Omics in the Red Palm Weevil Rhynchophorus ferrugineus (Olivier) (Coleoptera: Curculionidae): A Bridge to the Pest
Abstract
:Simple Summary
Abstract
1. Introduction
2. R. ferrugineus Is the Most Damaging Insect Pest of Palms in the World
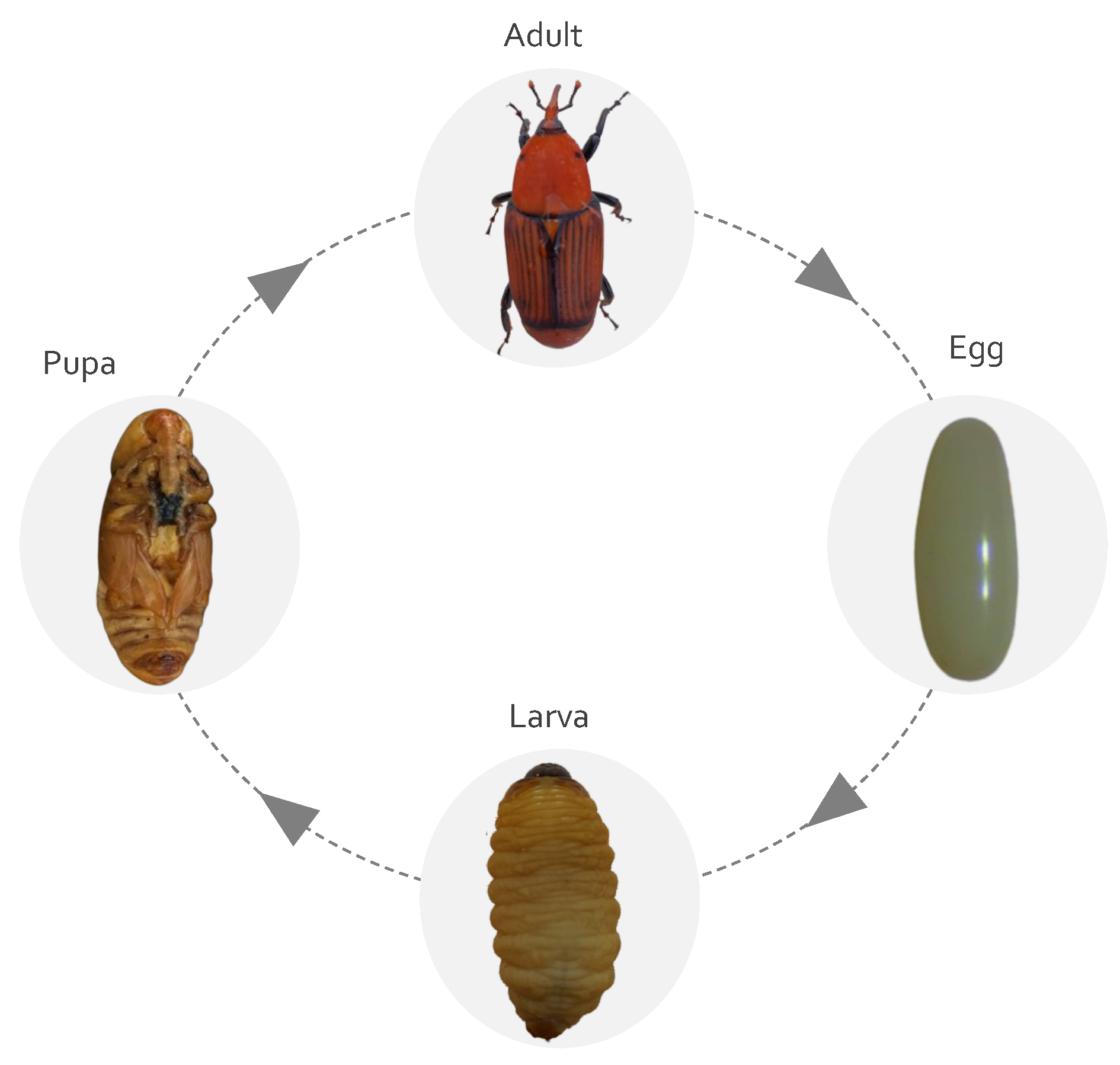
2.1. Life Cycle and Biology
2.2. Pest Status
2.3. Current Management Measures
3. Omics Technologies and Their Impact on Pest Science
3.1. Omics in Beetles
3.2. Large-Scale RNA-seq Gene Discovery Studies in the RPW
3.3. Whole Genome Sequencing in the RPW
3.4. Re-Analysis of Available Transcriptomic and Genomic Resources Using BUSCO
3.5. Metagenomics of Gut Microbiota in the RPW
3.6. Repetitive DNA in the RPW Genome
3.7. Mitochondrial Genome of the RPW
4. Opportunities and Challenges of Practical Uses of RPW Omics Research for Management Approaches
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chao, C.T.; Krueger, R.R. The date palm (Phoenix dactylifera L.): Overview of biology, uses, and cultivation. HortScience 2007, 42, 1077–1082. [Google Scholar] [CrossRef] [Green Version]
- Rugman-Jones, P.F.; Hoddle, C.D.; Hoddle, M.S.; Stouthamer, R. The lesser of two weevils: Molecular-genetics of pest palm weevil populations confirm Rhynchophorus vulneratus (Panzer 1798) as a valid species distinct from R. ferrugineus (Olivier 1790), and reveal the global extent of both. PLoS ONE 2013, 8, e78379. [Google Scholar] [CrossRef] [PubMed]
- El-Shafie, H.A.F.; Faleiro, J.R. Red palm weevil Rhynchophorus ferrugineus (Coleoptera: Curculionidae): Global invasion, current management options, challenges and future prospects. In Invasive Species-Introduction Pathways, Economic Impact, and Possible Management Options; IntechOpen: London, UK, 2020. [Google Scholar]
- Faleiro, J.R.; Ferry, M.; Yaseen, T.; Al-Dobai, S. Research Paper (Integrated Management: Insects) Overview of the gaps, challenges and prospects of red palm weevil management. Arab. J. Plant Prot. 2019, 37, 170. [Google Scholar] [CrossRef]
- Kassem, H.S.; Alotaibi, B.A.; Ahmed, A.; Aldosri, F.O. Sustainable management of the red palm weevil: The nexus between farmers’ adoption of integrated pest management and their knowledge of symptoms. Sustainability 2020, 12, 9647. [Google Scholar] [CrossRef]
- Turusov, V.; Rakitsky, V.; Tomatis, L. Dichlorodiphenyltrichloroethane (DDT): Ubiquity, persistence, and risks. Environ. Health Perspect. 2002, 110, 125–128. [Google Scholar] [CrossRef] [Green Version]
- Mafra-Neto, A.; Fettig, C.J.; Munson, A.S.; Rodriguez-Saona, C.; Holdcraft, R.; Faleiro, J.R.; El-Shafie, H.; Reinke, M.; Bernardi, C.; Villagran, K.M. Development of specialized pheromone and lure application technologies (SPLAT®) for management of coleopteran pests in agricultural and forest systems. In Biopesticides: State of The Art And Future Opportunities; ACS Publications: Washington, DC, USA, 2014; pp. 211–242. [Google Scholar]
- Rehman, G.; Mamoon-ur Rashid, M. Evaluation of entomopathogenic nematodes against red palm weevil, Rhynchophorus ferrugineus (Olivier)(Coleoptera: Curculionidae). Insects 2022, 13, 733. [Google Scholar] [CrossRef]
- Bait-Suwailam, M.M. Numerical Assessment of Red Palm Weevil Detection Mechanism in Palm Trees Using CSRR Microwave Sensors. Prog. Electromag. Res. Lett. 2021, 11, 63–71. [Google Scholar] [CrossRef]
- Saleh, R.A.A.; Ertunç, H.M. Development of a Neural Network Model for Recognizing Red Palm Weevil Insects Based on Image Processing. Kocaeli J. Sci. Eng. 2022, 5, 1–4. [Google Scholar] [CrossRef]
- Martin, B.; Shaby, S.M.; Premi, M.G. Studies on acoustic activity of red palm weevil the deadly pest on coconut crops. Procedia Mater. Sci. 2015, 10, 455–466. [Google Scholar] [CrossRef] [Green Version]
- Hoddle, M.S.; Hoddle, C.D.; Milosavljević, I. Quantification of the life time flight capabilities of the south American palm weevil, Rhynchophorus palmarum (L.) (Coleoptera: Curculionidae). Insects 2021, 12, 126. [Google Scholar] [CrossRef]
- Faleiro, J. A review of the issues and management of the red palm weevil Rhynchophorus ferrugineus (Coleoptera: Rhynchophoridae) in coconut and date palm during the last one hundred years. Int. J. Trop. Insect Sci. 2006, 26, 135–154. [Google Scholar]
- Nirula, K. Investigation on the pests of coconut palm, Part-IV. Rhynchophorus ferrugineus. Indian Coconut J. 1956, 9, 229–237. [Google Scholar]
- Giblin-Davis, R.M.; Faleiro, J.R.; Jacas, J.A.; Peña, J.E.; Vidyasagar, P. Biology and management of the red palm weevil, Rhynchophorus ferrugineus. Potential Invasive Pests Agric. Crop. 2013, 3, 1. [Google Scholar]
- León-Quinto, T.; Serna, A. Cryoprotective Response as Part of the Adaptive Strategy of the Red Palm Weevil, Rhynchophorus ferrugineus, against Low Temperatures. Insects 2022, 13, 134. [Google Scholar] [CrossRef] [PubMed]
- Hoddle, M.; Hoddle, C.; Faleiro, J.; El-Shafie, H.; Jeske, D.; Sallam, A. How far can the red palm weevil (Coleoptera: Curculionidae) fly? Computerized flight mill studies with field-captured weevils. J. Econ. Entomol. 2015, 108, 2599–2609. [Google Scholar] [CrossRef]
- Al-Dosary, N.M.; Al-Dobai, S.; Faleiro, J.R. Review on the management of red palm weevil Rhynchophorus ferrugineus Olivier in date palm Phoenix dactylifera L. Emir. J. Food Agric. 2016, 28, 34–44. [Google Scholar] [CrossRef]
- Chaya, M. Defining a programme and strategy for the management and containment of Red Palm Weevil. Int. Pest Control 2017, 59, 290. [Google Scholar]
- Kurdi, H.; Al-Aldawsari, A.; Al-Turaiki, I.; Aldawood, A.S. Early detection of red palm weevil, Rhynchophorus ferrugineus (olivier), infestation using data mining. Plants 2021, 10, 95. [Google Scholar] [CrossRef]
- Abraham, V.; Shuaibi, M.A.; Faleiro, J.; Abozuhairah, R.; Vidyasagar, P.S. An integrated management approach for red palm weevil Rhynchophorus ferrugineus Oliv. a key pest of date palm in the Middle East. J. Agric. Mar. Sci. 1998, 3, 77–83. [Google Scholar] [CrossRef] [Green Version]
- Faleiro, J.; Abdallah, A.B.; El-Bellaj, M.; Al-Ajlan, A.; Oihabi, A. Threat of the red palm weevil, Rhynchophorus ferrugineus (Olivier) to date palm plantations in North Africa. Arab. J. Plant Prot. 2012, 30, 274–280. [Google Scholar]
- Milosavljević, I.; El-Shafie, H.A.; Faleiro, J.R.; Hoddle, C.D.; Lewis, M.; Hoddle, M.S. Palmageddon: The wasting of ornamental palms by invasive palm weevils, Rhynchophorus spp. J. Pest Sci. 2019, 92, 143–156. [Google Scholar] [CrossRef]
- Rasool, K.G.; Khan, M.A.; Aldawood, A.S.; Tufail, M.; Mukhtar, M.; Takeda, M. Identification of proteins modulated in the date palm stem infested with red palm weevil (Rhynchophorus ferrugineus Oliv.) using two dimensional differential gel electrophoresis and mass spectrometry. Int. J. Mol. Sci. 2015, 16, 19326–19346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El-Sabea, A.M.; Faleiro, J.; Abo-El-Saad, M.M. The threat of red palm weevil Rhynchophorus ferrugineus to date plantations of the Gulf region in the Middle-East: An economic perspective. Outlooks Pest Manag. 2009, 20, 131–134. [Google Scholar] [CrossRef]
- Mohammed, M.E.; El-Shafie, H.A.; Alhajhoj, M.R. Recent trends in the early detection of the invasive red palm weevil, rhynchophorus ferrugineus (olivier). In Invasive Species-Introduction Pathways, Economic Impact, and Possible Management Options; IntechOpen: London, UK, 2020. [Google Scholar]
- Denholm, I.; Devine, G.; Williamson, M. Insecticide resistance on the move. Science 2002, 297, 2222–2223. [Google Scholar] [CrossRef]
- Wakil, W.; Yasin, M.; Qayyum, M.A.; Ghazanfar, M.U.; Al-Sadi, A.M.; Bedford, G.O.; Kwon, Y.J. Resistance to commonly used insecticides and phosphine fumigant in red palm weevil, Rhynchophorus ferrugineus (Olivier) in Pakistan. PLoS ONE 2018, 13, e0192628. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.X.; Su, Z.P.; Liu, H.H.; Lu, S.P.; Ma, B.; Zhao, Y.; Hou, Y.M.; Shi, Z.H. The Effect of Gut Bacteria on the Physiology of Red Palm Weevil, Rhynchophorus ferrugineus Olivier and Their Potential for the Control of This Pest. Insects 2021, 12, 594. [Google Scholar] [CrossRef]
- Lee, K.N. Appraising adaptive management. In Biological Diversity; CRC Press: Boca Raton, FL, USA, 2001; pp. 3–26. [Google Scholar]
- Faleiro, J.; Kumar, J.A. A rapid decision sampling plan for implementing area-wide management of the red palm weevil, Rhynchophorus ferrugineus, in coconut plantations of India. J. Insect Sci. 2008, 8, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Faleiro, J.; Abdallah, A.B.; Kumar, J.A.; Shagagh, A.; Al Abdan, S. Sequential sampling plan for area-wide management of Rhynchophorus ferrugineus (Olivier) in date palm plantations of Saudi Arabia. Int. J. Trop. Insect Sci. 2010, 30, 145–153. [Google Scholar] [CrossRef]
- El-Shafie, H.; Faleiro, J. Optimizing components of pheromone-baited trap for the management of red palm weevil, Rhynchophorus ferrugineus (Coleoptera: Curculionidae) in date palm agro-ecosystem. J. Plant Dis. Prot. 2017, 124, 279–287. [Google Scholar] [CrossRef]
- Massoud, M.; Sallam, A.; Faleiro, J.; Al-Abdan, S. Geographic information system-based study to ascertain the spatial and temporal spread of red palm weevil Rhynchophorus ferrugineus (Coleoptera: Curculionidae) in date plantations. Int. J. Trop. Insect Sci. 2012, 32, 108–115. [Google Scholar] [CrossRef]
- Faleiro, J.; Al-Shawaf, A. IPM of red palm weevil. In Date Palm Pests and Diseases: Integrated Management Guide; ICARDA: Beirut, Lebanon, 2018; p. 179. [Google Scholar]
- Dembilio, Ó.; Riba, J.M.; Gamón, M.; Jacas, J.A. Mobility and efficacy of abamectin and imidacloprid against Rhynchophorus ferrugineus in Phoenix canariensis by different application methods. Pest Manag. Sci. 2015, 71, 1091–1098. [Google Scholar] [CrossRef] [PubMed]
- Gomez, S.; Ferry, M. A simple and low cost injection technique to protect efficiently ornamental Phoenix against the red palm weevil during one year. Arab. J. Plant Prot. 2019, 37, 124–129. [Google Scholar] [CrossRef]
- Massa, R.; Panariello, G.; Migliore, M.; Pinchera, D.; Schettino, F.; Griffo, R.; Martano, M.; Power, K.; Maiolino, P.; Caprio, E. Research Paper (Control: Insects) Microwave heating: A promising and eco-compatible solution to fight the spread of red palm weevil. J. Plant Prot. 2019, 37, 143–148. [Google Scholar]
- Martano, M.; Massa, R.; Restucci, B.; Caprio, E.; Griffo, R.; Power, K.; Maiolino, P. Microwaves Induce Histological Alteration of Ovaries and Testis in Rhynchophorus ferrugineus Oliv. (Coleoptera: Curculionidae). Agronomy 2023, 13, 420. [Google Scholar] [CrossRef]
- Sutherst, R.W.; Constable, F.; Finlay, K.J.; Harrington, R.; Luck, J.; Zalucki, M.P. Adapting to crop pest and pathogen risks under a changing climate. Wiley Interdiscip. Rev. Clim. Change 2011, 2, 220–237. [Google Scholar] [CrossRef]
- Brevik, K.; Lindström, L.; McKay, S.D.; Chen, Y.H. Transgenerational effects of insecticides—Implications for rapid pest evolution in agroecosystems. Curr. Opin. Insect Sci. 2018, 26, 34–40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richards, S.; Gibbs, R.A.; Weinstock, G.M.; Brown, S.J.; Denell, R.; Beeman, R.W.; Gibbs, R.; Bucher, G.; Friedrich, M.; Grimmelikhuijzen, C.; et al. The genome of the model beetle and pest Tribolium castaneum. Nature 2008, 452, 949–955. [Google Scholar]
- Morey, M.; Fernández-Marmiesse, A.; Castiñeiras, D.; Fraga, J.M.; Couce, M.L.; Cocho, J.A. A glimpse into past, present, and future DNA sequencing. Mol. Genet. Metab. 2013, 110, 3–24. [Google Scholar] [CrossRef]
- Kingan, S.B.; Heaton, H.; Cudini, J.; Lambert, C.C.; Baybayan, P.; Galvin, B.D.; Durbin, R.; Korlach, J.; Lawniczak, M.K. A high-quality de novo genome assembly from a single mosquito using PacBio sequencing. Genes 2019, 10, 62. [Google Scholar] [CrossRef] [Green Version]
- Keeling, C.I.; Yuen, M.; Liao, N.Y.; T Docking, R.; Chan, S.K.; Taylor, G.A.; Palmquist, D.L.; Jackman, S.D.; Nguyen, A.; Li, M.; et al. Draft genome of the mountain pine beetle, Dendroctonus ponderosae Hopkins, a major forest pest. Genome Biol. 2013, 14, 1–20. [Google Scholar] [CrossRef] [Green Version]
- McKenna, D.D.; Scully, E.D.; Pauchet, Y.; Hoover, K.; Kirsch, R.; Geib, S.M.; Mitchell, R.F.; Waterhouse, R.M.; Ahn, S.J.; Arsala, D.; et al. Genome of the Asian longhorned beetle (Anoplophora glabripennis), a globally significant invasive species, reveals key functional and evolutionary innovations at the beetle–plant interface. Genome Biol. 2016, 17, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sayadi, A.; Martinez Barrio, A.; Immonen, E.; Dainat, J.; Berger, D.; Tellgren-Roth, C.; Nystedt, B.; Arnqvist, G. The genomic footprint of sexual conflict. Nat. Ecol. Evol. 2019, 3, 1725–1730. [Google Scholar] [CrossRef] [Green Version]
- Parisot, N.; Vargas-Chávez, C.; Goubert, C.; Baa-Puyoulet, P.; Balmand, S.; Beranger, L.; Blanc, C.; Bonnamour, A.; Boulesteix, M.; Burlet, N.; et al. The transposable element-rich genome of the cereal pest Sitophilus oryzae. BMC Biol. 2021, 19, 1–28. [Google Scholar] [CrossRef]
- Filipović, I.; Rašić, G.; Hereward, J.; Gharuka, M.; Devine, G.J.; Furlong, M.J.; Etebari, K. A high-quality de novo genome assembly based on nanopore sequencing of a wild-caught coconut rhinoceros beetle (Oryctes rhinoceros). BMC Genom. 2022, 23, 426. [Google Scholar] [CrossRef]
- Oppert, B.; Muszewska, A.; Steczkiewicz, K.; Šatović-Vukšić, E.; Plohl, M.; Fabrick, J.A.; Vinokurov, K.S.; Koloniuk, I.; Johnston, J.S.; Smith, T.P.; et al. The genome of Rhyzopertha dominica (Fab.)(Coleoptera: Bostrichidae): Adaptation for success. Genes 2022, 13, 446. [Google Scholar] [CrossRef]
- Fu, C.; Long, W.; Luo, C.; Nong, X.; Xiao, X.; Liao, H.; Li, Y.; Chen, Y.; Yu, J.; Cheng, S.; et al. Chromosome-Level Genome Assembly of Cyrtotrachelus buqueti and Mining of Its Specific Genes. Front. Ecol. Evol. 2021, 9, 693. [Google Scholar] [CrossRef]
- Mittapalli, O.; Bai, X.; Mamidala, P.; Rajarapu, S.P.; Bonello, P.; Herms, D.A. Tissue-specific transcriptomics of the exotic invasive insect pest emerald ash borer (Agrilus planipennis). PLoS ONE 2010, 5, e13708. [Google Scholar] [CrossRef] [Green Version]
- Lao, S.H.; Huang, X.H.; Huang, H.J.; Liu, C.W.; Zhang, C.X.; Bao, Y.Y. Genomic and transcriptomic insights into the cytochrome P450 monooxygenase gene repertoire in the rice pest brown planthopper, Nilaparvata lugens. Genomics 2015, 106, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Song, K.; Teng, H.; Zhang, B.; Li, W.; Xue, H.; Yang, X. Endogenous cellulolytic enzyme systems in the longhorn beetle Mesosa myops (Insecta: Coleoptera) studied by transcriptomic analysis. Acta Biochim. Biophys. Sin. 2015, 47, 741–748. [Google Scholar] [CrossRef] [Green Version]
- Tarver, M.R.; Huang, Q.; de Guzman, L.; Rinderer, T.; Holloway, B.; Reese, J.; Weaver, D.; Evans, J.D. Transcriptomic and functional resources for the small hive beetle Aethina tumida, a worldwide parasite of honey bees. Genom. Data 2016, 9, 97–99. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Zhang, X.W.; Pan, L.L.; Liu, W.F.; Wang, D.P.; Zhang, G.Y.; Yin, Y.X.; Yin, A.; Jia, S.G.; Yu, X.G.; et al. A large-scale gene discovery for the red palm weevil Rhynchophorus ferrugineus (Coleoptera: Curculionidae). Insect Sci. 2013, 20, 689–702. [Google Scholar] [CrossRef] [PubMed]
- Yin, A.; Pan, L.; Zhang, X.; Wang, L.; Yin, Y.; Jia, S.; Liu, W.; Xin, C.; Liu, K.; Yu, X.; et al. Transcriptomic study of the red palm weevil Rhynchophorus ferrugineus embryogenesis. Insect Sci. 2015, 22, 65–82. [Google Scholar] [CrossRef]
- Yan, W.; Liu, L.; Qin, W.; Li, C.; Peng, Z. Transcriptomic identification of chemoreceptor genes in the red palm weevil Rhynchophorus ferrugineus. Genet. Mol. Res. 2015, 14, 7469–7480. [Google Scholar] [CrossRef]
- Sánchez-Gracia, A.; Vieira, F.; Rozas, J. Molecular evolution of the major chemosensory gene families in insects. Heredity 2009, 103, 208–216. [Google Scholar] [CrossRef]
- Andersson, M.N.; Grosse-Wilde, E.; Keeling, C.I.; Bengtsson, J.M.; Yuen, M.; Li, M.; Hillbur, Y.; Bohlmann, J.; Hansson, B.S.; Schlyter, F. Antennal transcriptome analysis of the chemosensory gene families in the tree killing bark beetles, Ips typographus and Dendroctonus ponderosae (Coleoptera: Curculionidae: Scolytinae). BMC Genom. 2013, 14, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Antony, B.; Soffan, A.; Jakše, J.; Abdelazim, M.M.; Aldosari, S.A.; Aldawood, A.S.; Pain, A. Identification of the genes involved in odorant reception and detection in the palm weevil Rhynchophorus ferrugineus, an important quarantine pest, by antennal transcriptome analysis. BMC Genom. 2016, 17, 1–22. [Google Scholar] [CrossRef] [Green Version]
- Antony, B.; Johny, J.; Abdelazim, M.M.; Jakše, J.; Al-Saleh, M.A.; Pain, A. Global transcriptome profiling and functional analysis reveal that tissue-specific constitutive overexpression of cytochrome P450s confers tolerance to imidacloprid in palm weevils in date palm fields. BMC Genom. 2019, 20, 1–23. [Google Scholar] [CrossRef]
- Zhang, H.; Bai, J.; Huang, S.; Liu, H.; Lin, J.; Hou, Y. Neuropeptides and G-protein coupled receptors (GPCRs) in the red palm weevil Rhynchophorus ferrugineus olivier (Coleoptera: Dryophthoridae). Front. Physiol. 2020, 11, 159. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Xu, D.; Zhuo, Z.; Hu, J.; Lu, B. SMRT sequencing of the full-length transcriptome of the Rhynchophorus ferrugineus (Coleoptera: Curculionidae). PeerJ 2020, 8, e9133. [Google Scholar] [CrossRef]
- Yang, H.; Xu, D.; Zhuo, Z.; Hu, J.; Lu, B. Transcriptome and gene expression analysis of Rhynchophorus ferrugineus (Coleoptera: Curculionidae) during developmental stages. PeerJ 2020, 8, e10223. [Google Scholar] [CrossRef]
- Engsontia, P.; Satasook, C. Genome-Wide Identification of the Gustatory Receptor Gene Family of the Invasive Pest, Red Palm Weevil, Rhynchophorus ferrugineus (Olivier, 1790). Insects 2021, 12, 611. [Google Scholar] [CrossRef]
- Whibley, A.; Kelley, J.L.; Narum, S.R. The Changing Face of Genome Assemblies: Guidance on Achieving High-Quality Reference Genomes. Mol. Ecol. Resour. 2021, 21, 641–652. [Google Scholar] [CrossRef]
- Hazzouri, K.M.; Sudalaimuthuasari, N.; Kundu, B.; Nelson, D.; Al-Deeb, M.A.; Le Mansour, A.; Spencer, J.J.; Desplan, C.; Amiri, K. The genome of pest Rhynchophorus ferrugineus reveals gene families important at the plant-beetle interface. Commun. Biol. 2020, 3, 1–14. [Google Scholar] [CrossRef]
- Wheat, C.W.; Wahlberg, N. Phylogenomic insights into the Cambrian explosion, the colonization of land and the evolution of flight in Arthropoda. Syst. Biol. 2013, 62, 93–109. [Google Scholar] [CrossRef] [Green Version]
- Dias, G.B.; Altammami, M.A.; El-Shafie, H.A.; Alhoshani, F.M.; Al-Fageeh, M.B.; Bergman, C.M.; Manee, M.M. Haplotype-resolved genome assembly enables gene discovery in the red palm weevil Rhynchophorus ferrugineus. Sci. Rep. 2021, 11, 1–14. [Google Scholar] [CrossRef]
- Bruna, T.; Hoff, K.J.; Lomsadze, A.; Stanke, M.; Borodovsky, M. BRAKER2: Automatic eukaryotic genome annotation with GeneMark-EP+ and AUGUSTUS supported by a protein database. NAR Genom. Bioinform. 2021, 3, lqaa108. [Google Scholar] [CrossRef]
- Manee, M.M.; Al-Shomrani, B.M.; Altammami, M.A.; El-Shafie, H.A.; Alsayah, A.A.; Alhoshani, F.M.; Alqahtani, F.H. Microsatellite Variation in the Most Devastating Beetle Pests (Coleoptera: Curculionidae) of Agricultural and Forest Crops. Int. J. Mol. Sci. 2022, 23, 9847. [Google Scholar] [CrossRef]
- Biemont, C. Within-species variation in genome size. Heredity 2008, 101, 297–298. [Google Scholar] [CrossRef]
- Nishimura, O.; Hara, Y.; Kuraku, S. gVolante for standardizing completeness assessment of genome and transcriptome assemblies. Bioinformatics 2017, 33, 3635–3637. [Google Scholar] [CrossRef] [Green Version]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [Green Version]
- Kriventseva, E.V.; Kuznetsov, D.; Tegenfeldt, F.; Manni, M.; Dias, R.; Simão, F.A.; Zdobnov, E.M. OrthoDB v10: Sampling the diversity of animal, plant, fungal, protist, bacterial and viral genomes for evolutionary and functional annotations of orthologs. Nucleic Acids Res. 2019, 47, D807–D811. [Google Scholar] [CrossRef] [Green Version]
- Luo, C.; Tsementzi, D.; Kyrpides, N.; Read, T.; Konstantinidis, K.T. Direct comparisons of Illumina vs. Roche 454 sequencing technologies on the same microbial community DNA sample. PLoS ONE 2012, 7, e30087. [Google Scholar]
- Montagna, M.; Chouaia, B.; Mazza, G.; Prosdocimi, E.M.; Crotti, E.; Mereghetti, V.; Vacchini, V.; Giorgi, A.; De Biase, A.; Longo, S.; et al. Effects of the diet on the microbiota of the red palm weevil (Coleoptera: Dryophthoridae). PLoS ONE 2015, 10, e0117439. [Google Scholar] [CrossRef] [Green Version]
- Chen, B.; Teh, B.S.; Sun, C.; Hu, S.; Lu, X.; Boland, W.; Shao, Y. Biodiversity and activity of the gut microbiota across the life history of the insect herbivore Spodoptera littoralis. Sci. Rep. 2016, 6, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Muhammad, A.; Fang, Y.; Hou, Y.; Shi, Z. The gut entomotype of red palm weevil Rhynchophorus Ferrugineus Olivier (Coleoptera: Dryophthoridae) and their effect on host nutrition metabolism. Front. Microbiol. 2017, 8, 2291. [Google Scholar] [CrossRef] [Green Version]
- Rosenberg, E.; Zilber-Rosenberg, I. Symbiosis and development: The hologenome concept. Birth Defects Res. Part Embryo Today Rev. 2011, 93, 56–66. [Google Scholar] [CrossRef]
- Jia, S.; Zhang, X.; Zhang, G.; Yin, A.; Zhang, S.; Li, F.; Wang, L.; Zhao, D.; Yun, Q.; Wang, J.; et al. Seasonally variable intestinal metagenomes of the red palm weevil (Rhynchophorus ferrugineus). Environ. Microbiol. 2013, 15, 3020–3029. [Google Scholar] [CrossRef] [Green Version]
- Tagliavia, M.; Messina, E.; Manachini, B.; Cappello, S.; Quatrini, P. The gut microbiota of larvae of Rhynchophorus ferrugineus Oliver (Coleoptera: Curculionidae). BMC Microbiol. 2014, 14, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Charlesworth, B.; Sniegowski, P.; Stephan, W. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 1994, 371, 215–220. [Google Scholar] [CrossRef]
- Shapiro, J.A.; von Sternberg, R. Why repetitive DNA is essential to genome function. Biol. Rev. 2005, 80, 227–250. [Google Scholar] [CrossRef] [Green Version]
- Lynch, M.; Walsh, B. The Origins of Genome Architecture; Sinauer Associates: Sunderland, MA, USA, 2007; Volume 98. [Google Scholar]
- Montiel, E.E.; Mora, P.; Rico-Porras, J.M.; Palomeque, T.; Lorite, P. Satellitome of the Red Palm Weevil, Rhynchophorus ferrugineus (Coleoptera: Curculionidae), the Most Diverse Among Insects. Front. Ecol. Evol. 2022, 10, 826808. [Google Scholar] [CrossRef]
- Ding, S.; Wang, S.; He, K.; Jiang, M.; Li, F. Large-scale analysis reveals that the genome features of simple sequence repeats are generally conserved at the family level in insects. BMC Genom. 2017, 18, 1–10. [Google Scholar] [CrossRef]
- Boore, J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef] [Green Version]
- Manee, M.M.; Alshehri, M.A.; Binghadir, S.A.; Aldhafer, S.H.; Alswailem, R.M.; Algarni, A.T.; Al-Shomrani, B.M.; Al-Fageeh, M.B. Comparative analysis of camelid mitochondrial genomes. J. Genet. 2019, 98, 1–12. [Google Scholar] [CrossRef]
- Zhang, Z.; Bi, G.; Liu, G.; Du, Q.; Zhao, E.; Yang, J.; Shang, E. Complete mitochondrial genome of Rhynchophorus ferrugineus. Mitochondrial DNA Part 2017, 28, 208–209. [Google Scholar] [CrossRef]
- Chevreux, B. MIRA: An Automated Genome and EST Assembler. Ph.D. Thesis, University of Heidelberg, Heidelberg, Germany, 2007. [Google Scholar]
- Abdel-Banat, B.; El-Shafie, H.A. Genomics Approaches for Insect Control and Insecticide Resistance Development in Date Palm. In The Date Palm Genome, Vol. 2; Springer: Cham, Switzerland, 2021; pp. 215–248. [Google Scholar]
- Vogel, E.; Santos, D.; Mingels, L.; Verdonckt, T.W.; Broeck, J.V. RNA interference in insects: Protecting beneficials and controlling pests. Front. Physiol. 2019, 9, 1912. [Google Scholar] [CrossRef] [Green Version]
- Kandul, N.P.; Liu, J.; Sanchez C, H.M.; Wu, S.L.; Marshall, J.M.; Akbari, O.S. Transforming insect population control with precision guided sterile males with demonstration in flies. Nat. Commun. 2019, 10, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Laudani, F.; Strano, C.P.; Edwards, M.G.; Malacrinò, A.; Campolo, O.; Abd El Halim, H.M.; Gatehouse, A.M.; Palmeri, V. RNAi-mediated gene silencing in Rhynchophorus ferrugineus (Oliver)(Coleoptera: Curculionidae). Open Life Sci. 2017, 12, 214–222. [Google Scholar] [CrossRef]
- Al-Ayedh, H.; Hussain, A.; Rizwan-ul Haq, M.; Al-Jabr, A.M. Status of insecticide resistance in field-collected populations of Rhynchophorus ferrugineus (Olivier)(Coleoptera: Curculionidae). Int. J. Agric. Biol. 2016, 18, 103–110. [Google Scholar] [CrossRef]
- El-Mergawy, R.A.; Faure, N.; Nasr, M.I.; Avand-Faghih, A.; Rochat, D.; Silvain, J.F. Mitochondrial genetic variation and invasion history of Red palm weevil, Rhynchophorus ferrugineus (Coleoptera: Curculionidae), in middle-east and Mediterranean basin. Int. J. Agric. Biol. 2011, 13, 631–637. [Google Scholar]
- Wang, G.; Hou, Y.; Zhang, X.; Zhang, J.; Li, J.; Chen, Z. Strong population genetic structure of an invasive species, Rhynchophorus ferrugineus (Olivier), in southern China. Ecol. Evol. 2017, 7, 10770–10781. [Google Scholar] [CrossRef]
- Sabit, H.; Abdel-Ghany, S.; Al-Dhafar, Z.; Said, O.A.; Al-Saeed, J.A.; Alfehaid, Y.A.; Osman, M.A. Molecular characterization and phylogenetic analysis of Rhynchophorus ferrugineus (Olivier) in Eastern Province, Saudi Arabia. Saudi J. Biol. Sci. 2021, 28, 5621–5630. [Google Scholar] [CrossRef]
- Abdel-Baky, N.F.; Aldeghairi, M.A.; Motawei, M.I.; Al-Shuraym, L.A.; Al-Nujiban, A.A.; Alharbi, M.T.; Rehan, M. Genetic Diversity of Palm Weevils, Rhynchophorus Species (Coleoptera: Curculionidae) by Mitochondrial COI Gene Sequences Declares a New Species, R. bilineatus in Qassim, Saudi Arabia. Arab. J. Sci. Eng. 2023, 48, 63–80. [Google Scholar] [CrossRef]
- Kroemer, J.A.; Bonning, B.C.; Harrison, R.L. Expression, delivery and function of insecticidal proteins expressed by recombinant baculoviruses. Viruses 2015, 7, 422–455. [Google Scholar] [CrossRef] [Green Version]
- Nikolouli, K.; Sassù, F.; Mouton, L.; Stauffer, C.; Bourtzis, K. Combining sterile and incompatible insect techniques for the population suppression of Drosophila suzukii. J. Pest Sci. 2020, 93, 647–661. [Google Scholar] [CrossRef] [Green Version]
- Al-Dous, E.K.; George, B.; Al-Mahmoud, M.E.; Al-Jaber, M.Y.; Wang, H.; Salameh, Y.M.; Al-Azwani, E.K.; Chaluvadi, S.; Pontaroli, A.C.; DeBarry, J.; et al. De novo genome sequencing and comparative genomics of date palm (Phoenix dactylifera). Nat. Biotechnol. 2011, 29, 521–527. [Google Scholar] [CrossRef]
- Al-Mssallem, I.S.; Hu, S.; Zhang, X.; Lin, Q.; Liu, W.; Tan, J.; Yu, X.; Liu, J.; Pan, L.; Zhang, T.; et al. Genome sequence of the date palm Phoenix dactylifera L. Nat. Commun. 2013, 4, 1–9. [Google Scholar] [CrossRef] [Green Version]
- El Hadrami, A.; Al-Khayri, J.M. Socioeconomic and traditional importance of date palm. Emir. J. Food Agric. 2012, 24, 371. [Google Scholar]


| Resources | BUSCO | ||||
|---|---|---|---|---|---|
| Complete | Single-Copy | Duplicated | Fragmented | Missing | |
| Transcriptomes | |||||
| Wang et al. [56] | 81.3 | 71.1 | 9.6 | 10.6 | 8.1 |
| Antony et al. [61] | 95.9 | 94.8 | 1.1 | 2.7 | 1.4 |
| Yang et al. [64] | 92.8 | 39.5 | 53.3 | 1.5 | 5.7 |
| Genomes | |||||
| Hazzouri et al. [68] | 93.3 | 55.6 | 37.7 | 2.2 | 4.5 |
| Dias et al. [70] | 98.6 | 96.5 | 2.1 | 1.1 | 0.3 |
| Manee et al. [72] | 94.6 | 92.2 | 1.7 | 4.1 | 1.3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manee, M.M.; Alqahtani, F.H.; Al-Shomrani, B.M.; El-Shafie, H.A.F.; Dias, G.B. Omics in the Red Palm Weevil Rhynchophorus ferrugineus (Olivier) (Coleoptera: Curculionidae): A Bridge to the Pest. Insects 2023, 14, 255. https://doi.org/10.3390/insects14030255
Manee MM, Alqahtani FH, Al-Shomrani BM, El-Shafie HAF, Dias GB. Omics in the Red Palm Weevil Rhynchophorus ferrugineus (Olivier) (Coleoptera: Curculionidae): A Bridge to the Pest. Insects. 2023; 14(3):255. https://doi.org/10.3390/insects14030255
Chicago/Turabian StyleManee, Manee M., Fahad H. Alqahtani, Badr M. Al-Shomrani, Hamadttu A. F. El-Shafie, and Guilherme B. Dias. 2023. "Omics in the Red Palm Weevil Rhynchophorus ferrugineus (Olivier) (Coleoptera: Curculionidae): A Bridge to the Pest" Insects 14, no. 3: 255. https://doi.org/10.3390/insects14030255
APA StyleManee, M. M., Alqahtani, F. H., Al-Shomrani, B. M., El-Shafie, H. A. F., & Dias, G. B. (2023). Omics in the Red Palm Weevil Rhynchophorus ferrugineus (Olivier) (Coleoptera: Curculionidae): A Bridge to the Pest. Insects, 14(3), 255. https://doi.org/10.3390/insects14030255








