Genome Res. 2014, 24, 1236–1250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yashiro, R.; Murota, Y.; Nishida, K.M.; Yamashiro, H.; Fujii, K.; Ogai, A.; Yamanaka, S.; Negishi, L.; Siomi, H.; Siomi, M.C. Piwi nuclear localization and its regulatory mechanism in Drosophila ovarian somatic cells. Cell Rep. 2018, 23, 3647–3657. [Google Scholar] [CrossRef]
- Vrettos, N.; Maragkakis, M.; Alexiou, P.; Mourelatos, Z. Kc167, a widely used Drosophila cell line, contains an active primary piRNA pathway. RNA 2017, 23, 108–118. [Google Scholar] [CrossRef]
- Siomi, M.C.; Sato, K.; Pezic, D.; Aravin, A.A. PIWI-interacting small RNAs: The vanguard of genome defence. Nat. Rev. Mol. Cell Biol. 2011, 12, 246–258. [Google Scholar] [CrossRef]
- Miesen, P.; Joosten, J.; van Rij, R.P. PIWIs go viral: Arbovirus-derived piRNAs in vector mosquitoes. PLoS Pathog. 2016, 12, e1006017. [Google Scholar] [CrossRef] [PubMed]
- Tassetto, M.; Kunitomi, M.; Whitfield, Z.J.; Dolan, P.T.; Sánchez-Vargas, I.; Garcia-Knight, M.; Ribiero, I.; Chen, T.; Olson, K.E.; Andino, R. Control of RNA viruses in mosquito cells through the acquisition of vDNA and endogenous viral elements. eLife 2019, 8, e41244. [Google Scholar] [CrossRef]
- Wu, Q.; Luo, Y.; Lu, R.; Lau, N.; Lai, E.C.; Li, W.-X.; Ding, S.-W. Virus discovery by deep sequencing and assembly of virus-derived small silencing RNAs. Proc. Natl. Acad. Sci. USA 2010, 107, 1606–1611. [Google Scholar] [CrossRef] [PubMed]
- Petit, M.; Mongelli, V.; Frangeul, L.; Blanc, H.; Jiggins, F.; Saleh, M.C. piRNA pathway is not required for antiviral defense in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2016, 113, E4218–E4227. [Google Scholar] [CrossRef]
- Vodovar, N.; Bronkhorst, A.W.; van Cleef, K.W.R.; Miesen, P.; Blanc, H.; van Rij, R.P.; Saleh, M.-C. Arbovirus-derived piRNAs exhibit a ping-pong signature in mosquito cells. PLoS ONE 2012, 7, e30861. [Google Scholar] [CrossRef]
- Campbell, C.L.; Black, W.C.; Hess, A.M.; Foy, B.D. Comparative genomics of small RNA regulatory pathway components in vector mosquitoes. BMC Genom. 2008, 9, 425. [Google Scholar]
- Marconcini, M.; Hernandez, L.; Iovino, G.; Houé, V.; Valerio, F.; Palatini, U.; Pischedda, E.; Crawford, J.E.; White, B.J.; Lin, T.; et al. Polymorphism analyses and protein modelling inform on functional specialization of Piwi clade genes in the arboviral vector Aedes albopictus. PLoS Negl. Trop. Dis. 2019, 13, e0007919. [Google Scholar] [CrossRef]
- Arensburger, P.; Hice, R.H.; Wright, J.A.; Craig, N.L.; Atkinson, P.W. The mosquito Aedes aegypti has a large genome size and high transposable element load but contains a low proportion of transposon-specific piRNAs. BMC Genom. 2011, 12, 606. [Google Scholar] [CrossRef] [Green Version]
- Girardi, E.; Miesen, P.; Pennings, B.; Frangeul, L.; Saleh, M.-C.; van Rij, R.P. Histone-derived piRNA biogenesis depends on the ping-pong partners Piwi5 and Ago3 in Aedes aegypti. Nucleic Acids Res. 2017, 45, 4881–4892. [Google Scholar] [PubMed]
- Palatini, U.; Masri, R.A.; Cosme, L.V.; Koren, S.; Thibaud-Nissen, F.; Biedler, J.K.; Krsticevic, F.; Johnston, J.S.; Halbach, R.; Crawford, J.E.; et al. Improved reference genome of the arboviral vector Aedes albopictus. Genome Biol. 2020, 21, 215. [Google Scholar] [CrossRef] [PubMed]
- ter Horst, A.M.; Nigg, J.C.; Dekker, F.M.; Falk, B.W. Endogenous viral elements are widespread in arthropod genomes and commonly give rise to PIWI-interacting RNAs. J. Virol. 2019, 93, e02124-18. [Google Scholar] [CrossRef] [PubMed]
- Pischedda, E.; Scolari, F.; Valerio, F.; Carballar-Lejarazú, R.; Catapano, P.L.; Waterhouse, R.M.; Bonizzoni, M. Insights into an unexplored component of the mosquito repeatome: Distribution and variability of viral sequences integrated into the genome of the arboviral vector Aedes Albopictus. Front. Genet. 2019, 10, 93. [Google Scholar] [CrossRef]
- Miesen, P.; Girardi, E.; van Rij, R.P. Distinct sets of PIWI proteins produce arbovirus and transposon-derived piRNAs in Aedes aegypti mosquito cells. Nucleic Acids Res. 2015, 43, 6545–6556. [Google Scholar] [CrossRef]
- Akbari, O.S.; Antoshechkin, I.; Amrhein, H.; Williams, B.; Diloreto, R.; Sandler, J.; Hay, B.A. The developmental transcriptome of the mosquito Aedes aegypti, an invasive species and major arbovirus vector. G3 2013, 3, 1493–1509. [Google Scholar] [CrossRef]
- Joosten, J.; Taşköprü, E.; Jansen, P.W.T.C.; Pennings, B.; Vermeulen, M.; Van Rij, R.P. PIWI proteomics identifies Atari and Pasilla as piRNA biogenesis factors in Aedes mosquitoes. Cell Rep. 2021, 35, 109073. [Google Scholar] [CrossRef]
- Joosten, J.; Overheul, G.J.; Van Rij, R.P.; Miesen, P. Endogenous piRNA-guided slicing triggers responder and trailer piRNA production from viral RNA in Aedes aegypti mosquitoes. Nucleic Acids Res. 2021, 49, 8886–8899. [Google Scholar] [CrossRef]
- Joosten, J.; Miesen, P.; Taşköprü, E.; Pennings, B.; Jansen, P.W.T.C.; Huynen, M.A.; Vermeulen, M.; Van Rij, R.P. The Tudor protein Veneno assembles the ping-pong amplification complex that produces viral piRNAs in Aedes mosquitoes. Nucleic Acids Res. 2019, 47, 2546–2559. [Google Scholar] [CrossRef] [PubMed]
- Schnettler, E.; Donald, C.L.; Human, S.; Watson, M.; Siu, R.W.C.; McFarlane, M.; Fazakerley, J.K.; Kohl, A.; Fragkoudis, R. Knockdown of piRNA pathway proteins results in enhanced Semliki Forest virus production in mosquito cells. J. Gen. Virol. 2013, 94, 1680–1689. [Google Scholar] [CrossRef] [Green Version]
- McFarlane, M.; Arias-Goeta, C.; Martin, E.; O’Hara, Z.; Lulla, A.; Mousson, L.; Rainey, S.M.; Misbah, S.; Schnettler, E.; Donald, C.L.; et al. Characterization of Aedes aegypti innate-immune pathways that limit Chikungunya virus replication. PLoS Negl. Trop. Dis. 2014, 8, e2994. [Google Scholar] [CrossRef] [PubMed]
- Varjak, M.; Maringer, K.; Watson, M.; Sreenu, V.B.; Fredericks, A.C.; Pondeville, E.; Donald, C.L.; Sterk, J.; Kean, J.; Vazeille, M.; et al. Aedes aegypti Piwi4 is a noncanonical PIWI protein involved in antiviral responses. mSphere 2017, 2, e00144-17. [Google Scholar] [CrossRef]
- Varjak, M.; Donald, C.L.; Mottram, T.J.; Sreenu, V.B.; Merits, A.; Maringer, K.; Schnettler, E.; Kohl, A. Characterization of the Zika virus induced small RNA response in Aedes aegypti cells. PLoS Negl. Trop. Dis. 2017, 11, e0006010. [Google Scholar] [CrossRef]
- Dietrich, I.; Jansen, S.; Fall, G.; Lorenzen, S.; Rudolf, M.; Huber, K.; Heitmann, A.; Schicht, S.; Ndiaye, E.H.; Watson, M.; et al. RNA interference restricts Rift Valley Fever virus in multiple insect systems. mSphere 2017, 2, e00090-17. [Google Scholar] [CrossRef]
- Varjak, M.; Dietrich, I.; Sreenu, V.B.; Till, B.E.; Merits, A.; Kohl, A.; Schnettler, E. Spindle-E acts antivirally against alphaviruses in mosquito cells. Viruses 2018, 10, 88. [Google Scholar] [CrossRef]
- Rosendo Machado, S.; van der Most, T.; Miesen, P. Genetic determinants of antiviral immunity in dipteran insects—Compiling the experimental evidence. Dev. Comp. Immunol. 2021, 119, 104010. [Google Scholar] [CrossRef] [PubMed]
- Goic, B.; Stapleford, K.A.; Frangeul, L.; Doucet, A.J.; Gausson, V.; Blanc, H.; Schemmel-Jofre, N.; Cristofari, G.; Lambrechts, L.; Vignuzzi, M.; et al. Virus-derived DNA drives mosquito vector tolerance to arboviral infection. Nat. Commun. 2016, 7, 12410. [Google Scholar] [CrossRef]
- Nag, D.K.; Brecher, M.; Kramer, L.D. DNA forms of arboviral RNA genomes are generated following infection in mosquito cell cultures. Virology 2016, 498, 164–171. [Google Scholar] [CrossRef]
- Poirier, E.Z.; Goic, B.; Tomé-Poderti, L.; Frangeul, L.; Boussier, J.; Gausson, V.; Blanc, H.; Vallet, T.; Loyd, H.; Levi, L.I.; et al. Dicer-2-dependent generation of viral DNA from defective genomes of RNA viruses modulates antiviral immunity in Insects. Cell Host Microbe 2018, 23, 353–365. [Google Scholar] [CrossRef]
- Parry, R.; Bishop, C.; De Hayr, L.; Asgari, S. Density-dependent enhanced replication of a densovirus in Wolbachia-infected Aedes cells is associated with production of piRNAs and higher virus-derived siRNAs. Virology 2019, 528, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Morazzani, E.M.; Wiley, M.R.; Murreddu, M.G.; Adelman, Z.N.; Myles, K.M. Production of virus-derived pingpong-dependent piRNA-like small RNAs in the mosquito soma. PLoS Pathog. 2012, 8, e1002470. [Google Scholar] [CrossRef]
- Gestuveo, R.J.; Parry, R.; Dickson, L.B.; Lequime, S.; Sreenu, V.B.; Arnold, M.J.; Khromykh, A.A.; Schnettler, E.; Lambrechts, L.; Varjak, M.; et al. Mutational analysis of Aedes aegypti Dicer 2 provides insights into the biogenesis of antiviral exogenous small interfering RNAs. PLoS Pathog. 2022, 18, e1010202. [Google Scholar] [CrossRef] [PubMed]
- Whitfield, Z.J.; Dolan, P.T.; Kunitomi, M.; Tassetto, M.; Seetin, M.G.; Oh, S.; Heiner, C.; Paxinos, E.; Andino, R. The diversity, structure, and function of heritable adaptive immunity sequences in the Aedes aegypti genome. Curr. Biol. 2017, 27, 3511–3519. [Google Scholar] [CrossRef] [PubMed]
- Palatini, U.; Miesen, P.; Carballar-Lejarazu, R.; Ometto, L.; Rizzo, E.; Tu, Z.; van Rij, R.P.; Bonizzoni, M. Comparative genomics shows that viral integrations are abundant and express piRNAs in the arboviral vectors Aedes aegypti and Aedes albopictus. BMC Genom. 2017, 18, 512. [Google Scholar] [CrossRef]
- Suzuki, Y.; Frangeul, L.; Dickson, L.B.; Blanc, H.; Verdier, Y.; Vinh, J.; Lambrechts, L.; Saleh, M.-C. Uncovering the repertoire of endogenous flaviviral elements in Aedes mosquito genomes. J. Virol. 2017, 91, e00571-17. [Google Scholar] [CrossRef]
- Crava, C.M.; Varghese, F.S.; Pischedda, E.; Halbach, R.; Palatini, U.; Marconcini, M.; Gasmi, L.; Redmond, S.; Afrane, Y.; Ayala, D.; et al. Population genomics in the arboviral vector Aedes aegypti reveals the genomic architecture and evolution of endogenous viral elements. Mol. Ecol. 2020, 30, 1594–1611. [Google Scholar] [CrossRef] [PubMed]
- Dezordi, F.Z.; Vasconcelos, C.R.D.S.; Rezende, A.M.; Wallau, G.L. In and outs of Chuviridae endogenous viral elements: Origin of a potentially new retrovirus and signature of ancient and ongoing arms race in mosquito genomes. Front. Genet. 2020, 11, 542437. [Google Scholar] [CrossRef] [PubMed]
- Marconcini, M.; Pischedda, E.; Houé, V.; Palatini, U.; Lozada-Chávez, N.; Sogliani, D.; Failloux, A.-B.; Bonizzoni, M. Profile of small RNAs, vDNA forms and viral integrations in late Chikungunya virus infection of Aedes albopictus mosquitoes. Viruses 2021, 13, 553. [Google Scholar] [CrossRef]
- Suzuki, Y.; Baidaliuk, A.; Miesen, P.; Frangeul, L.; Crist, A.B.; Merkling, S.H.; Fontaine, A.; Lequime, S.; Moltini-Conclois, I.; Blanc, H.; et al. Non-retroviral endogenous viral element limits cognate virus replication in Aedes aegypti ovaries. Curr. Biol. 2020, 30, 3495–3506. [Google Scholar] [CrossRef] [PubMed]
- Ophinni, Y.; Palatini, U.; Hayashi, Y.; Parrish, N.F. piRNA-guided CRISPR-like immunity in Eukaryotes. Trends Immunol. 2019, 40, 998–1010. [Google Scholar] [CrossRef] [PubMed]
- Blair, C.D.; Olson, K.E.; Bonizzoni, M. The widespread occurrence and potential biological roles of endogenous viral elements in insect genomes. Curr. Issues Mol. Biol. 2020, 34, 13–30. [Google Scholar] [CrossRef] [PubMed]
- Rückert, C.; Prasad, A.N.; Garcia-Luna, S.M.; Robison, A.; Grubaugh, N.D.; Weger-Lucarelli, J.; Ebel, G.D. Small RNA responses of Culex mosquitoes and cell lines during acute and persistent virus infection. Insect Biochem. Mol. Biol. 2019, 109, 13–23. [Google Scholar] [CrossRef]
- Göertz, G.P.; Miesen, P.; Overheul, G.J.; van Rij, R.P.; van Oers, M.M.; Pijlman, G.P. Mosquito small RNA responses to West Nile and insect-specific virus infections in Aedes and Culex mosquito cells. Viruses 2019, 11, 271. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Srivastav, S.P.; Gamez, S.; Dayama, G.; Feitosa-Suntheimer, F.; Patterson, E.I.; Johnson, R.M.; Matson, E.M.; Gold, A.S.; Brackney, D.E.; et al. A mosquito small RNA genomics resource reveals dynamic evolution and host responses to viruses and transposons. Genome Res. 2021, 31, 512–528. [Google Scholar] [CrossRef] [PubMed]
- Olson, K.E.; Bonizzoni, M. Nonretroviral integrated RNA viruses in arthropod vectors: An occasional event or something more? Curr. Opin. Insect Sci. 2017, 22, 45–53. [Google Scholar] [CrossRef]
- Öhlund, P.; Hayer, J.; Hesson, J.C.; Blomström, A.L. Small RNA response to infection of the insect-specific Lammi virus and Hanko virus in an Aedes albopictus cell line. Viruses 2021, 13, 2181. [Google Scholar] [CrossRef]
- Palatini, U.; Contreras, C.A.; Gasmi, L.; Bonizzoni, M. Endogenous viral elements in mosquito genomes: Current knowledge and outstanding questions. Curr. Opin. Insect Sci. 2022, 49, 22–30. [Google Scholar] [CrossRef]
- Halbach, R.; Miesen, P.; Joosten, J.; Taşköprü, E.; Rondeel, I.; Pennings, B.; Vogels, C.B.F.; Merkling, S.H.; Koenraadt, C.J.; Lambrechts, L.; et al. A satellite repeat-derived piRNA controls embryonic development of Aedes. Nature 2020, 580, 274–277. [Google Scholar] [CrossRef]
- Betting, V.; Joosten, J.; Halbach, R.; Thaler, M.; Miesen, P.; van Rij, R.P. A piRNA-lncRNA regulatory network initiates responder and trailer piRNA formation during mosquito embryonic development. RNA 2021, 27, 1155–1172. [Google Scholar] [CrossRef]
- Williams, A.E.; Shrivastava, G.; Gittis, A.G.; Ganesan, S.; Martin-Martin, I.; Valenzuela Leon, P.C.; Olson, K.E.; Calvo, E. Aedes aegypti Piwi4 structural features are necessary for RNA binding and nuclear localization. Int. J. Mol. Sci. 2021, 22, 12733. [Google Scholar] [CrossRef]
- Handler, D.; Meixner, K.; Pizka, M.; Lauss, K.; Schmied, C.; Gruber, F.S.; Brennecke, J. The Genetic Makeup of the Drosophila piRNA Pathway. Mol. Cell 2013, 50, 762–777. [Google Scholar] [CrossRef]
- Kawaoka, S.; Hayashi, N.; Suzuki, Y.; Abe, H.; Sugano, S.; Tomari, Y.; Shimada, T.; Katsuma, S. The Bombyx ovary-derived cell line endogenously expresses PIWI/PIWI-interacting RNA complexes. RNA 2009, 15, 1258–1264. [Google Scholar] [CrossRef] [PubMed]
- Dowling, D.; Pauli, T.; Donath, A.; Meusemann, K.; Podsiadlowski, L.; Petersen, M.; Peters, R.S.; Mayer, C.; Liu, S.; Zhou, X.; et al. Phylogenetic origin and oiversification of RNAi pathway genes in insects. Genome Biol. Evol. 2016, 8, 3784–3793. [Google Scholar]
- Kawaoka, S.; Minami, K.; Katsuma, S.; Mita, K.; Shimada, T. Developmentally synchronized expression of two Bombyx mori Piwi subfamily genes, SIWI and BmAGO3 in germ-line cells. Biochem. Biophys. Res. Commun. 2008, 367, 755–760. [Google Scholar] [CrossRef] [PubMed]
- Xiol, J.; Spinelli, P.; Laussmann, M.A.; Hornolka, D.; Yang, Z.; Cora, E.; Couté, Y.; Conn, S.; Kadlec, J.; Sachidanandam, R.; et al. RNA clamping by Vasa assembles a piRNA amplifier complex on transposon transcripts. Cell 2014, 157, 1698–1711. [Google Scholar] [CrossRef]
- Tatsuke, T.; Zhu, L.; Li, Z.; Mitsunobu, H.; Yoshimura, K.; Mon, H.; Lee, J.M.; Kusakabe, T. Roles of Piwi proteins in transcriptional regulation mediated by HP1s in cultured silkworm cells. PLoS ONE 2014, 9, e92313. [Google Scholar] [CrossRef]
- Kawaoka, S.; Hara, K.; Shoji, K.; Kobayashi, M.; Shimada, T.; Sugano, S.; Tomari, Y.; Suzuki, Y.; Katsuma, S. The comprehensive epigenome map of piRNA clusters. Nucl. Acid Res. 2013, 41, 1581–1590. [Google Scholar] [CrossRef] [PubMed]
- Patil, A.A.; Tatsuke, T.; Mon, H.; Lee, J.M.; Morokuma, D.; Hino, M.; Kusakabe, T. Molecular characterization of mitochondrial Zucchini and its relation to nuage-piRNA pathway components in Bombyx mori ovary-derived BmN4 cells. Biochem. Biophys. Res. Commun. 2017, 493, 971–978. [Google Scholar] [CrossRef]
- Kawaoka, S.; Izumi, N.; Katsuma, S.; Tomari, Y. 3′ End formation of PIWI-interacting RNAs in vitro. Mol. Cell 2011, 43, 1015–1022. [Google Scholar] [CrossRef]
- Nishida, K.M.; Iwasaki, Y.W.; Murota, Y.; Nagao, A.; Mannen, T.; Kato, Y.; Siomi, H.; Siomi, M.C. Respective functions of two distinct Siwi complexes assembled during PIWI-interacting RNA biogenesis in Bombyx germ cells. Cell Rep. 2015, 10, 193–203. [Google Scholar] [CrossRef]
- Nishida, K.M.; Sakakibara, K.; Sumiyoshi, T.; Yamazaki, H.; Mannen, T.; Kawamura, T.; Kodama, T.; Siomi, M.C. Siwi levels reversibly regulate secondary piRISC biogenesis by affecting Ago3 body morphology in Bombyx mori. EMBO J. 2020, 39, e105130. [Google Scholar] [CrossRef] [PubMed]
- Izumi, N.; Shoji, K.; Suzuki, Y.; Katsuma, S.; Tomari, Y. Zucchini consensus motifs determine the mechanism of pre-piRNA production. Nature 2020, 578, 311–316. [Google Scholar] [CrossRef]
- Izumi, N.; Shoji, K.; Sakaguchi, Y.; Honda, S.; Kirino, Y.; Suzuki, T.; Katsuma, S.; Tomari, Y. Identification and functional analysis of the pre-piRNA 3′ Trimmer in silkworms. Cell 2016, 164, 962–973. [Google Scholar] [CrossRef] [PubMed]
- Czech, B.; Hannon, G.J. A Happy 3′ Ending to the piRNA Maturation Story. Cell 2016, 164, 838–840. [Google Scholar] [CrossRef]
- Nishida, K.M.; Sakakibara, K.; Iwasaki, Y.W.; Yamada, H.; Murakami, R.; Murota, Y.; Kawamura, T.; Kodama, T.; Siomi, H.; Siomi, M.C. Hierarchical roles of mitochondrial Papi and Zucchini in Bombyx germline piRNA biogenesis. Nature 2018, 555, 260–264. [Google Scholar] [CrossRef] [PubMed]
- Murakami, R.; Sumiyoshi, T.; Negishi, L.; Siomi, M.C. DEAD-box polypeptide 43 facilitates piRNA amplification by actively liberating RNA from Ago3-piRISC. EMBO Rep. 2021, 22, e51313. [Google Scholar] [CrossRef]
- Xiao, Y.; Ke, A. PIWI Takes a Giant Step. Cell 2016, 167, 310–312. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, N.; Nishimasu, H.; Sakakibara, K.; Nishida, K.M.; Hirano, T.; Ishitani, R.; Siomi, H.; Siomi, M.C.; Nureki, O. Crystal structure of silkworm PIWI-clade Argonaute Siwi bound to piRNA. Cell 2016, 167, 484–497. [Google Scholar] [CrossRef]
- Xiol, J.; Cora, E.; Koglgruber, R.; Chuma, S.; Subramanian, S.; Hosokawa, M.; Reuter, M.; Yang, Z.; Berninger, P.; Palencia, A.; et al. A Role for Fkbp6 and the chaperone machinery in piRNA amplification and transposon silencing. Mol. Cell 2012, 47, 970–979. [Google Scholar] [CrossRef]
- Namba, Y.; Iwasaki, Y.W.; Nishida, K.M.; Nishihara, H.; Sumiyoshi, T.; Siomi, M.C. Maelstrom functions in the production of Siwi-piRISC capable of regulating transposons in Bombyx germ cells. iScience 2022, 25, 103914. [Google Scholar] [CrossRef]
- Honda, S.; Kirino, Y.; Maragkakis, M.; Alexiou, P.; Ohtaki, A.; Murali, R.; Mourelatos, Z.; Kirino, Y. Mitochondrial protein BmPAPI modulates the length of mature piRNAs. RNA 2013, 19, 1405–1418. [Google Scholar] [CrossRef] [Green Version]
- Pandey, R.R.; Tokuzawa, Y.; Yang, Z.; Hayashi, E.; Ichisaka, T.; Kajita, S.; Asano, Y.; Kunieda, T.; Sachidanandam, R.; Chuma, S.; et al. Tudor domain containing 12 (TDRD12) is essential for secondary PIWI interacting RNA biogenesis in mice. Proc. Natl. Acad. Sci. USA 2013, 110, 16492–16497. [Google Scholar] [CrossRef]
- Patil, A.A.; Tatsuke, T.; Mon, H.; Lee, J.M.; Morokuma, D.; Hino, M.; Kusakabe, T. Characterization of Armitage and Yb containing granules and their relationship to nuage in ovary-derived cultured silkworm cell. Biochem. Biophys. Res. Commun. 2017, 490, 134–140. [Google Scholar] [CrossRef]
- Chung, P.Y.; Shoji, K.; Izumi, N.; Tomari, Y. Dynamic subcellular compartmentalization ensures fidelity of piRNA biogenesis in silkworms. EMBO Rep. 2021, 22, e51342. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Yang, Y.; Zhang, H.; Farley, G.; Wang, J.; Quarles, K.A.; Weng, Z.; Zamore, P.D. The genome of the Hi5 germ cell line from Trichoplusia ni, an agricultural pest and novel model for small RNA biology. eLife 2018, 7, e31628. [Google Scholar] [CrossRef] [PubMed]
- Kawaoka, S.; Hayashi, N.; Katsuma, S.; Kishino, H.; Kohara, Y.; Mita, K.; Shimada, T. Bombyx small RNAs: Genomic defense system against transposons in the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 2008, 38, 1058–1065. [Google Scholar] [CrossRef] [PubMed]
- Tatsuke, T.; Sakashita, K.; Masaki, Y.; Lee, J.M.; Kawaguchi, Y.; Kusakabe, T. The telomere-specific non-LTR retrotransposons SART1 and TRAS1 are suppressed by Piwi subfamily proteins in the silkworm, Bombyx mori. Cell Mol. Biol. Lett. 2010, 15, 118–133. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; You, L.; Yan, D.; James, A.A.; Huang, Y.; Tan, A. Bombyx mori histone methyltransferase BmAsh2 is essential for silkworm piRNA-mediated sex determination. PLoS Genet. 2018, 14, e1007245. [Google Scholar] [CrossRef] [PubMed]
- Swevers, L.; Liu, J.; Huvenne, H.; Smagghe, G. Search for limiting factors in the RNAi pathway in silkmoth tissues and the Bm5 cell line: The RNA binding proteins R2D2 and Translin. PLoS ONE 2011, 6, e20250. [Google Scholar] [CrossRef]
- Kawaoka, S.; Arai, Y.; Kadota, K.; Suzuki, Y.; Hara, K.; Sugano, S.; Shimizu, K.; Tomari, Y.; Shimada, T.; Katsuma, S. Zygotic amplification of secondary piRNAs during silkworm embryogenesis. RNA 2011, 17, 1401–1407. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Zhou, Q.; Yu, C.; Wang, X.; Hu, S.; Yu, J.; Yu, X. Transposable-element associated small RNAs in Bombyx mori genome. PLoS ONE 2012, 7, e36599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osanai-Futahashi, M.; Suetsugu, Y.; Mita, K.; Fujiwara, H. Genome-wide screening and characterization of transposable elements and their distribution analysis in the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 2008, 38, 1046–1057. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.E.; Zhang, H.H.; Xia, T.; Han, M.J.; Shen, Y.H.; Zhang, Z. BmTEdb: A collective database of transposable elements in the silkworm genome. Database 2013, 2013, bat055. [Google Scholar] [CrossRef] [PubMed]
- Kawamoto, M.; Jouraku, A.; Toyoda, A.; Yokoi, K.; Minakuchi, Y.; Katsuma, S.; Fujiyama, A.; Kiuchi, T.; Yamamoto, K.; Shimada, T. High-quality genome assembly of the silkworm, Bombyx mori. Insect Biochem Mol Biol. 2019, 107, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Kawaoka, S.; Kadota, K.; Arai, Y.; Suzuki, Y.; Fujii, T.; Abe, H.; Yasukochi, Y.; Mita, K.; Sugano, S.; Shimizu, K.; et al. The silkworm W chromosome is a source of female-enriched piRNAs. RNA 2011, 17, 2144–2151. [Google Scholar] [CrossRef]
- Honda, S.; Loher, P.; Morichika, K.; Shigematsu, M.; Kawamura, T.; Kirino, Y.; Rigoutsos, I.; Kirino, Y. Increasing cell density globally enhances the biogenesis of Piwi-interacting RNAs in Bombyx mori germ cells. Sci. Rep. 2017, 7, 4110. [Google Scholar] [CrossRef]
- Shigematsu, M.; Kawamura, T.; Morichika, K.; Izumi, N.; Kiuchi, T.; Honda, S.; Pliatsika, V.; Matsubara, R.; Rigoutsos, I.; Katsuma, S.; et al. RNase κ promotes robust piRNA production by generating 2′,3′-cyclic phosphate-containing precursors. Nat. Commun. 2021, 12, 4498. [Google Scholar] [CrossRef]
- Kawaoka, S.; Mitsutake, H.; Kiuchi, T.; Kobayashi, M.; Yoshikawa, M.; Suzuki, Y.; Sugano, S.; Shimada, T.; Kobayashi, J.; Tomari, Y.; et al. A role for transcription from a piRNA cluster in de novo piRNA production. RNA 2011, 18, 265–273. [Google Scholar] [CrossRef] [PubMed]
- Si, H.; Cao, Y.; Zhu, H.; Li, D.; Lv, Z.; Sheng, Q.; Nie, Z. Transposable element Bm1645 is a source of BmAGO2-associated small RNAs that affect its expression in Bombyx mori. BMC Genom. 2017, 18, 201. [Google Scholar] [CrossRef] [PubMed]
- Katsuma, S.; Kiuchi, T.; Kawamoto, M.; Fujimoto, T.; Sahara, K. Unique sex determination system in the silkworm, Bombyx mori: Current status and beyond. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2018, 94, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Katsuma, S.; Kawamoto, M.; Kiuchi, T. Guardian small RNAs and sex determination. RNA Biol. 2014, 11, 1238–1242. [Google Scholar] [CrossRef] [Green Version]
- Marec, F. Developmental genetics: Female silkworms have the sex factor. Nature 2014, 509, 570–571. [Google Scholar] [CrossRef]
- Matson, C.K.; Zarkower, D. Sex and the singular DM domain: Insights into sexual regulation, evolution and plasticity. Nat. Rev. Genet. 2012, 13, 163–174. [Google Scholar] [CrossRef]
- Chen, K.; Yu, Y.; Yang, D.; Yang, X.; Tang, L.; Liu, Y.; Luo, X.; Walters, J.R.; Liu, Z.; Xu, J.; et al. Gtsf1 is essential for proper female sex determination and transposon silencing in the silkworm, Bombyx mori. PLoS Genet. 2020, 16, e1009194. [Google Scholar] [CrossRef] [PubMed]
- Arif, A.; Bailey, S.; Izumi, N.; Anzelon, T.A.; Ozata, D.M.; Andersson, C.; Gainetdinov, I.; MacRae, I.J.; Tomari, Y.; Zamore, P.D. GTSF1 accelerates target RNA cleavage by PIWI-clade Argonaute proteins. Nature 2022, 608, 618–625. [Google Scholar] [CrossRef]
- Yang, X.; Chen, K.; Wang, Y.; Yang, D.; Huang, Y. The sex determination cascade in the silkworm. Genes 2021, 12, 315. [Google Scholar] [CrossRef]
- Visser, S.; Voleníková, A.; Nguyen, P.; Verhulst, E.C.; Marec, F. A conserved role of the duplicated Masculinizer gene in sex determination of the Mediterranean flour moth, Ephestia kuehniella. PLoS Genet. 2021, 17, e1009420. [Google Scholar] [CrossRef]
- Lee, J.; Kiuchi, T.; Kawamoto, M.; Shimada, T.; Katsuma, S. Identification and functional analysis of a Masculinizer orthologue in Trilocha varians (Lepidoptera: Bombycidae). Insect Mol. Biol. 2015, 24, 561–569. [Google Scholar] [CrossRef]
- Katsuma, S.; Tanaka, S.; Omuro, N.; Takabuchi, L.; Daimon, T.; Imanishi, S.; Yamahita, S.; Iwanaga, M.; Mita, K.; Maeda, S.; et al. Novel Macula-like virus identified in Bombyx mori cultured cells. J. Virol. 2005, 79, 5577–5584. [Google Scholar] [CrossRef]
- Li, T.-C.; Scotti, P.D.; Miyamura, T.; Takeda, N. Latent infection of a new alphanodavirus in an insect cell line. J. Virol. 2007, 81, 10890–10896. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Galvin, T.A.; Glasner, D.R.; Shaheduzzaman, S.; Khan, A.S. Identification of a novel rhabdovirus in Spodoptera frugiperda cell lines. J. Virol. 2014, 88, 6576–6585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swevers, L.; Ioannidis, K.; Kolovou, M.; Zografidis, A.; Labropoulou, V.; Santos, D.; Wynant, N.; Vanden Broeck, J.; Wang, L.; Capelle, K.; et al. Persistent RNA virus infection of lepidopteran cell lines: Interactions with the RNAi machinery. J. Insect Physiol. 2016, 93–94, 81–93. [Google Scholar] [CrossRef] [PubMed]
- Santos, D.; Mingels, L.; Vogel, E.; Wang, L.; Christiaens, O.; Cappelle, K.; Wynant, N.; Gansemans, Y.; Van Nieuwerburgh, F.; Smagghe, G.; et al. Generation of virus- and dsRNA-derived siRNAs with species-dependent length in insects. Viruses 2019, 11, 738. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Cappelle, K.; Santos, D.; Vanden Broeck, J.; Smagghe, G.; Swevers, L. Short-term persistence precedes pathogenic infection: Infection kinetics of cricket paralysis virus in silkworm-derived Bm5 cells. J. Insect Physiol. 2019, 115, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Santos, D.; Verdonckt, T.W.; Mingels, L.; Van den Brande, S.; Geens, B.; Van Nieuwerburgh, F.; Kolliopoulou, A.; Swevers, L.; Wynant, N.; Vanden Broeck, J. PIWI proteins play an antiviral role in Lepidopteran cell lines. Viruses 2022, 14, 1442. [Google Scholar] [CrossRef] [PubMed]
- Katsuma, S.; Kawamoto, M.; Shoji, K.; Aizawa, T.; Kiuchi, T.; Izumi, N.; Ogawa, M.; Mashiko, T.; Kawasaki, H.; Sugano, S.; et al. Transcriptome profiling reveals infection strategy of an insect maculavirus. DNA Res. 2018, 25, 277–286. [Google Scholar] [CrossRef]
- Santos, D.; Wynant, N.; Van den Brande, S.; Verdonckt, T.W.; Mingels, L.; Peeters, P.; Kolliopoulou, A.; Swevers, L.; Vanden Broeck, J. Insights into RNAi-based antiviral immunity in Lepidoptera: Acute and persistent infections in Bombyx mori and Trichoplusia ni cell lines. Sci Rep. 2018, 8, 2423. [Google Scholar] [CrossRef] [PubMed]
- Zografidis, A.; Van Nieuwerburgh, F.; Kolliopoulou, A.; Apostolou-Karampelis, K.; Head, S.R.; Deforce, D.; Smagghe, G.; Swevers, L. Viral small-RNA analysis of Bombyx mori larval midgut during persistent and pathogenic cytoplasmic polyhedrosis virus infection. J. Virol. 2015, 89, 11473–11486. [Google Scholar] [CrossRef] [PubMed]
- Kolliopoulou, A.; Van Nieuwerburgh, F.; Stravopodis, D.J.; Deforce, D.; Swevers, L.; Smagghe, G. Transcriptome analysis of Bombyx larval midgut during persistent and pathogenic cytoplasmic polyhedrosis virus infection. PLoS ONE 2015, 10, e0121447. [Google Scholar] [CrossRef]
- Mehrabadi, M.; Hussain, M.; Matindoost, L.; Asgari, S. The baculovirus antiapoptotic p35 protein functions as an inhibitor of the host RNA interference antiviral response. J. Virol. 2015, 89, 8182–8192. [Google Scholar] [CrossRef] [PubMed]
- Feng, M.; Kolliopoulou, A.; Zhou, Y.H.; Fei, S.G.; Xia, J.M.; Swevers, L.; Sun, J.C. The piRNA response to BmNPV infection in the silkworm fat body and midgut. Insect Sci. 2021, 28, 662–679. [Google Scholar] [CrossRef]
- Xia, J.; Fei, S.; Wu, H.; Yang, Y.; Yu, W.; Zhang, M.; Guo, Y.; Swevers, L.; Sun, J.; Feng, M. The piRNA pathway is required for nucleopolyhedrovirus replication in Lepidoptera. Insect Sci. 2022, Epub ahead of print. [Google Scholar] [CrossRef]
- Kong, X.; Wei, G.; Chen, N.; Zhao, S.; Shen, Y.; Zhang, J.; Li, Y.; Zeng, X.; Wu, X. Dynamic chromatin accessibility profiling reveals changes in host genome organization in response to baculovirus infection. PLoS Pathog. 2020, 16, e1008633. [Google Scholar] [CrossRef] [PubMed]
- Young, J.C.; MacKinnon, E.A.; Faulkner, P. The architecture of the virogenic stroma in isolated nuclei of Spodoptera frugiperda cells in vitro Infected by Autographa californica Nuclear Polyhedrosis Virus. J. Struct. Biol. 1993, 110, 141–153. [Google Scholar] [CrossRef]
- Feng, M.; Ren, F.; Zhou, Y.; Zhang, N.; Lu, Q.; Swevers, L.; Sun, J. Correlation in expression between LTR Retrotransposons and potential host cis-targets during infection of Antherea pernyi with ApNPV Baculovirus. Viruses 2019, 11, 421. [Google Scholar] [CrossRef] [PubMed]
- Geisler, C.; Jarvis, D.L. Rhabdovirus-like endogenous viral elements in the genome of Spodoptera frugiperda insect cells are actively transcribed: Implications for adventitious virus detection. Biologicals 2014, 44, 219–225. [Google Scholar] [CrossRef]
- Sobala, A.; Hutvagner, G. Transfer RNA-derived fragments: Origins, processing, and functions. Wiley Interdiscip. Rev. RNA 2011, 2, 853–862. [Google Scholar] [CrossRef]
- Honda, S.; Kawamura, T.; Loher, P.; Morichika, K.; Rigoutsos, I.; Kirino, Y. The biogenesis pathway of tRNA-derived piRNAs in Bombyx germ cells. Nucleic Acids Res. 2017, 45, 9108–9120. [Google Scholar] [CrossRef]
- Harrison, M.C.; Jongepier, E.; Robertson, H.M.; Arning, N.; Bitard-Feildel, T.; Chao, H.; Childers, C.P.; Dinh, H.; Doddapaneni, H.; Dugan, S.; et al. Hemimetabolous genomes reveal molecular basis of termite eusociality. Nat. Ecol. Evol. 2018, 2, 557–566. [Google Scholar] [CrossRef] [PubMed]
- Llonga, N.; Ylla, G.; Bau, J.; Belles, X.; Piulachs, M.-D. Diversity of piRNA expression patterns during the ontogeny of the German cockroach. J. Exp. Zool. (Mol. Dev. Evol.) 2018, 330, 288–295. [Google Scholar] [CrossRef]
- Elsner, D.; Meusemann, K.; Korb, J. Longevity and transposon defense, the case of termite reproductives. Proc. Natl. Acad. Sci. USA 2018, 115, 5504–5509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Fang, X.; Yang, P.; Jiang, X.; Jiang, F.; Zhao, D.; Li, B.; Cui, F.; Wei, J.; Ma, C.; et al. The locust genome provides insight into swarm formation and long-distance flight. Nat. Commun. 2014, 5, 2957. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Chen, S.; Yang, P.; Ma, Z.; Kang, L. Characterization and comparative profiling of the small RNA transcriptomes in two phases of locust. Genome Biol. 2009, 10, R6. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, X.; Kang, L. A k-mer scheme to predict piRNAs and characterize locust piRNAs. Bioinformatics 2011, 27, 771–776. [Google Scholar] [CrossRef]
- Wang, H.; Jiang, F.; Liu, X.; Liu, Q.; Fu, Y.; Li, R.; Hou, L.; Zhang, J.; He, J.; Kang, L. Piwi/piRNAs control food intake by promoting neuropeptide F expression in locusts. EMBO Rep. 2022, 23, e50851. [Google Scholar] [CrossRef]
- Jiang, F.; Zhang, J.; Liu, Q.; Liu, X.; Wang, H.; He, J.; Kang, L. Long-read direct RNA sequencing by 5′-Cap capturing reveals the impact of Piwi on the widespread exonization of transposable elements in locusts. RNA Biol. 2019, 16, 950–959. [Google Scholar] [CrossRef]
- Van Wielendaele, P.; Dillen, S.; Zels, S.; Badisco, L.; Vanden Broeck, J. Regulation of feeding by neuropeptide F in the desert locust, Schistocerca gregaria. Insect Biochem. Mol. Biol. 2013, 43, 102–114. [Google Scholar] [CrossRef]
- Teixeira, F.K.; Okuniewska, M.; Malone, C.D.; Coux, R.X.; Rio, D.C.; Lehmann, R. piRNA-mediated regulation of transposon alternative splicing in the soma and germ line. Nature 2017, 552, 268–272. [Google Scholar] [CrossRef]
- Chen, W.; Hasegawa, D.K.; Kaur, N.; Kliot, A.; Pinheiro, P.V.; Luan, J.; Stensmyr, M.C.; Zheng, Y.; Liu, W.; Sun, H.; et al. The draft genome of whitefly Bemisia tabaci MEAM1, a global crop pest, provides novel insights into virus transmission, host adaptation, and insecticide resistance. BMC Biol. 2016, 14, 110. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.; Chen, C.; Yang, Z.; Guo, L.; Yang, X.; Wang, D.; Chen, M.; Huang, J.; Wen, Y.; Zeng, Y.; et al. Genome sequencing of the sweetpotato whitefly Bemisia tabaci MED/Q. Gigascience 2017, 6, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Shamimuzzaman, M.; Hasegawa, D.K.; Chen, W.; Simmons, A.M.; Fei, Z.; Ling, K.S. Genome-wide profiling of piRNAs in the whitefly Bemisia tabaci reveals cluster distribution and association with begomovirus transmission. PLoS ONE 2019, 14, e0213149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mondal, M.; Brown, J.K.; Flynt, A. Exploiting somatic piRNAs in Bemisia tabaci enables novel gene silencing through RNA feeding. Life Sci. Alliance 2020, 3, e202000731. [Google Scholar] [CrossRef] [PubMed]
- Flynt, A.S. Insecticidal RNA interference, thinking beyond long dsRNA. Pest Manag. Sci. 2021, 77, 2179–2187. [Google Scholar] [CrossRef]
- Nigg, J.C.; Kuo, Y.W.; Falk, B.W. Endogenous viral element-derived Piwi-interacting RNAs (piRNAs) are not required for production of ping-pong-dependent piRNAs from Diaphorina citri densovirus. mBio 2019, 11, e02209-20. [Google Scholar] [CrossRef]
- Saha, S.; Hosmani, P.S.; Villalobos-Ayala, K.; Miller, S.; Shippy, T.; Flores, M.; Rosendale, A.; Cordola, C.; Bell, T.; Mann, H.; et al. Improved annotation of the insect vector of citrus greening disease: Biocuration by a diverse genomics community. Database 2017, 2017, bax032. [Google Scholar] [CrossRef]
- Srinivasan, D.G.; Brisson, J.A. Aphids: A model for polyphenism and epigenetics. Genet. Res. Int. 2012, 2012, 431531. [Google Scholar] [CrossRef]
- Le Trionnaire, G.; Wucher, V.; Tagu, D. Genome expression control during the photoperiodic response of aphids. Physiol. Entomol. 2013, 38, 117–125. [Google Scholar] [CrossRef]
- Ogawa, K.; Miura, T. Aphid polyphenisms: Trans-generational developmental regulation through viviparity. Front. Physiol. 2014, 5, 1. [Google Scholar] [CrossRef]
- International Aphid Genomics Consortium Genome sequence of the pea aphid Acyrthosiphon pisum. PLoS Biol. 2010, 8, e1000313.
- Fernández, R.; Marcet-Houben, M.; Legeai, F.; Richard, G.; Robin, S.; Wucher, V.; Pegueroles, C.; Gabaldón, T.; Tagu, D. Selection following gene duplication shapes recent genome evolution in the pea aphid Acyrthosiphon pisum. Mol. Biol. Evol. 2020, 37, 2601–2615. [Google Scholar] [CrossRef]
- Lu, H.L.; Tanguy, S.; Rispe, C.; Gauthier, J.P.; Walsh, T.; Gordon, K.; Edwards, O.; Tagu, D.; Chang, C.C.; Jaubert-Possamai, S. Expansion of genes encoding piRNA-associated argonaute proteins in the pea aphid: Diversification of expression profiles in different plastic morphs. PLoS ONE 2011, 6, e28051. [Google Scholar] [CrossRef] [PubMed]
- Miura, T.; Braendle, C.; Shingleton, A.; Sisk, G.; Kambhampati, S.; Stern, D.L. A comparison of parthenogenetic and sexual embryogenesis of the pea aphid Acyrthosiphon pisum (Hemiptera: Aphidoidea). J. Exp. Zool. B Mol. Dev. Evol. 2003, 295, 59–81. [Google Scholar] [CrossRef]
- Chang, C.C.; Lin, G.W.; Cook, C.E.; Horng, S.B.; Lee, H.J.; Huang, T.Y. Apvasa marks germ-cell migration in the parthenogenetic pea aphid Acyrthosiphon pisum (Hemiptera: Aphidoidea). Dev. Genes Evol. 2007, 217, 275–287. [Google Scholar] [CrossRef]
- Gallot, A.; Shigenobu, S.; Hashiyama, T.; Jaubert-Possamai, S.; Tagu, D. Sexual and asexual oogenesis require the expression of unique and shared sets of genes in the insect Acyrthosiphon pisum. BMC Genom. 2012, 13, 76. [Google Scholar] [CrossRef] [PubMed]
- Grantham, M.E.; Brisson, J.A. Extensive differential splicing underlies phenotypically plastic aphid morphs. Mol. Biol. Evol. 2018, 35, 1934–1946. [Google Scholar] [CrossRef]
- Couvillion, M.T.; Lee, S.R.; Hogstad, B.; Malone, C.D.; Tonkin, L.A.; Sachidanandam, R.; Hannon, G.J.; Collins, K. Sequence, biogenesis, and function of diverse small RNA classes bound to the Piwi family proteins of Tetrahymena thermophila. Genes Dev. 2009, 23, 2016–2032. [Google Scholar] [CrossRef] [PubMed]
- Schurko, A.M.; Logsdon, J.M., Jr.; Eads, B.D. Meiosis genes in Daphnia pulex and the role of parthenogenesis in genome evolution. BMC Evol. Biol. 2009, 9, 78. [Google Scholar] [CrossRef] [PubMed]
- Brito, T.; Julio, A.; Berni, M.; de Castro Poncio, L.; Bernardes, E.S.; Araujo, H.; Sammeth, M.; Pane, A. Transcriptomic and functional analyses of the piRNA pathway in the Chagas disease vector Rhodnius prolixus. PLoS Negl. Trop. Dis. 2018, 12, e0006760. [Google Scholar] [CrossRef]
- Mesquita, R.D.; Vionette-Amaral, R.J.; Lowenberger, C.; Rivera-Pomar, R.; Monteiro, F.A.; Minx, P.; Spieth, J.; Carvalho, A.B.; Panzera, F.; Lawson, D.; et al. Genome of Rhodnius prolixus, an insect vector of Chagas disease, reveals unique adaptations to hematophagy and parasite infection. Proc. Natl. Acad. Sci. USA 2015, 112, 14936–14941. [Google Scholar] [CrossRef]
- Brito, T.F.; Coelho, V.L.; Cardoso, M.A.; Brito, I.A.A.; Berni, M.A.; Zenk, F.L.; Iovino, N.; Pane, A. Transovarial transmission of a core virome in the Chagas disease vector Rhodnius prolixus. PLoS Pathog. 2021, 17, e1009780. [Google Scholar] [CrossRef]
- Tomoyasu, Y.; Miller, S.C.; Tomita, S.; Schoppmeier, M.; Grossmann, D.; Bucher, G. Exploring systemic RNA interference in insects: A genome-wide survey for RNAi genes in Tribolium. Genome Biol. 2008, 9, R10. [Google Scholar] [CrossRef]
- Ninova, M.; Ronshaugen, M.; Griffiths-Jones, S. MicroRNA evolution, expression, and function during short germband development in Tribolium castaneum. Genome Res. 2016, 26, 85–96. [Google Scholar] [CrossRef] [Green Version]
- Ninova, M.; Griffiths-Jones, S.; Ronshaugen, M. Abundant expression of somatic transposon-derived piRNAs throughout Tribolium castaneum embryogenesis. Genome Biol. 2017, 18, 184. [Google Scholar] [CrossRef] [PubMed]
- Tribolium Genome Sequencing Consortium; Richards, S.; Gibbs, R.A.; Weinstock, G.M.; Brown, S.J.; Denell, R.; Beeman, R.W.; Gibbs, R.; Beeman, R.W.; Brown, S.J.; et al. The genome of the model beetle and pest Tribolium castaneum. Nature 2008, 452, 949–955. [Google Scholar] [PubMed]
- Parisot, N.; Vargas-Chávez, C.; Goubert, C.; Baa-Puyoulet, P.; Balmand, S.; Beranger, L.; Blanc, C.; Bonnamour, A.; Boulesteix, M.; Burlet, N.; et al. The transposable element-rich genome of the cereal pest Sitophilus oryzae. BMC Biol. 2021, 19, 241. [Google Scholar] [CrossRef] [PubMed]
- Liao, Z.; Jia, Q.; Li, F.; Han, Z. Identification of two piwi genes and their expression profile in honeybee, Apis mellifera. Arch. Insect Biochem. Physiol. 2010, 74, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Elsik, C.G.; Worley, K.C.; Bennett, A.K.; Beye, M.; Camara, F.; Childers, C.P.; de Graaf, D.C.; Debyser, G.; Deng, J.; Devreese, B.; et al. Finding the missing honey bee genes: Lessons learned from a genome upgrade. BMC Genom. 2014, 15, 86. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Ashby, R.; Ying, H.; Maleszka, R.; Forêt, S. Contrasting sex-and caste-dependent piRNA profiles in the transposon depleted haplodiploid honeybee Apis mellifera. Genome Biol. Evol. 2017, 9, 1341–1356. [Google Scholar] [CrossRef] [PubMed]
- Watson, O.T.; Buchmann, G.; Young, P.; Lo, K.; Remnant, E.J.; Yagound, B.; Shambrook, M.; Hill, A.F.; Oldroyd, B.P.; Ashe, A. Abundant small RNAs in the reproductive tissues and eggs of the honey bee, Apis mellifera. BMC Genom. 2022, 23, 257. [Google Scholar] [CrossRef] [PubMed]
- Fung, E.; Hill, K.; Hogendoorn, K.; Glatz, R.V.; Napier, K.R.; Bellgard, M.I.; Barrero, R.A. De novo assembly of honey bee RNA viral genomes by tapping into the innate insect antiviral response pathway. J. Invertebr. Pathol. 2018, 152, 38–47. [Google Scholar] [CrossRef]
- Kondo, H.; Chiba, S.; Maruyama, K.; Andika, I.B.; Suzuki, N. A novel insect-infecting virga/nege-like virus group and its pervasive endogenization into insect genomes. Virus Res. 2019, 262, 37–47. [Google Scholar] [CrossRef]
- Maori, E.; Tanne, E.; Sela, I. Reciprocal sequence exchange between non-retro viruses and hosts leading to the appearance of new host phenotypes. Virology 2007, 362, 342–349. [Google Scholar] [CrossRef] [Green Version]
- Parhad, S.S.; Theurkauf, W.E. Rapid evolution and conserved function of the piRNA pathway. Open Biol. 2019, 9, 180181. [Google Scholar] [CrossRef] [PubMed]
- Dönertas, D.; Sienski, G.; Brennecke, J. Drosophila Gtsf1 is an essential component of the Piwi-mediated transcriptional silencing complex. Genes Dev. 2013, 27, 1693–1705. [Google Scholar] [CrossRef]
- Haley, B.; Zamore, P.D. Kinetic analysis of the RNAi enzyme complex. Nat. Struct. Mol. Biol. 2004, 11, 599–606. [Google Scholar] [CrossRef]
- Wessels, H.H.; Lebedeva, S.; Hirsekorn, A.; Wurmus, R.; Akalin, A.; Mukherjee, N.; Ohler, U. Global identification of functional microRNA-mRNA interactions in Drosophila. Nat. Commun. 2019, 10, 1626. [Google Scholar] [CrossRef] [PubMed]
- Anzelon, T.A.; Chowdhury, S.; Hughes, S.M.; Xiao, Y.; Lander, G.C.; MacRae, I.J. Structural basis for piRNA targeting. Nature 2021, 597, 285–289. [Google Scholar] [CrossRef]
- Chandradoss, S.D.; Schirle, N.T.; Szczepaniak, M.; MacRae, I.J.; Joo, C. A dynamic search process underlies microRNA targeting. Cell 2015, 162, 96–107. [Google Scholar] [CrossRef] [PubMed]
- Jonas, S.; Izaurralde, E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 2015, 16, 421–433. [Google Scholar] [CrossRef]
- Sarkar, A.; Volff, J.N.; Vaury, C. piRNAs and their diverse roles: A transposable element-driven tactic for gene regulation? FASEB J. 2017, 31, 436–446. [Google Scholar] [CrossRef] [PubMed]
- Darricarrère, N.; Liu, N.; Watanabe, T.; Lin, H. Function of Piwi, a nuclear Piwi/Argonaute protein, is independent of its slicer activity. Proc. Natl Acad. Sci. USA 2013, 110, 1297–1302. [Google Scholar] [CrossRef]
- Dennis, C.; Zanni, V.; Brasset, E.; Eymery, A.; Zhang, L.; Mteirek, R.; Jensen, S.; Rong, Y.S.; Vaury, C. “Dot COM”, a nuclear transit center for the primary piRNA pathway in Drosophila. PLoS ONE 2013, 8, e72752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomes, E.; Shorter, J. The molecular language of membraneless organelles. J. Biol. Chem. 2019, 294, 7115–7127. [Google Scholar] [CrossRef] [PubMed]
- Dennis, C.; Brasset, E.; Sarkar, A.; Vaury, C. Export of piRNA precursors by EJC triggers assembly of cytoplasmic Yb-body in Drosophila. Nat. Commun. 2016, 7, 13739. [Google Scholar] [CrossRef] [PubMed]
- Hirakata, S.; Siomi, M.C. Assembly and function of gonad-specific non-membranous organelles in Drosophila piRNA biogenesis. Noncoding RNA 2019, 5, 52. [Google Scholar] [CrossRef] [PubMed]
- Siomi, M.C.; Mannen, T.; Siomi, H. How does the royal family of Tudor rule the PIWI-interacting RNA pathway? Genes Dev. 2010, 24, 636–646. [Google Scholar] [CrossRef] [PubMed]
- Sato, K.; Iwasaki, Y.W.; Shibuya, A.; Carninci, P.; Tsuchizawa, Y.; Ishizu, H.; Siomi, M.C.; Siomi, H. Krimper enforces an antisense bias on piRNA pools by binding AGO3 in the Drosophila germline. Mol. Cell. 2015, 59, 553–563. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Wang, J.; Xu, J.; Zhang, Z.; Koppetsch, B.S.; Schultz, N.; Vreven, T.; Meignin, C.; Davis, I.; Zamore, P.D.; et al. UAP56 couples piRNA clusters to the perinuclear transposon silencing machinery. Cell 2012, 151, 871–884. [Google Scholar] [CrossRef]
- Pandey, R.R.; Homolka, D.; Chen, K.M.; Sachidanandam, R.; Fauvarque, M.O.; Pillai, R.S. Recruitment of Armitage and Yb to a transcript triggers its phased processing into primary piRNAs in Drosophila ovaries. PLoS Genet. 2017, 13, e1006956. [Google Scholar] [CrossRef]
- Teefy, B.B.; Siebert, S.; Cazet, J.F.; Lin, H.; Juliano, C.E. PIWI-piRNA pathway-mediated transposable element repression in Hydra somatic stem cells. RNA 2020, 26, 550–563. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Taylor, D.H.; van Wolfswinkel, J.C. PIWI-mediated control of tissue-specific transposons is essential for somatic cell differentiation. Cell Rep. 2021, 37, 109776. [Google Scholar] [CrossRef] [PubMed]
- Molina, M.D.; Cebrià, F. Decoding stem cells: An overview on planarian stem cell heterogeneity and lineage progression. Biomolecules 2021, 11, 1532. [Google Scholar] [CrossRef]
- van Wolfswinkel, J.C. Piwi and potency: PIWI proteins in animal stem cells and regeneration. Integr. Comp. Biol. 2014, 54, 700–713. [Google Scholar] [CrossRef]
- Sousa-Victor, P.; Ayyaz, A.; Hayashi, R.; Qi, Y.; Madden, D.T.; Lunyak, V.V.; Jasper, H. Piwi is required to limit exhaustion of aging somatic stem cells. Cell Rep. 2017, 20, 2527–2537. [Google Scholar] [CrossRef] [PubMed]
- Lin, K.Y.; Wang, W.D.; Lin, C.H.; Rastegari, E.; Su, Y.H.; Chang, Y.T.; Liao, Y.F.; Chang, Y.C.; Pi, H.; Yu, B.Y.; et al. Piwi reduction in the aged niche eliminates germline stem cells via Toll-GSK3 signaling. Nat. Commun. 2020, 11, 3147. [Google Scholar] [CrossRef] [PubMed]
- Jeong, H.; Park, K.H.; Lee, Y.; Jeong, A.; Choi, S.; Kim, K.W. The regulation and role of piRNAs and PIWI proteins in cancer. Processes 2021, 9, 1208. [Google Scholar] [CrossRef]
- Tsai, K.; Cullen, B.R. Epigenetic and epitranscriptomic regulation of viral replication. Nat. Rev. Microbiol. 2018, 18, 559–570. [Google Scholar] [CrossRef]
- Perera, B.P.U.; Tsai, Z.T.; Colwell, M.L.; Jones, T.R.; Goodrich, J.M.; Wang, K.; Sartor, M.A.; Faulk, C.; Dolinoy, D.C. Somatic expression of piRNA and associated machinery in the mouse identifies short, tissue-specific piRNA. Epigenetics 2019, 14, 504–521. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Jin, B.; Liu, P.; Li, J.; Chen, X.; Gu, J. piRNA profiling of Dengue virus type 2-infected Asian tiger mosquito and midgut tissues. Viruses 2018, 10, 213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Species | Tissue | Process |
|---|---|---|
| Drosophila melanogaster (Diptera) | Ovary | Transposon control (transcriptional silencing): somatic cells Transposon control (posttranscriptional silencing): germline cells Stem cell function Telomere maintenance (germline cells) mRNA/piRNA transport and localization (nurse cell-oocyte complex) |
| Testis | Stem cell function Spermatocyte differentiation Regulation of fertility | |
| Somatic tissues | Transposon control Stem cell function | |
| Embryo | Maternal mRNA degradation (early embryogenesis) | |
| Aedes aegypti Aedes albopictus (Diptera) | Aag2 cells Somatic tissues | Transposon control (posttranscriptional silencing) Antiviral defense Immunological memory? |
| Embryo | Maternal mRNA degradation (early embryogenesis) | |
| Bombyx mori (Lepidoptera) | BmN4 cells | Transposon control (posttranscriptional silencing) Antiviral defense? Proviral role during baculovirus infection? Stress response? |
| Embryo | Maternal mRNA degradation (early embryogenesis) Sex determination | |
| Blatella germanica (Blattodea) | Embryo | Maternal mRNA degradation (early embryogenesis)? |
| Macrotermes bellicosus (Blattodea) | Whole body | Transposon control (related to caste differentiation) |
| Locusta migratoria (Orthoptera) | Somatic tissues | Transposon control Splicing regulation |
| Bemisia tabaci (Hemiptera) | Somatic tissues | Transposon control |
| Diaphorina citri (Hemiptera) | Somatic tissues | Antiviral defense? |
| Acyrthosiphon pisum (Hemiptera) | Ovary | Oocyte differentiation? Parthenogenetic embryogenesis? |
| Rhodnius prolixus (Hemiptera) | Ovary | Oocyte differentiation |
| Tribolium castaneum (Coleoptera) | Embryo | Maternal mRNA degradation (early embryogenesis) |
| Apis mellifera (Hymenoptera) | Whole body | Transposon control (related to caste differentiation) |
| Embryo | Maternal mRNA degradation (early embryogenesis)? |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santos, D.; Feng, M.; Kolliopoulou, A.; Taning, C.N.T.; Sun, J.; Swevers, L. What Are the Functional Roles of Piwi Proteins and piRNAs in Insects? Insects 2023, 14, 187. https://doi.org/10.3390/insects14020187
Santos D, Feng M, Kolliopoulou A, Taning CNT, Sun J, Swevers L. What Are the Functional Roles of Piwi Proteins and piRNAs in Insects? Insects. 2023; 14(2):187. https://doi.org/10.3390/insects14020187
Chicago/Turabian StyleSantos, Dulce, Min Feng, Anna Kolliopoulou, Clauvis N. T. Taning, Jingchen Sun, and Luc Swevers. 2023. "What Are the Functional Roles of Piwi Proteins and piRNAs in Insects?" Insects 14, no. 2: 187. https://doi.org/10.3390/insects14020187
APA StyleSantos, D., Feng, M., Kolliopoulou, A., Taning, C. N. T., Sun, J., & Swevers, L. (2023). What Are the Functional Roles of Piwi Proteins and piRNAs in Insects? Insects, 14(2), 187. https://doi.org/10.3390/insects14020187







